Inanimate Surfaces as a Source of Hospital Infections Caused by Fungi, Bacteria and Viruses with Particular Emphasis on SARS-CoV-2
Abstract
1. Introduction
2. Methods
3. Discussion
3.1. Factors Influencing the Persistence of Pathogens
3.2. Viruses
3.3. Bacteria
3.4. Fungi
3.5. Hygiene and Disinfection of Surfaces
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Klevens, R.M.; Edwards, J.R.; Richards, C.L., Jr.; Horan, T.C.; Gaynes, R.P.; Pollock, D.A.; Cardo, D.M. Estimating Health Care-Associated Infections and Deaths in U.S. Hospitals, 2002. Public Health Rep. 2007, 122, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Daneman, N.; Sarwar, S.; Fowler, R.A.; Cuthbertson, B.H.; SuDDICU Canadian Study Group. Effect of Selective Decontamination on Antimicrobial Resistance in Intensive Care Units: A Systematic Review and Meta-Analysis. Lancet Infect. Dis. 2013, 13, 328–341. [Google Scholar] [CrossRef]
- Zaragoza, R.; Ramírez, P.; López-Pueyo, M.J. Infección nosocomial en las unidades de cuidados intensivos. Enferm. Infecc. Microbiol. Clin. 2014, 32, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Schwebke, I.; Kampf, G. How Long Do Nosocomial Pathogens Persist on Inanimate Surfaces? A Systematic Review. BMC Infect. Dis. 2006, 6, 130. [Google Scholar] [CrossRef] [PubMed]
- Wißmann, J.E.; Kirchhoff, L.; Brüggemann, Y.; Todt, D.; Steinmann, J.; Steinmann, E. Persistence of Pathogens on Inanimate Surfaces: A Narrative Review. Microorganisms 2021, 9, 343. [Google Scholar] [CrossRef] [PubMed]
- Moccia, G.; Motta, O.; Pironti, C.; Proto, A.; Capunzo, M.; De Caro, F. An Alternative Approach for the Decontamination of Hospital Settings. J. Infect. Public Health 2020, 13, 2038–2044. [Google Scholar] [CrossRef]
- Trudel-Ferland, M.; Jubinville, E.; Jean, J. Persistence of Hepatitis A Virus RNA in Water, on Non-Porous Surfaces, and on Blueberries. Front. Microbiol. 2021, 12, 618352. [Google Scholar] [CrossRef]
- Dancer, S.J.; King, M.-F. Systematic Review on Use, Cost and Clinical Efficacy of Automated Decontamination Devices. Antimicrob. Resist. Infect. Control 2021, 10, 34. [Google Scholar] [CrossRef]
- Mao, N.; An, C.K.; Guo, L.Y.; Wang, M.; Guo, L.; Guo, S.R.; Long, E.S. Transmission Risk of Infectious Droplets in Physical Spreading Process at Different Times: A Review. Build. Environ. 2020, 185, 107307. [Google Scholar] [CrossRef]
- Suleyman, G.; Alangaden, G.; Bardossy, A.C. The Role of Environmental Contamination in the Transmission of Nosocomial Pathogens and Healthcare-Associated Infections. Curr. Infect. Dis. Rep. 2018, 20, 12. [Google Scholar] [CrossRef]
- Kramer, A.; Assadian, O. Survival of Microorganisms on Inanimate Surfaces. In Use of Biocidal Surfaces for Reduction of Healthcare Acquired Infections; Springer International Publishing: Berlin/Heidelberg, Germany, 2014; pp. 7–26. [Google Scholar]
- Tang, J.W. The Effect of Environmental Parameters on the Survival of Airborne Infectious Agents. J. R. Soc. Interface 2009, 6, S737–S746. [Google Scholar] [CrossRef] [PubMed]
- Igo, M.J.; Schaffner, D.W. Quantifying the Influence of Relative Humidity, Temperature, and Diluent on the Survival and Growth of Enterobacter Aerogenes. J. Food Prot. 2019, 82, 2135–2147. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Bang, J.; Beuchat, L.R.; Ryu, J.-H. Fate of Enterobacter sakazakii Attached to or in Biofilms on Stainless Steel upon Exposure to Various Temperatures or Relative Humidities. J. Food Prot. 2008, 71, 940–945. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.P.; Avery, L.M.; Killham, K.; Jones, D.L. Persistence of Escherichia coli O157 on Farm Surfaces under Different Environmental Conditions. J. Appl. Microbiol. 2005, 98, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Levin, P.D.; Shatz, O.; Sviri, S.; Moriah, D.; Or-Barbash, A.; Sprung, C.L.; Moses, A.E.; Block, C. Contamination of Portable Radiograph Equipment with Resistant Bacteria in the ICU. Chest 2009, 136, 426–432. [Google Scholar] [CrossRef]
- Vickery, K.; Deva, A.; Jacombs, A.; Allan, J.; Valente, P.; Gosbell, I.B. Presence of Biofilm Containing Viable Multiresistant Organisms despite Terminal Cleaning on Clinical Surfaces in an Intensive Care Unit. J. Hosp. Infect. 2012, 80, 52–55. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilm Formation: A Clinically Relevant Microbiological Process. Clin. Infect. Dis. 2001, 33, 1387–1392. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Sinde, E.; Carballo, J. Attachment of Salmonella spp. and Listeria monocytogenes to Stainless Steel, Rubber and Polytetrafluorethylene: The Influence of Free Energy and the Effect of Commercial Sanitizers. Food Microbiol. 2000, 17, 439–447. [Google Scholar] [CrossRef]
- Lagha, R.; Bellon-Fontaine, M.-N.; Renault, M.; Briandet, R.; Herry, J.-M.; Mrabet, B.; Bakhrouf, A.; Chehimi, M.M. Impact of Long-Term Starvation on Adhesion to and Biofilm Formation on Stainless Steel 316 L and Gold Surfaces of Salmonella Enterica Serovar Typhimurium. Ann. Microbiol. 2015, 65, 399–409. [Google Scholar] [CrossRef]
- Hota, B. Contamination, Disinfection, and Cross-Colonization: Are Hospital Surfaces Reservoirs for Nosocomial Infection? Clin. Infect. Dis. 2004, 39, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Boyce, J.M. Environmental Contamination Makes an Important Contribution to Hospital Infection. J. Hosp. Infect. 2007, 65, 50–54. [Google Scholar] [CrossRef]
- Murphy, T.F.; Brauer, A.L. Expression of Urease by Haemophilus Influenzae during Human Respiratory Tract Infection and Role in Survival in an Acid Environment. BMC Microbiol. 2011, 11, 183. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.D.; DeLeo, F.R. Staphylococcus aureus Protein A Promotes Immune Suppression. MBio 2013, 4, e00764-13. [Google Scholar] [CrossRef] [PubMed]
- Mahl, M.C.; Sadler, C. Virus Survival on Inanimate Surfaces. Can. J. Microbiol. 1975, 21, 819–823. [Google Scholar] [CrossRef]
- World Health Organization. First Data on Stability and Resistance of SARS Coronavirus Compiled by Members of WHO Laboratory Network. Available online: http://www.who.int/csr/sars/survival_2003_05_04/en/index.html (accessed on 28 February 2021).
- Sizun, J.; Yu, M.W.; Talbot, P.J. Survival of Human Coronaviruses 229E and OC43 in Suspension and after Drying Onsurfaces: A Possible Source Ofhospital-Acquired Infections. J. Hosp. Infect. 2000, 46, 55–60. [Google Scholar] [CrossRef]
- Casanova, L.M.; Jeon, S.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Effects of Air Temperature and Relative Humidity on Coronavirus Survival on Surfaces. Appl. Environ. Microbiol. 2010, 76, 2712–2717. [Google Scholar] [CrossRef]
- Eggers, M.; Schwebke, I.; Suchomel, M.; Fotheringham, V.; Gebel, J.; Meyer, B.; Morace, G.; Roedger, H.J.; Roques, C.; Visa, P.; et al. The European Tiered Approach for Virucidal Efficacy Testing-Rationale for Rapidly Selecting Disinfectants against Emerging and Re-Emerging Viral Diseases. Euro. Surveill. 2021, 26, 2000708. [Google Scholar] [CrossRef]
- Tarka, P.; Nitsch-Osuch, A. Evaluating the Virucidal Activity of Disinfectants According to European Union Standards. Viruses 2021, 13, 534. [Google Scholar] [CrossRef]
- King, A.M.Q.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; Knowles, N.J.; et al. Changes to Taxonomy and the International Code of Virus Classification and Nomenclature Ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2018, 163, 2601–2631. [Google Scholar] [CrossRef]
- SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html (accessed on 24 June 2022).
- Rabi, F.A.; Al Zoubi, M.S.; Kasasbeh, G.A.; Salameh, D.M.; Al-Nasser, A.D. SARS-CoV-2 and Coronavirus Disease 2019: What We Know so Far. Pathogens 2020, 9, 231. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Zhao, K.; Shi, Z.-L.; Zhou, P. Bat Coronaviruses in China. Viruses 2019, 11, 210. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Holmes, E.C.; Goldstein, S.A.; Rasmussen, A.L.; Robertson, D.L.; Crits-Christoph, A.; Wertheim, J.O.; Anthony, S.J.; Barclay, W.S.; Boni, M.F.; Doherty, P.C.; et al. The origins of SARS-CoV-2: A critical review. Cell 2021, 184, 4848–4856. [Google Scholar] [CrossRef] [PubMed]
- DeCapprio, D.; Gartner, J.; McCall, C.J.; Burgess, T.; Kothari, S.; Sayed, S. Building a COVID-19 Vulnerability Index. arXiv 2020, arXiv:2003.07347. [Google Scholar] [CrossRef]
- Lai, C.-C.; Shih, T.-P.; Ko, W.-C.; Tang, H.-J.; Hsueh, P.-R. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Coronavirus Disease-2019 (COVID-19): The Epidemic and the Challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A Familial Cluster of Pneumonia Associated with the 2019 Novel Coronavirus Indicating Person-to-Person Transmission: A Study of a Family Cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef]
- Aydogdu, M.O.; Altun, E.; Chung, E.; Ren, G.; Homer-Vanniasinkam, S.; Chen, B.; Edirisinghe, M. Surface Interactions and Viability of Coronaviruses. J. R. Soc. Interface 2021, 18, 20200798. [Google Scholar] [CrossRef]
- Aboubakr, H.A.; Sharafeldin, T.A.; Goyal, S.M. Stability of SARS-CoV-2 and Other Coronaviruses in the Environment and on Common Touch Surfaces and the Influence of Climatic Conditions: A Review. Transbound. Emerg. Dis. 2021, 68, 296–312. [Google Scholar] [CrossRef]
- Balaraman, V.; Drolet, B.S.; Mitzel, D.N.; Wilson, W.C.; Owens, J.; Gaudreault, N.N.; Meekins, D.A.; Bold, D.; Trujillo, J.D.; Noronha, L.E.; et al. Mechanical Transmission of SARS-CoV-2 by House Flies. Parasit. Vectors 2021, 14, 214. [Google Scholar] [CrossRef]
- Balaraman, V.; Drolet, B.S.; Gaudreault, N.N.; Wilson, W.C.; Owens, J.; Bold, D.; Swanson, D.A.; Jasperson, D.C.; Noronha, L.E.; Richt, J.A.; et al. Susceptibility of Midge and Mosquito Vectors to SARS-CoV-2. J. Med. Entomol. 2021, 58, 1948–1951. [Google Scholar] [CrossRef] [PubMed]
- Valsamatzi-Panagiotou, A.; Penchovsky, R. Environmental Factors Influencing the Transmission of the Coronavirus 2019: A Review. Environ. Chem. Lett. 2022, 20, 1603–1610. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.; Walsh, K.; Willcox, M.; Morgan, P.; Nichols, J. The COVID-19 Pandemic: Important Considerations for Contact Lens Practitioners. Cont. Lens Anterior Eye 2020, 43, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Vasickova, P.; Pavlik, I.; Verani, M.; Carducci, A. Issues Concerning Survival of Viruses on Surfaces. Food Environ. Virol. 2010, 2, 24–34. [Google Scholar] [CrossRef]
- Chin, A.W.H.; Chu, J.T.S.; Perera, M.R.A.; Hui, K.P.Y.; Yen, H.-L.; Chan, M.C.W.; Peiris, M.; Poon, L.L.M. Stability of SARS-CoV-2 in Different Environmental Conditions. Lancet Microbe 2020, 1, e10. [Google Scholar] [CrossRef]
- Siddiquie, R.Y.; Agrawal, A.; Joshi, S.S. Surface Alterations to Impart Antiviral Properties to Combat COVID-19 Transmission. Trans Indian Natl. Acad. Eng. 2020, 5, 343–347. [Google Scholar] [CrossRef]
- Wolff, M.H.; Sattar, S.A.; Adegbunrin, O.; Tetro, J. Environmental Survival and Microbicide Inactivation of Coronaviruses. In Coronaviruses with Special Emphasis on First Insights Concerning SARS; Birkhäuser: Basel, Switzerland, 2005; pp. 201–212. [Google Scholar]
- WHO Surface Sampling of Coronavirus Disease (COVID-19): A Practical “How to” Protocol for Health Care and Public Health Professionals. Who.int. Available online: https://www.who.int/publications/i/item/surface-sampling-of-coronavirus-disease-(-covid-19)-a-practical-how-to-protocol-for-health-care-and-public-health-professionals (accessed on 9 June 2022).
- Biryukov, J.; Boydston, J.A.; Dunning, R.A.; Yeager, J.J.; Wood, S.; Reese, A.L.; Ferris, A.; Miller, D.; Weaver, W.; Zeitouni, N.E.; et al. Increasing Temperature and Relative Humidity Accelerates Inactivation of SARS-CoV-2 on Surfaces. mSphere 2020, 5, e00441-20. [Google Scholar] [CrossRef]
- Ratnesar-Shumate, S.; Williams, G.; Green, B.; Krause, M.; Holland, B.; Wood, S.; Bohannon, J.; Boydston, J.; Freeburger, D.; Hooper, I.; et al. Simulated Sunlight Rapidly Inactivates SARS-CoV-2 on Surfaces. J. Infect. Dis. 2020, 222, 214–222. [Google Scholar] [CrossRef]
- Colaneri, M.; Seminari, E.; Novati, S.; Asperges, E.; Biscarini, S.; Piralla, A.; Percivalle, E.; Cassaniti, I.; Baldanti, F.; Bruno, R.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 RNA Contamination of Inanimate Surfaces and Virus Viability in a Health Care Emergency Unit. Clin. Microbiol. Infect. 2020, 26, 1094.e1–1094.e5. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of Coronaviruses on Inanimate Surfaces and Their Inactivation with Biocidal Agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Rawlinson, S.; Ciric, L.; Cloutman-Green, E. COVID-19 Pandemic–Let’s Not Forget Surfaces. J. Hosp. Infect. 2020, 105, 790–791. [Google Scholar] [CrossRef]
- Ren, S.-Y.; Wang, W.-B.; Hao, Y.-G.; Zhang, H.-R.; Wang, Z.-C.; Chen, Y.-L.; Gao, R.-D. Stability and Infectivity of Coronaviruses in Inanimate Environments. World J. Clin. Cases 2020, 8, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Henrickson, K.J. Parainfluenza Viruses. Clin. Microbiol. Rev. 2003, 16, 242–264. [Google Scholar] [CrossRef] [PubMed]
- Perry, K.A.; Coulliette, A.D.; Rose, L.J.; Shams, A.M.; Edwards, J.R.; Noble-Wang, J.A. Persistence of Influenza A (H1N1) Virus on Stainless Steel Surfaces. Appl. Environ. Microbiol. 2016, 82, 3239–3245. [Google Scholar] [CrossRef] [PubMed]
- Glass, R.I.; Parashar, U.D.; Estes, M.K. Norovirus Gastroenteritis. N. Engl. J. Med. 2009, 361, 1776–1785. [Google Scholar] [CrossRef]
- Lopman, B.; Gastañaduy, P.; Park, G.W.; Hall, A.J.; Parashar, U.D.; Vinjé, J. Environmental Transmission of Norovirus Gastroenteritis. Curr. Opin. Virol. 2012, 2, 96–102. [Google Scholar] [CrossRef]
- Kuusi, M.; Nuorti, J.P.; Maunula, L.; Tran Minh, N.N.; Ratia, M.; Karlsson, J.; Von Bonsdorff, C.-H. A Prolonged Outbreak of Norwalk-like Calicivirus (NLV) Gastroenteritis in a Rehabilitation Centre Due to Environmental Contamination. Epidemiol. Infect. 2002, 129, 133–138. [Google Scholar] [CrossRef]
- Kuchar, E.; Nitsch-Osuch, A.; Szenborn, L.; Ołdak, E. Rotaviruses as a cause of nosocomial infections in Poland--systematic review with metaanalysis of 11 studies. Przegl. Epidemiol. 2012, 66, 409–415. [Google Scholar]
- Smith, M.J.; Clark, H.F.; Lawley, D.; Bell, L.M.; Hodinka, R.L.; DiStefano, D.J.; Kulnis, G.; Zaoutis, T.E.; Coffin, S.E. The Clinical and Molecular Epidemiology of Community- and Healthcare-Acquired Rotavirus Gastroenteritis. Pediatr. Infect. Dis. J. 2008, 27, 54–58. [Google Scholar] [CrossRef]
- Gervasi, G.; Capanna, A.; Mita, V.; Zaratti, L.; Franco, E. Nosocomial Rotavirus Infection: An up to Date Evaluation of European Studies. Hum. Vaccin. Immunother. 2016, 12, 2413–2418. [Google Scholar] [CrossRef] [PubMed]
- Goldmann, D.A. Blood-Borne Pathogens and Nosocomial Infections. J. Allergy Clin. Immunol. 2002, 110, S21–S26. [Google Scholar] [CrossRef]
- Sin, W.W.; Lin, A.W.; Chan, K.C.; Wong, K.H. Management of Health Care Workers Following Occupational Exposure to Hepatitis B, Hepatitis C, and Human Immunodeficiency Virus. Hong Kong Med. J. 2016, 22, 472–477. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Askarian, M.; Yadollahi, M.; Kuochak, F.; Danaei, M.; Vakili, V.; Momeni, M. Precautions for Health Care Workers to Avoid Hepatitis B and C Virus Infection. Int. J. Occup. Environ. Med. 2011, 2, 191–198. [Google Scholar] [PubMed]
- Katzenberger, R.H.; Rösel, A.; Vonberg, R.-P. Bacterial Survival on Inanimate Surfaces: A Field Study. BMC Res. Notes 2021, 14, 97. [Google Scholar] [CrossRef]
- Kramer, A.; Guggenbichler, P.; Heldt, P.; Jünger, M.; Ladwig, A.; Thierbach, H.; Weber, U.; Daeschlein, G. Hygienic Relevance and Risk Assessment of Antimicrobial-Impregnated Textiles. Curr. Probl. Dermatol. 2006, 33, 78–109. [Google Scholar] [CrossRef]
- Woodford, N.; Turton, J.F.; Livermore, D.M. Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 736–755. [Google Scholar] [CrossRef]
- Paredes-Sabja, D.; Shen, A.; Sorg, J.A. Clostridium difficile Spore Biology: Sporulation, Germination, and Spore Structural Proteins. Trends Microbiol. 2014, 22, 406–416. [Google Scholar] [CrossRef]
- Jump, R.L.P.; Pultz, M.J.; Donskey, C.J. Vegetative Clostridium Difficile Survives in Room Air on Moist Surfaces and in Gastric Contents with Reduced Acidity: A Potential Mechanism to Explain the Association between Proton Pump Inhibitors and C. Difficile -Associated Diarrhea? Antimicrob. Agents Chemother. 2007, 51, 2883–2887. [Google Scholar] [CrossRef]
- Metcalf, D.S.; Costa, M.C.; Dew, W.M.V.; Weese, J.S. Clostridium difficile in Vegetables, Canada: Clostridium difficile in Vegetables. Lett. Appl. Microbiol. 2010, 51, 600–602. [Google Scholar] [CrossRef]
- Otter, J.A.; French, G.L. Survival of Nosocomial Bacteria and Spores on Surfaces and Inactivation by Hydrogen Peroxide Vapor. J. Clin. Microbiol. 2009, 47, 205–207. [Google Scholar] [CrossRef] [PubMed]
- Neely, A.N. A Survey of Gram-Negative Bacteria Survival on Hospital Fabrics and Plastics. J. Burn Care Rehabil. 2000, 21, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Esteves, D.C.; Pereira, V.C.; Souza, J.M.; Keller, R.; Simões, R.D.; Winkelstroter Eller, L.K.; Rodrigues, M.V.P. Influence of Biological Fluids in Bacterial Viability on Different Hospital Surfaces and Fomites. Am. J. Infect. Control 2016, 44, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Pathogen Safety Data Sheets: Infectious Substances–Klebsiella spp. Available online: https://www.canada.ca/en/public-health/services/laboratory-biosafety-biosecurity/pathogen-safety-data-sheets-risk-assessment/klebsiella.html (accessed on 15 April 2022).
- Lanjri, S.; Uwingabiye, J.; Frikh, M.; Abdellatifi, L.; Kasouati, J.; Maleb, A.; Bait, A.; Lemnouer, A.; Elouennass, M. In Vitro Evaluation of the Susceptibility of Acinetobacter Baumannii Isolates to Antiseptics and Disinfectants: Comparison between Clinical and Environmental Isolates. Antimicrob. Resist. Infect. Control 2017, 6, 36. [Google Scholar] [CrossRef]
- Siroli, L.; Patrignani, F.; Serrazanetti, D.I.; Chiavari, C.; Benevelli, M.; Grazia, L.; Lanciotti, R. Survival of Spoilage and Pathogenic Microorganisms on Cardboard and Plastic Packaging Materials. Front. Microbiol. 2017, 8, 2606. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.; Bloomfield, S.F. The Survival and Transfer of Microbial Contamination via Cloths, Hands and Utensils. J. Appl. Bacteriol. 1990, 68, 271–278. [Google Scholar] [CrossRef]
- Public Health Agency of Canada. Pathogen Safety Data Sheets: Infectious Substances–Escherichia coli, enterohemorrhagic. Canada.ca. Available online: https://www.canada.ca/en/public-health/services/laboratory-biosafety-biosecurity/pathogen-safety-data-sheets-risk-assessment/escherichia-coli-enterohemorrhagic.html (accessed on 8 June 2022).
- Waman, P.; Swapnil, S.; Krishna, K.; Rajshree, C.; Chetan, K.; Naresh, B.; Udit, P.; Ram, P.; Ghanshyam, L.B. Future prospects of plasma treatment technology for disinfection. In Engineering Technology of the 21st Century, 1st ed.; Roy, A.K., Ed.; New India Publishing Agency: New Delhi, India, 2015; Chapter 10. [Google Scholar]
- Pina Pérez, M.C.; Rodrigo Aliaga, D.; Saucedo Reyes, D.; Martínez López, A. Pressure Inactivation Kinetics of Enterobacter sakazakii in Infant Formula Milk. J. Food Prot. 2007, 70, 2281–2289. [Google Scholar] [CrossRef]
- Weterings, V.; Veenemans, J.; Kleefman, A.; den Bergh, M.K.; Mulder, P.; Verhulst, C.; Willemsen, I.; Kluytmans, J. Evaluation of an in Vitro Model with a Novel Statistical Approach to Measure Differences in Bacterial Survival of Extended-Spectrum β-Lactamase-Producing Escherichia coli on an Inanimate Surface. Antimicrob. Resist. Infect. Control 2019, 8, 106. [Google Scholar] [CrossRef]
- Koca, O.; Altoparlak, U.; Ayyildiz, A.; Kaynar, H. Persistence of Nosocomial Pathogens on Various Fabrics. Eurasian J. Med. 2012, 44, 28–31. [Google Scholar] [CrossRef]
- Nasr, A.M.; Mostafa, M.S.; Arnaout, H.H.; Elshimy, A.A.A. The Effect of Exposure to Sub-Inhibitory Concentrations of Hypochlorite and Quaternary Ammonium Compounds on Antimicrobial Susceptibility of Pseudomonas aeruginosa. Am. J. Infect. Control 2018, 46, e57–e63. [Google Scholar] [CrossRef]
- Wagenvoort, J.H.; Sluijsmans, W.; Penders, R.J. Better Environmental Survival of Outbreak vs. Sporadic MRSA Isolates. J. Hosp. Infect. 2000, 45, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Chaibenjawong, P.; Foster, S.J. Desiccation Tolerance in Staphylococcus aureus. Arch. Microbiol. 2011, 193, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Neely, A.N.; Maley, M.P. Survival of Enterococci and Staphylococci on Hospital Fabrics and Plastic. J. Clin. Microbiol. 2000, 38, 724–726. [Google Scholar] [CrossRef] [PubMed]
- Moesby, L.; Hansen, E.W.; Christensen, J.D.; Høyer, C.H.; Juhl, G.L.; Olsen, H.B. Dry and Moist Heat Sterilisation Cannot Inactivate Pyrogenicity of Gram Positive Microorganisms. Eur. J. Pharm. Sci. 2005, 26, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Ayliffe, G.A.J. Control of Staphylococcus aureus and Enterococcal Infections. In Disinfection, Sterilization, and Preservation, 5th ed.; Block, S.S., Ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2001; pp. 491–504. [Google Scholar]
- Broukhanski, G.; Budylowski, P. Laboratory Plasticware–Use at Your Own Risk: Suitability of Microcentrifuge Tubes for Spores’ Analysis of Clostridium difficile. Anaerobe 2019, 55, 61–66. [Google Scholar] [CrossRef]
- Weber, D.J.; Anderson, D.J.; Sexton, D.J.; Rutala, W.A. Role of the Environment in the Transmission of Clostridium difficile in Health Care Facilities. Am. J. Infect. Control 2013, 41, S105–S110. [Google Scholar] [CrossRef]
- Weber, D.J.; Rutala, W.A.; Miller, M.B.; Huslage, K.; Sickbert-Bennett, E. Role of Hospital Surfaces in the Transmission of Emerging Health Care-Associated Pathogens: Norovirus, Clostridium difficile, and Acinetobacter species. Am. J. Infect. Control 2010, 38, S25–S33. [Google Scholar] [CrossRef]
- Lessa, F.C.; Gould, C.V.; McDonald, L.C. Current Status of Clostridium difficile Infection Epidemiology. Clin. Infect. Dis. 2012, 55, S65–S70. [Google Scholar] [CrossRef]
- Soyfoo, R.; Shaw, K. How to Cut Clostridium difficile Infection. Nurs. Times 2008, 104, 42–44. [Google Scholar]
- Claro, T.; Cahill, O.J.; O’Connor, N.; Daniels, S.; Humphreys, H. Cold-Air Atmospheric Pressure Plasma against Clostridium difficile Spores: A Potential Alternative for the Decontamination of Hospital Inanimate Surfaces. Infect. Control Hosp. Epidemiol. 2015, 36, 742–744. [Google Scholar] [CrossRef]
- Claro, T.; Daniels, S.; Humphreys, H. Detecting Clostridium difficile Spores from Inanimate Surfaces of the Hospital Environment: Which Method Is Best? J. Clin. Microbiol. 2014, 52, 3426–3428. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Ou, Q.; Lin, J.; Peng, Y.; Yao, Z. A Meta-Analysis of the Rates of Staphylococcus aureus and Methicillin-Resistant S Aureus Contamination on the Surfaces of Environmental Objects That Health Care Workers Frequently Touch. Am. J. Infect. Control 2017, 45, 421–429. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, L.M.; Calfee, D.P.; Miller, L.G.; Pineles, L.; Magder, L.S.; Johnson, J.K.; Morgan, D.J.; Harris, A.D. Optimizing Contact Precautions to Curb the Spread of Antibiotic-Resistant Bacteria in Hospitals: A Multicenter Cohort Study to Identify Patient Characteristics and Healthcare Personnel Interactions Associated with Transmission of Methicillin-Resistant Staphylococcus aureus. Clin. Infect. Dis. 2019, 69, S171–S177. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Jones, R.M.; Li, Y. Exploring Surface Cleaning Strategies in Hospital to Prevent Contact Transmission of Methicillin-Resistant Staphylococcus aureus. BMC Infect. Dis. 2017, 17, 85. [Google Scholar] [CrossRef]
- Cimolai, N. MRSA and the Environment: Implications for Comprehensive Control Measures. Eur. J. Clin. Microbiol. Infect. Dis. 2008, 27, 481–493. [Google Scholar] [CrossRef]
- Hardy, K.J.; Oppenheim, B.A.; Gossain, S.; Gao, F.; Hawkey, P.M. A Study of the Relationship between Environmental Contamination with Methicillin-Resistant Staphylococcus aureus (MRSA) and Patients’ Acquisition of MRSA. Infect. Control Hosp. Epidemiol. 2006, 27, 127–132. [Google Scholar] [CrossRef]
- Aldeyab, M.A.; McElnay, J.C.; Elshibly, S.M.; Hughes, C.M.; McDowell, D.A.; McMahon, M.A.S.; Scott, M.G.; Kearney, M.P. Evaluation of the Efficacy of a Conventional Cleaning Regimen in Removing Methicillin-Resistant Staphylococcus aureus from Contaminated Surfaces in an Intensive Care Unit. Infect. Control Hosp. Epidemiol. 2009, 30, 304–306. [Google Scholar] [CrossRef]
- Davis, M.F.; Iverson, S.A.; Baron, P.; Vasse, A.; Silbergeld, E.K.; Lautenbach, E.; Morris, D.O. Household Transmission of Meticillin-Resistant Staphylococcus aureus and Other Staphylococci. Lancet Infect. Dis. 2012, 12, 703–716. [Google Scholar] [CrossRef]
- Dietze, B.; Rath, A.; Wendt, C.; Martiny, H. Survival of MRSA on Sterile Goods Packaging. J. Hosp. Infect. 2001, 49, 255–261. [Google Scholar] [CrossRef]
- Hu, D.-L.; Nakane, A. Mechanisms of Staphylococcal Enterotoxin-Induced Emesis. Eur. J. Pharmacol. 2014, 722, 95–107. [Google Scholar] [CrossRef]
- Domon, H.; Uehara, Y.; Oda, M.; Seo, H.; Kubota, N.; Terao, Y. Poor Survival of Methicillin-Resistant Staphylococcus aureus on Inanimate Objects in the Public Spaces. Microbiologyopen 2016, 5, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Greene, C.; Wu, J.; Rickard, A.H.; Xi, C. Evaluation of the Ability of Acinetobacter Baumannii to Form Biofilms on Six Different Biomedical Relevant Surfaces. Lett. Appl. Microbiol. 2016, 63, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Chapartegui-González, I.; Lázaro-Díez, M.; Bravo, Z.; Navas, J.; Icardo, J.M.; Ramos-Vivas, J. Acinetobacter Baumannii Maintains Its Virulence after Long-Time Starvation. PLoS ONE 2018, 13, e0201961. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, T.; Okubo, T.; Enoeda, Y.; Yano, R.; Nakamura, S.; Thapa, J.; Yamaguchi, H. Effect of Thermal Control of Dry Fomites on Regulating the Survival of Human Pathogenic Bacteria Responsible for Nosocomial Infections. PLoS ONE 2019, 14, e0226952. [Google Scholar] [CrossRef]
- Drees, M.; Snydman, D.R.; Schmid, C.H.; Barefoot, L.; Hansjosten, K.; Vue, P.M.; Cronin, M.; Nasraway, S.A.; Golan, Y. Prior Environmental Contamination Increases the Risk of Acquisition of Vancomycin-Resistant Enterococci. Clin. Infect. Dis. 2008, 46, 678–685. [Google Scholar] [CrossRef]
- Harris, A.D.; Jackson, S.S.; Robinson, G.; Pineles, L.; Leekha, S.; Thom, K.A.; Wang, Y.; Doll, M.; Pettigrew, M.M.; Johnson, J.K. Pseudomonas aeruginosa Colonization in the Intensive Care Unit: Prevalence, Risk Factors, and Clinical Outcomes. Infect. Control Hosp. Epidemiol. 2016, 37, 544–548. [Google Scholar] [CrossRef]
- Garvey, M.I.; Bradley, C.W.; Tracey, J.; Oppenheim, B. Continued Transmission of Pseudomonas aeruginosa from a Wash Hand Basin Tap in a Critical Care Unit. J. Hosp. Infect. 2016, 94, 8–12. [Google Scholar] [CrossRef]
- Badiee, P.; Alborzi, A.; Joukar, M. Molecular Assay to Detect Nosocomial Fungal Infections in Intensive Care Units. Eur. J. Intern. Med. 2011, 22, 611–615. [Google Scholar] [CrossRef]
- Katiyar, S.; Edlind, T. New Locus for Candida Glabrata Sequence-Based Strain Typing Provides Evidence for Nosocomial Transmission. J. Clin. Microbiol. 2021, 59, e02933-20. [Google Scholar] [CrossRef]
- Pristov, K.E.; Ghannoum, M.A. Resistance of Candida to Azoles and Echinocandins Worldwide. Clin. Microbiol. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef]
- Shields, R.K.; Kline, E.G.; Healey, K.R.; Kordalewska, M.; Perlin, D.S.; Nguyen, M.H.; Clancy, C.J. Spontaneous Mutational Frequency and FKS Mutation Rates Vary by Echinocandin Agent against Candida Glabrata. Antimicrob. Agents Chemother. 2019, 63, e01692-18. [Google Scholar] [CrossRef] [PubMed]
- Schelenz, S.; Hagen, F.; Rhodes, J.L.; Abdolrasouli, A.; Chowdhary, A.; Hall, A.; Ryan, L.; Shackleton, J.; Trimlett, R.; Meis, J.F.; et al. First Hospital Outbreak of the Globally Emerging Candida Auris in a European Hospital. Antimicrob. Resist. Infect. Control 2016, 5, 35. [Google Scholar] [CrossRef]
- Traoré, O.; Springthorpe, V.S.; Sattar, S.A. A Quantitative Study of the Survival of Two Species of Candida on Porous and Non-Porous Environmental Surfaces and Hands. J. Appl. Microbiol. 2002, 92, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Sakita, K.M.; Faria, D.R.; da Silva, E.M.; Tobaldini-Valério, F.K.; Kioshima, E.S.; Svidzinski, T.I.E.; Bonfim-Mendonça, P. de S. Healthcare Workers’ Hands as a Vehicle for the Transmission of Virulent Strains of Candida spp.: A Virulence Factor Approach. Microb. Pathog. 2017, 113, 225–232. [Google Scholar] [CrossRef]
- Paluchowska, P.; Tokarczyk, M.; Bogusz, B.; Skiba, I.; Budak, A. Molecular Epidemiology of Candida Albicans and Candida Glabrata Strains Isolated from Intensive Care Unit Patients in Poland. Mem. Inst. Oswaldo Cruz 2014, 109, 436–441. [Google Scholar] [CrossRef]
- Gerson, S.L.; Parker, P.; Jacobs, M.R.; Creger, R.; Lazarus, H.M. Aspergillosis Due to Carpet Contamination. Infect. Control Hosp. Epidemiol. 1994, 15, 221–223. [Google Scholar] [CrossRef] [PubMed]
- Cristina, M.L.; Sartini, M.; Spagnolo, A.M. Health Care-Acquired Aspergillosis and Air Conditioning Systems. J. Prev. Med. Hyg. 2009, 50, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Neely, A.N.; Orloff, M.M. Survival of Some Medically Important Fungi on Hospital Fabrics and Plastics. J. Clin. Microbiol. 2001, 39, 3360–3361. [Google Scholar] [CrossRef]
- Prigione, V.; Lingua, G.; Marchisio, V.F. Development and Use of Flow Cytometry for Detection of Airborne Fungi. Appl. Environ. Microbiol. 2004, 70, 1360–1365. [Google Scholar] [CrossRef][Green Version]
- Baudisch, C.; Assadian, O.; Kramer, A. Evaluation of Errors and Limits of the 63-Μm House-Dust-Fraction Method, a Surrogate to Predict Hidden Moisture Damage. BMC Res. Notes 2009, 2, 218. [Google Scholar] [CrossRef]
- Biswal, M.; Rudramurthy, S.M.; Jain, N.; Shamanth, A.S.; Sharma, D.; Jain, K.; Yaddanapudi, L.N.; Chakrabarti, A. Controlling a Possible Outbreak of Candida Auris Infection: Lessons Learnt from Multiple Interventions. J. Hosp. Infect. 2017, 97, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Rutala, W.A.; Weber, D.J. Disinfection and Sterilization in Health Care Facilities: An Overview and Current Issues. Infect. Dis. Clin. North. Am. 2016, 30, 609–637. [Google Scholar] [CrossRef] [PubMed]
- Dubberke, E.R.; Reske, K.A.; Noble-Wang, J.; Thompson, A.; Killgore, G.; Mayfield, J.; Camins, B.; Woeltje, K.; McDonald, J.R.; McDonald, L.C.; et al. Prevalence of Clostridium difficile Environmental Contamination and Strain Variability in Multiple Health Care Facilities. Am. J. Infect. Control 2007, 35, 315–318. [Google Scholar] [CrossRef] [PubMed]
- WHO. Infection Prevention and Control during Health Care When Novel Coronavirus (nCoV) Infection Is Suspected. Who.int. Available online: https://www.who.int/publications/i/item/10665-331495 (accessed on 9 June 2022).
- Dev Kumar, G.; Mishra, A.; Dunn, L.; Townsend, A.; Oguadinma, I.C.; Bright, K.R.; Gerba, C.P. Biocides and Novel Antimicrobial Agents for the Mitigation of Coronaviruses. Front. Microbiol. 2020, 11, 1351. [Google Scholar] [CrossRef] [PubMed]
- Sakudo, A.; Yagyu, Y.; Onodera, T. Disinfection and Sterilization Using Plasma Technology: Fundamentals and Future Perspectives for Biological Applications. Int. J. Mol. Sci. 2019, 20, 5216. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xu, R.; Zhao, Y.; Liu, D.; Liu, Z.; Wang, X.; Chen, H.; Kong, M.G. Gas Plasma Pre-Treatment Increases Antibiotic Sensitivity and Persister Eradication in Methicillin-Resistant Staphylococcus aureus. Front. Microbiol. 2018, 9, 537. [Google Scholar] [CrossRef]
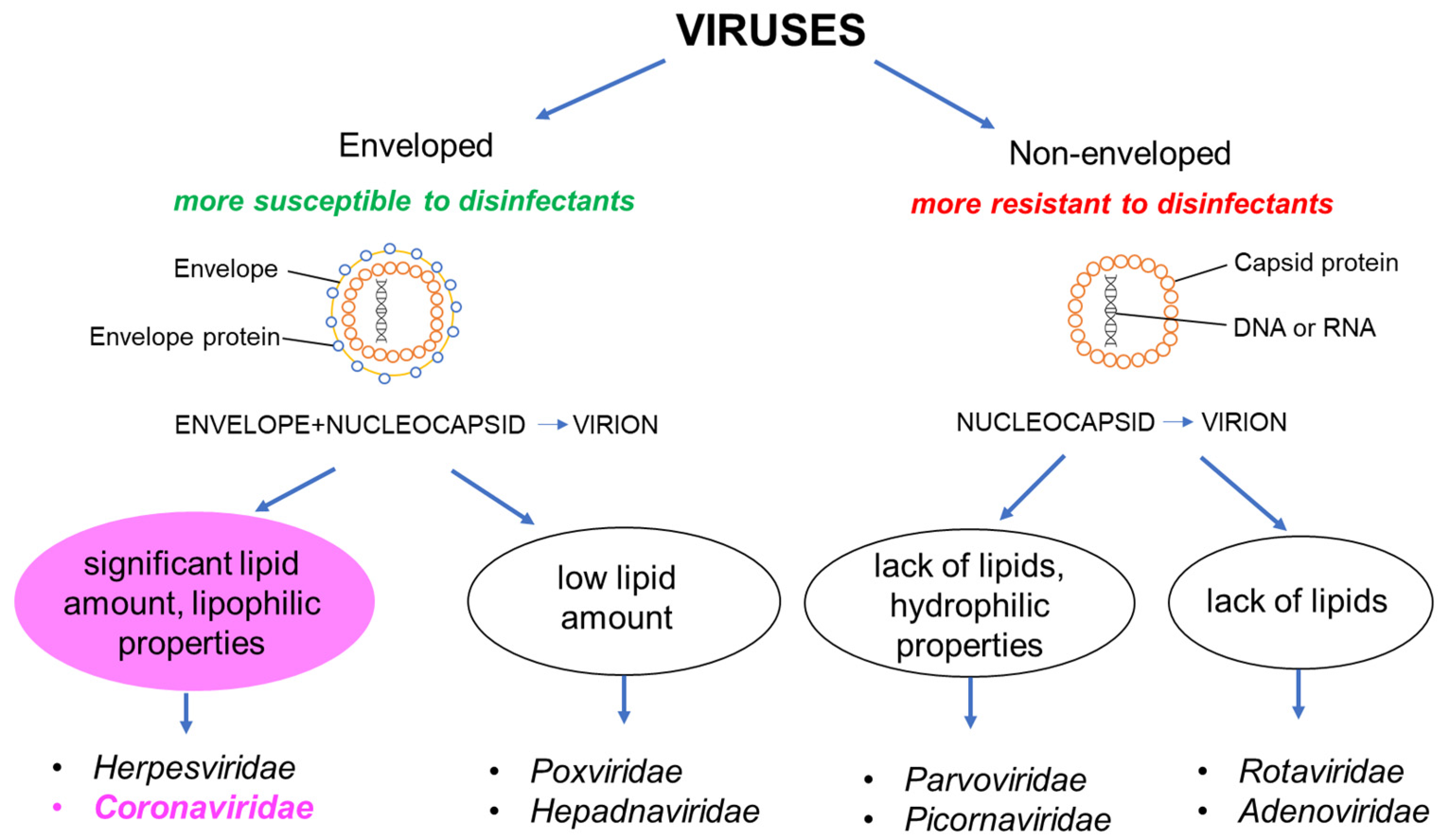
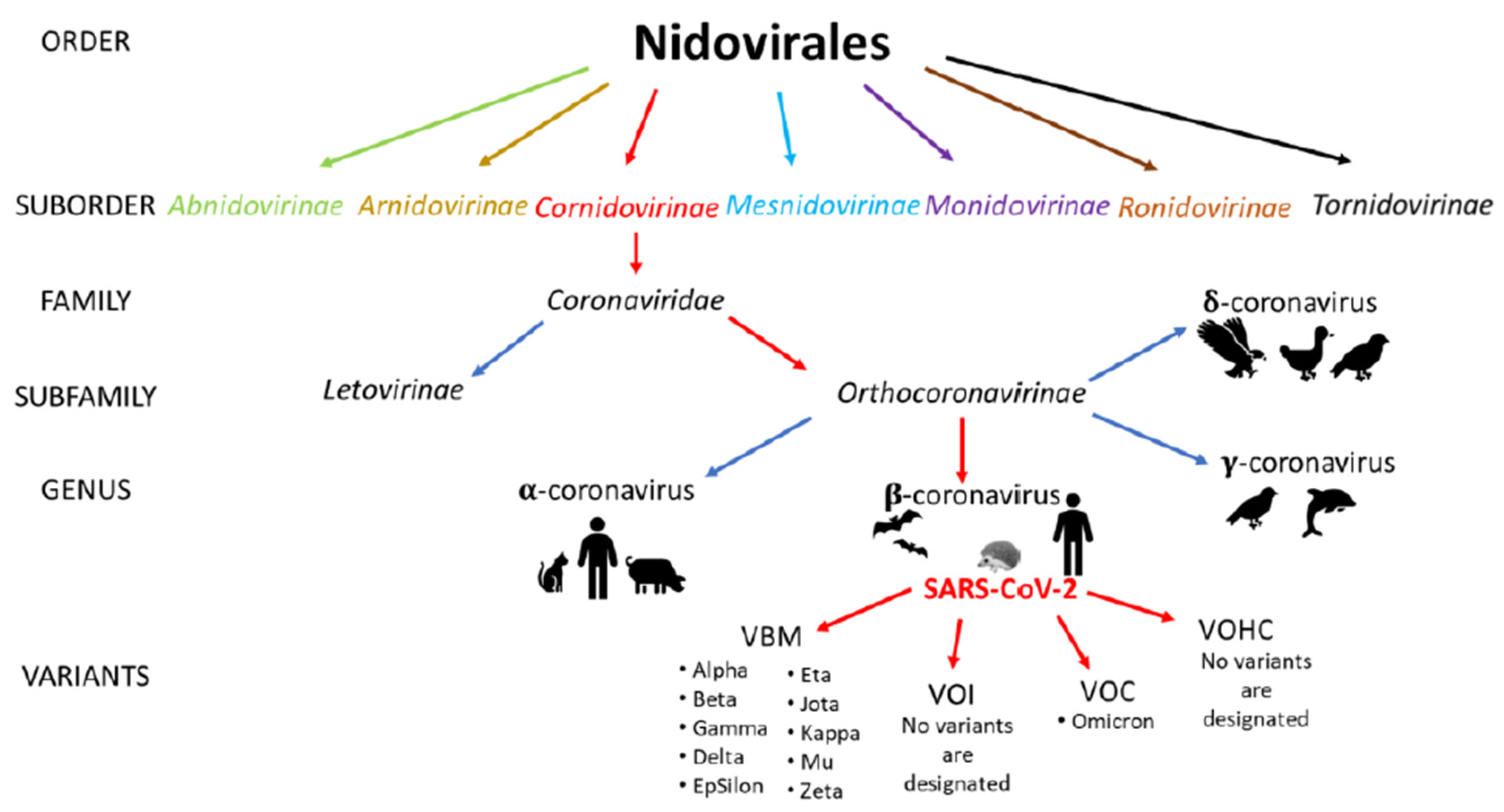
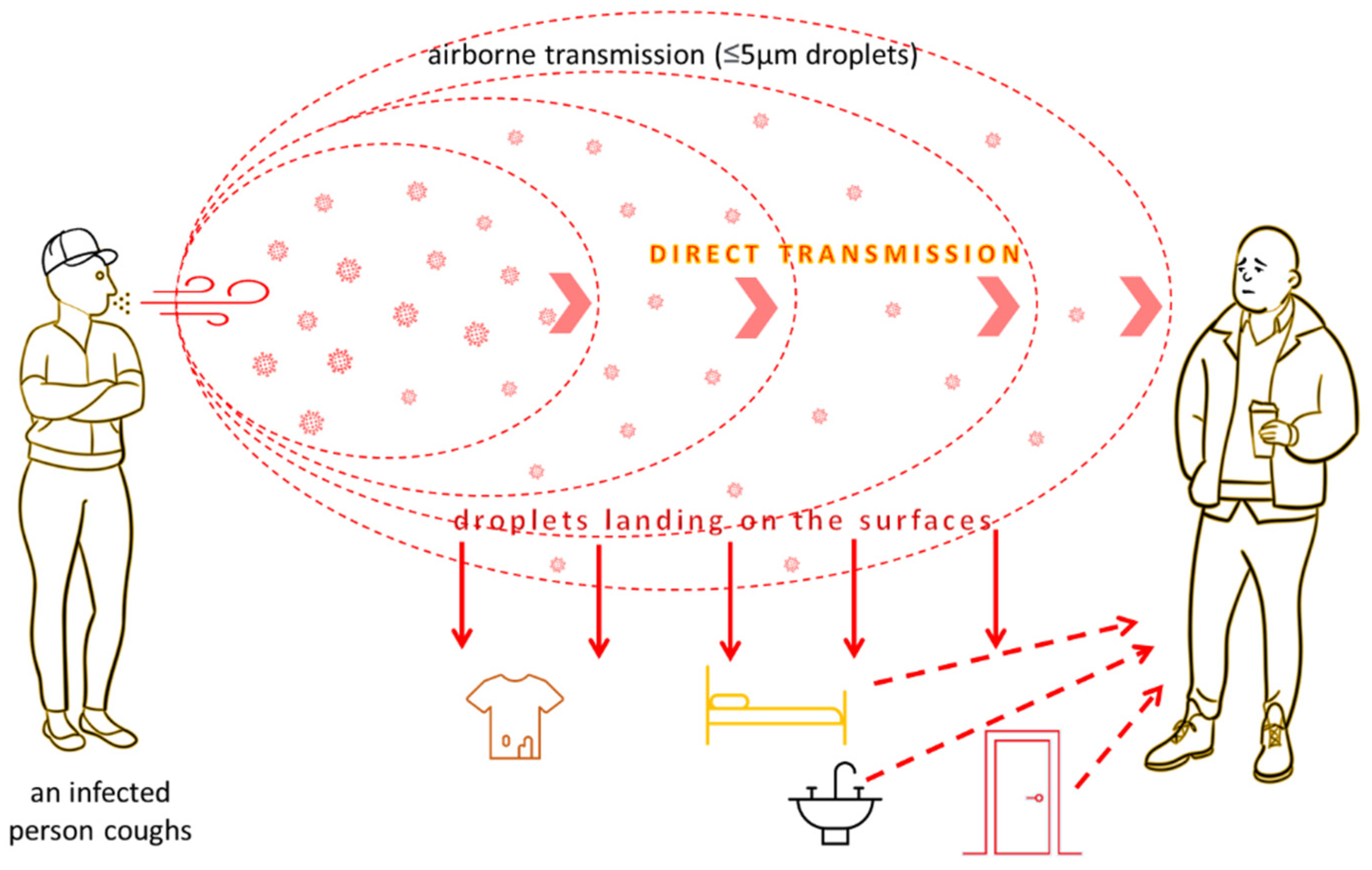

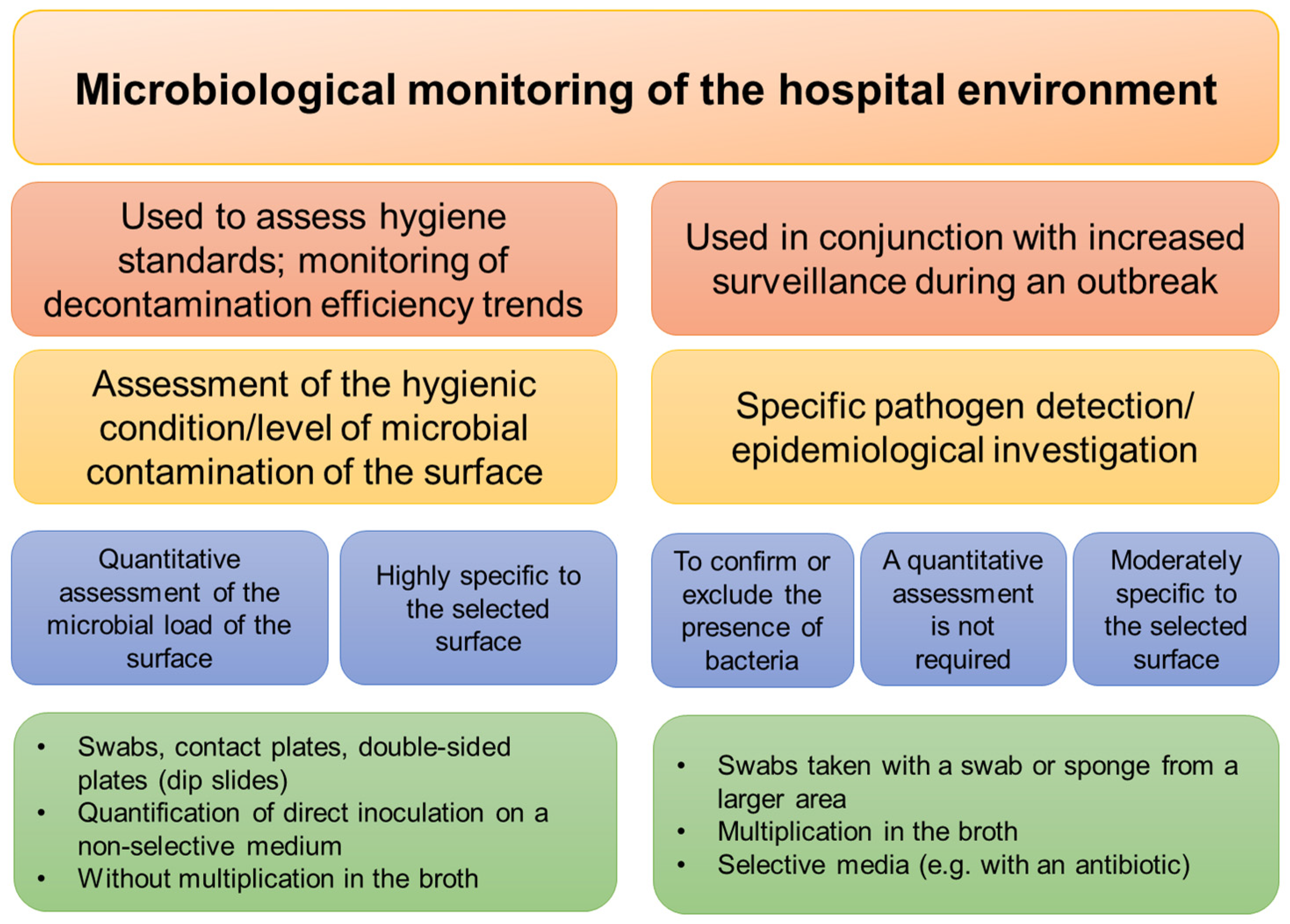
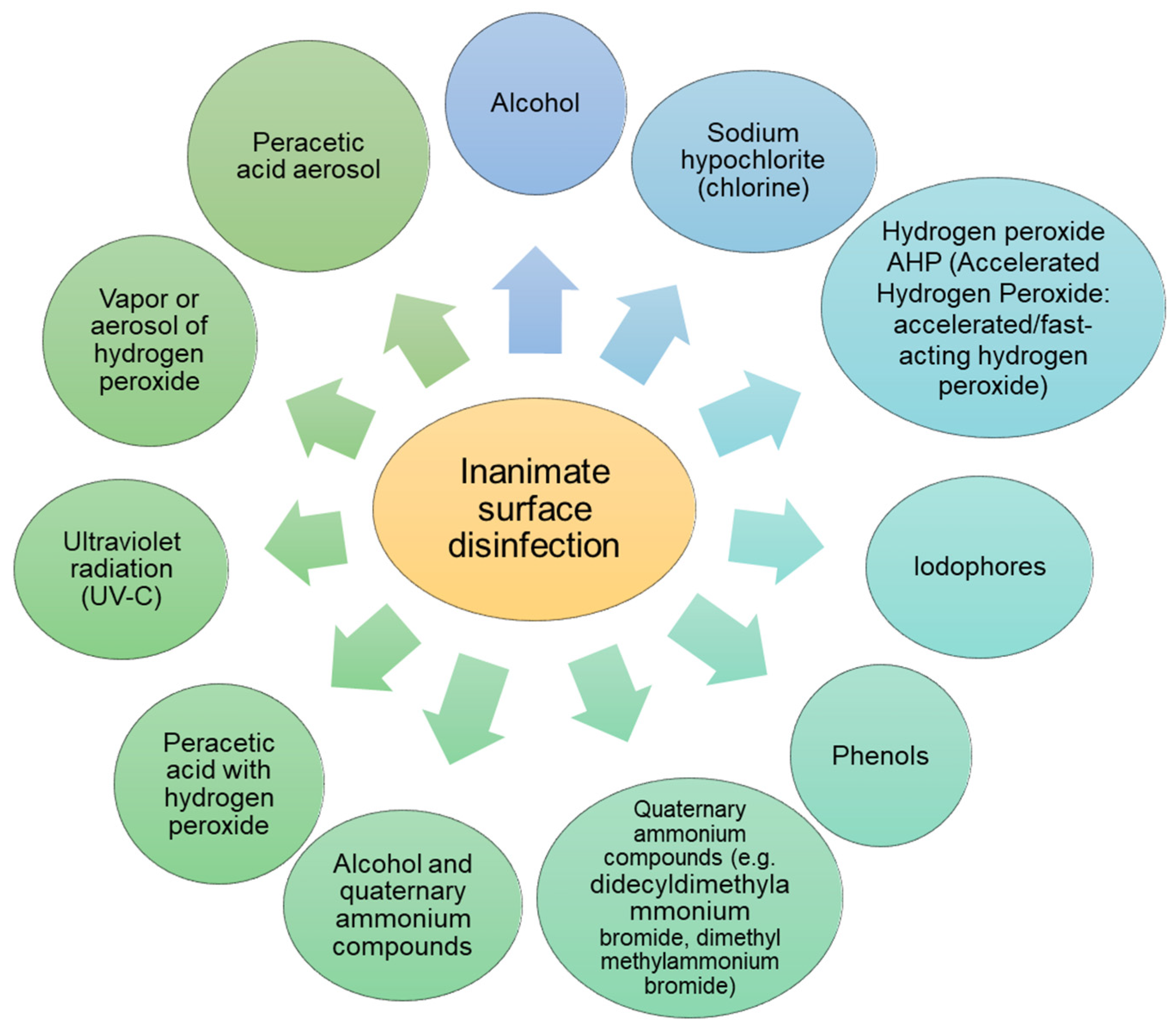
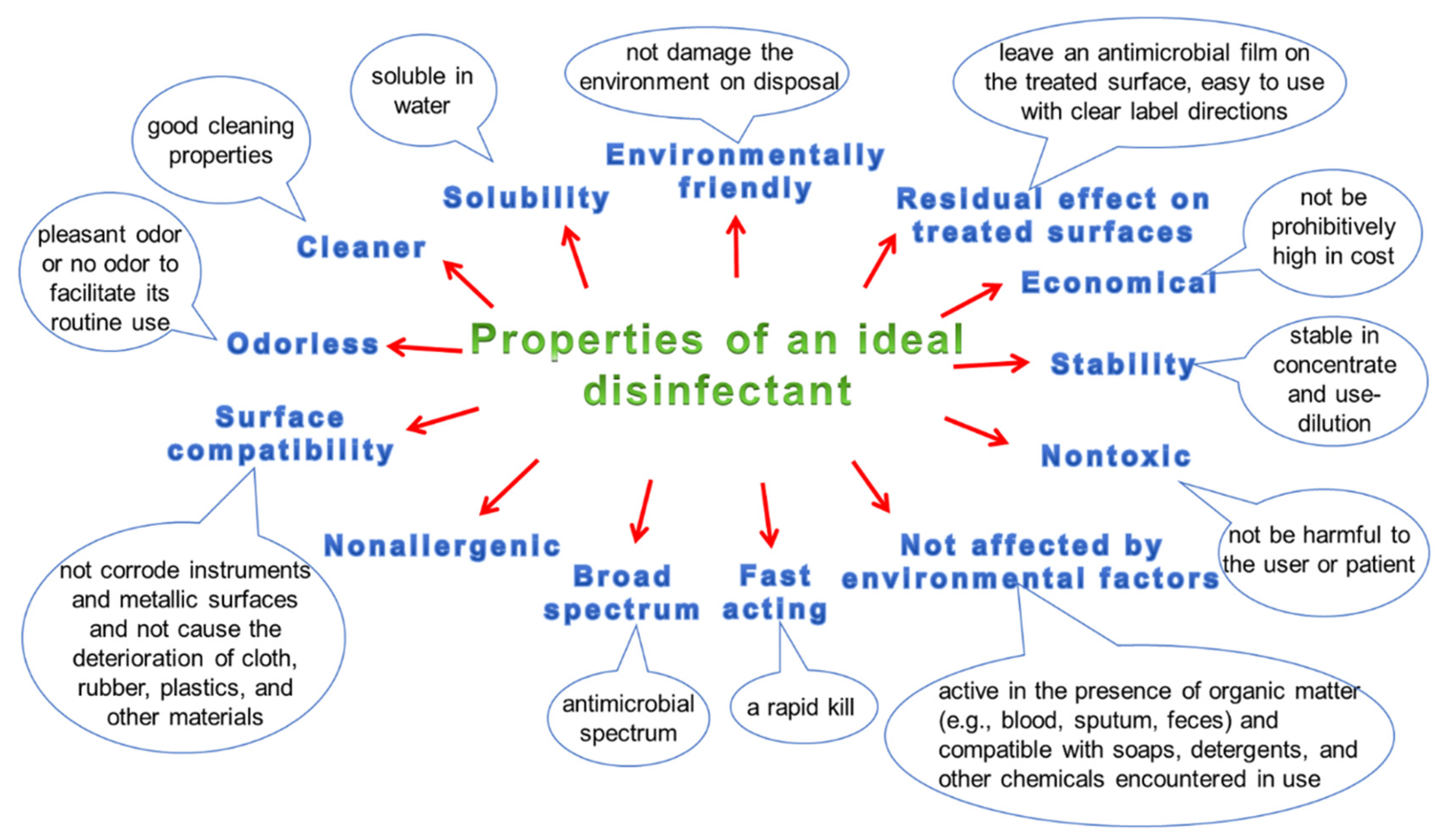
| Pathogen | Material | Survival on Surfaces | Susceptibility/Resistance to Disinfectants | Physical Inactivation | Ref. |
|---|---|---|---|---|---|
| Clostridium difficile | Stainless steel | >6 weeks | Clostridium spores are resistant to ethyl and propyl alcohols; high level disinfectants such as 2% glutaraldehyde, 8% formaldehyde and 20 ppm sodium hypochlorite can kill spores within 20 min | inactivated by moist heat at 121 °C for 15–30 min | [73,74,75] |
| Glass | 15 min | ||||
| Flooring material | 5 months | ||||
| Klebsiella pneumoniae | Plastics surfaces | 9–32 days | Gram-negative bacteria are susceptible to disinfectants including phenolic compounds, alcohols (70% ethanol), hypochlorites (1% sodium hypochlorite), glutaraldehyde, iodines (0.075 g/L) and formaldehyde (18.5 g/L; 5% formalin in water). | Reduction in the growth and metabolic activity at temperatures > 35 °C and significant growth reduction at 60 °C; | [76,77,78,79] |
| Stainless steel | 3–6 weeks | ||||
| Ceramics/Flooring material | 2 weeks | ||||
| fabrics | <1 h–4 weeks | ||||
| Acinetobacter sp. | Glass | 7–20 days | Susceptible to disinfectants such as povidone-iodine, 0.5% chlorhexidine digluconate, 70% ethyl alcohol and didecyl dimethyl ammonium chloride in combination with N-(3-aminopropyl)-N-dodecylpropane-1, 3-diamine; glutaraldehyde-based product has a high-level disinfection claim of 5 min at 35 °C. | successfully survived at −20 to 44 °C; inactivated by moist heat at 70 °C for 30 min and dry heat (160–170 °C for 1–2 h) | [80] |
| Fabrics | 25 days | ||||
| Paper | 6 days | ||||
| Escherichia coli | Glass | 1–≥14 days | Susceptible to many disinfectants—1% sodium hypochlorite, 70% ethanol, phenolics, glutaraldehyde, iodines, formaldehyde | Heat sensitive, inactivated by moist heat (121 °C for at least 15 min) and dry heat (160–170 °C for at least 1 h) Dielectric-barrier discharges (DBDs) plasma | [77,78,81,82,83,84] |
| Steel | 14–>60 days | ||||
| Fabrics | 4 h–>8 weeks | ||||
| Plastics surfaces | 24 h–>300 days | ||||
| Flooring materials | 1 h–>8 weeks | ||||
| Enterobacter sakazakii | stainless steel | 1–24 days | Susceptible to 70–80% ethanol, 1% sodium hypochlorite, formaldehyde, glutaraldehyde, hydrogen peroxide, iodines, peracetic acid, and quaternary ammonium compounds | Inactivated by pulsed electric fields and high hydrostatic pressure; can persist longer at higher relative humidity and low temperature | [12,85] |
| Enterobacter cloacae | stainless steel | 1–24 days | Susceptible to 70–80% ethanol, 1% sodium hypochlorite, formaldehyde, glutaraldehyde, hydrogen peroxide, iodines, peracetic acid, and quaternary ammonium compounds | Can persist longer at higher relative humidity and low temperature; inactivated by moist heat (121 °C for 15 min–30 min) and dry heat (160–170 °C for 1–2 h) | [12,86] |
| Pseudomonas aeruginosa | fabrics | 1 h–>8 weeks | Susceptibility has been shown for 1% sodium hypochlorite, 70% ethanol, 2% glutaraldehyde, and formaldehyde | Inactivation and sterilization by moist heat at 121 °C for 15 min or longer, dry heat at 170–250 °C or higher for 30 min or more; can persist longer at higher relative humidity and low temperature | [77,78,82,87,88] |
| Plastics surfaces | 9 h–10 days | ||||
| Flooring materials | 1 h–>8 weeks | ||||
| Stainless steel | 5 days | ||||
| Staphylococcus aureus (MRSA) | Glass | 15–25 days | Susceptible to 70% ethanol, clorhexidine, 1% sodium hypochlorite, 2% glutaraldehyde, 0.25% benzalkonium chloride, and formaldehyde | Can persist longer at low humidity; can be inactivated by dry heat (160–170 °C for 1 h) but not to moist heat treatment | [77,78,82,89,90] |
| fabrics | 1–>70 days | ||||
| Plastics surfaces | 21 days–>3 years | ||||
| Flooring materials | >4 h–8 weeks | ||||
| Stainless steel | 6 h–>6 weeks | ||||
| polyethylene | 90–1097 days | ||||
| Staphylococcus aureus (VRSA) | fabrics | 1–2 weeks | Susceptible to 70% ethanol, clorhexidine, 1% sodium hypochlorite, 2% glutaraldehyde, 0.25% benzalkonium chloride, and formaldehyde | Can persist longer at low humidity, grow in a pH of 4.2 to 9.3 and in salt concentrations of up to 15%; can be inactivated by dry heat (160–170 °C for 1 h) but not to moist heat treatment | [91,92] |
| polyethylene | >90 days | ||||
| countertop | 2-month | ||||
| Enterococcus sp. | fabrics | 5 to 7 days | Susceptible to 70% isopropyl alcohol, 70% ethanol, 5.25% sodium hypochlorite, phenolic and quaternary ammonia compounds, and glutaraldehyde. Resistant to 3% hydrogen peroxide | Enterococci are killed by temperatures in excess of 80 °C | [91,93] |
| Plastics surfaces | 1 day | ||||
| polyethylene | 5 days–2 months |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jabłońska-Trypuć, A.; Makuła, M.; Włodarczyk-Makuła, M.; Wołejko, E.; Wydro, U.; Serra-Majem, L.; Wiater, J. Inanimate Surfaces as a Source of Hospital Infections Caused by Fungi, Bacteria and Viruses with Particular Emphasis on SARS-CoV-2. Int. J. Environ. Res. Public Health 2022, 19, 8121. https://doi.org/10.3390/ijerph19138121
Jabłońska-Trypuć A, Makuła M, Włodarczyk-Makuła M, Wołejko E, Wydro U, Serra-Majem L, Wiater J. Inanimate Surfaces as a Source of Hospital Infections Caused by Fungi, Bacteria and Viruses with Particular Emphasis on SARS-CoV-2. International Journal of Environmental Research and Public Health. 2022; 19(13):8121. https://doi.org/10.3390/ijerph19138121
Chicago/Turabian StyleJabłońska-Trypuć, Agata, Marcin Makuła, Maria Włodarczyk-Makuła, Elżbieta Wołejko, Urszula Wydro, Lluis Serra-Majem, and Józefa Wiater. 2022. "Inanimate Surfaces as a Source of Hospital Infections Caused by Fungi, Bacteria and Viruses with Particular Emphasis on SARS-CoV-2" International Journal of Environmental Research and Public Health 19, no. 13: 8121. https://doi.org/10.3390/ijerph19138121
APA StyleJabłońska-Trypuć, A., Makuła, M., Włodarczyk-Makuła, M., Wołejko, E., Wydro, U., Serra-Majem, L., & Wiater, J. (2022). Inanimate Surfaces as a Source of Hospital Infections Caused by Fungi, Bacteria and Viruses with Particular Emphasis on SARS-CoV-2. International Journal of Environmental Research and Public Health, 19(13), 8121. https://doi.org/10.3390/ijerph19138121











