Bio-Enhanced Degradation Strategies for Fluoroquinolones in the Sewage Sludge Composting Stage: Molecular Modification and Resistance Gene Regulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Source of FQs and Biodegradation Enzymes in Sewage Sludge Composting Stage
2.2. Characterization of FQs Molecular Biodegradability in Sewage Sludge Composting Stage—Molecular Docking Method
2.3. Characterization of the Combined Biodegradability of Thermophilic Bacteria of FQs Sewage Sludge Composting Stage—Entropy Weight Method and Index Normalization Method
2.4. Construction of 3D-QSAR Models of FQs Biodegradability for Single and Combined Thermophilic Bacteria—CoMFA Method
2.5. Optimization of External Environmental Conditions for Combined Biodegradability Improvement of FQs Sewage Sludge Composting Stage under Multiple Scenarios
2.5.1. Characterization of Key Proteins Affecting the Combined Biodegradability of FQs Sewage Sludge Composting Stage—Protein–Protein Docking
2.5.2. Screening of External Environmental Conditions Affecting the Combined Biodegradability of FQs Sewage Sludge Composting Stage and Setting of Degradation Scenarios
2.5.3. Optimization of External Environmental Conditions for Combined Biodegradability Improvement of FQ Sewage Sludge Composting Stage—MD Simulation
3. Results and Discussion
3.1. Quantitative Analysis of Combined Biodegradation Levels of Single Thermophilic Bacteria and Thermophilic Group to FQs in Sewage Sludge Composting Stage
3.2. Construction and Evaluation of the Biodegradability 3D-QSAR Models of FQs by Thermophilic Bacteria in Sewage Sludge Composting Stage
3.3. Molecular Modification of FQs with Improved Biodegradation and Prediction of Relevant Properties for FQ Derivatives
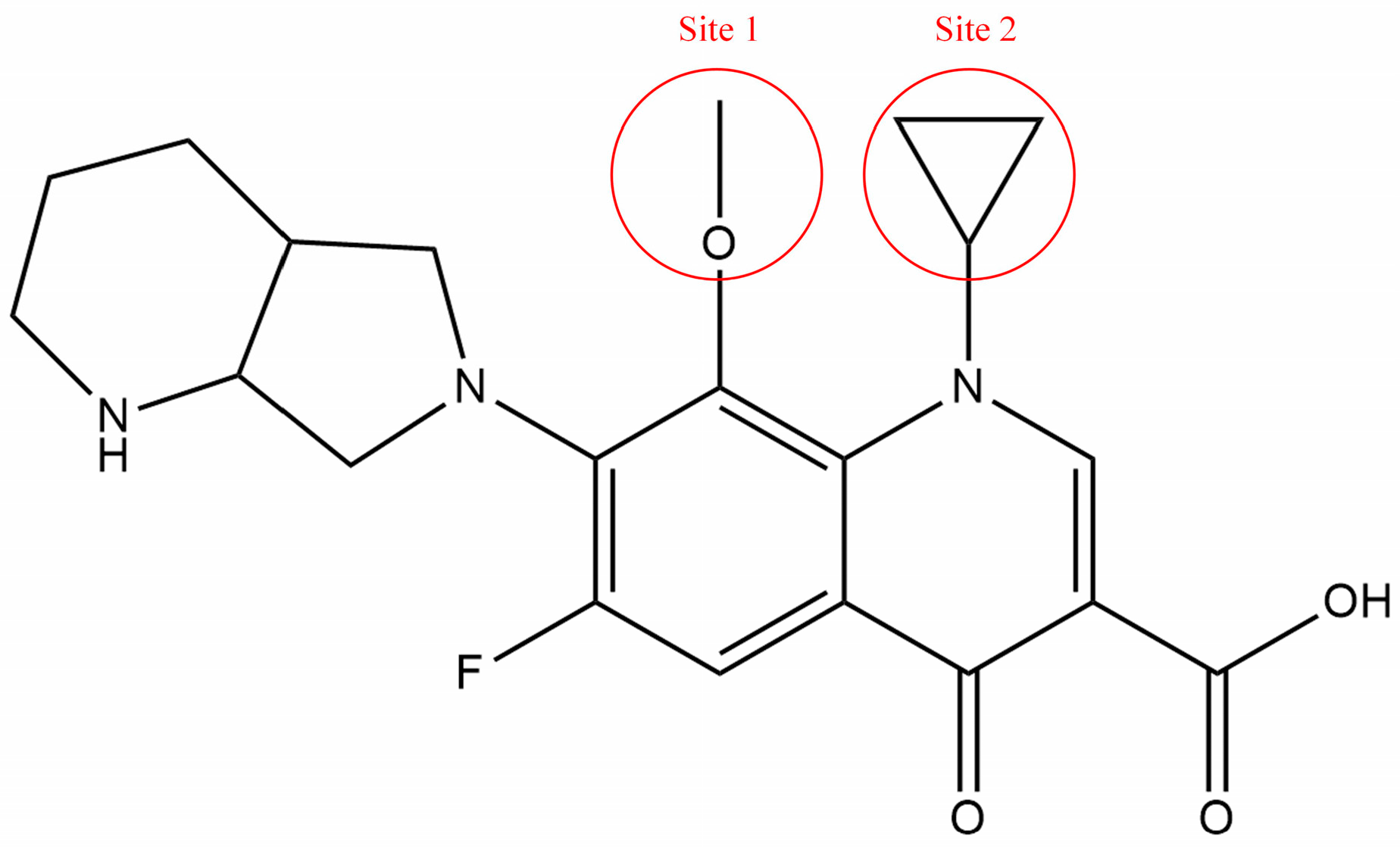
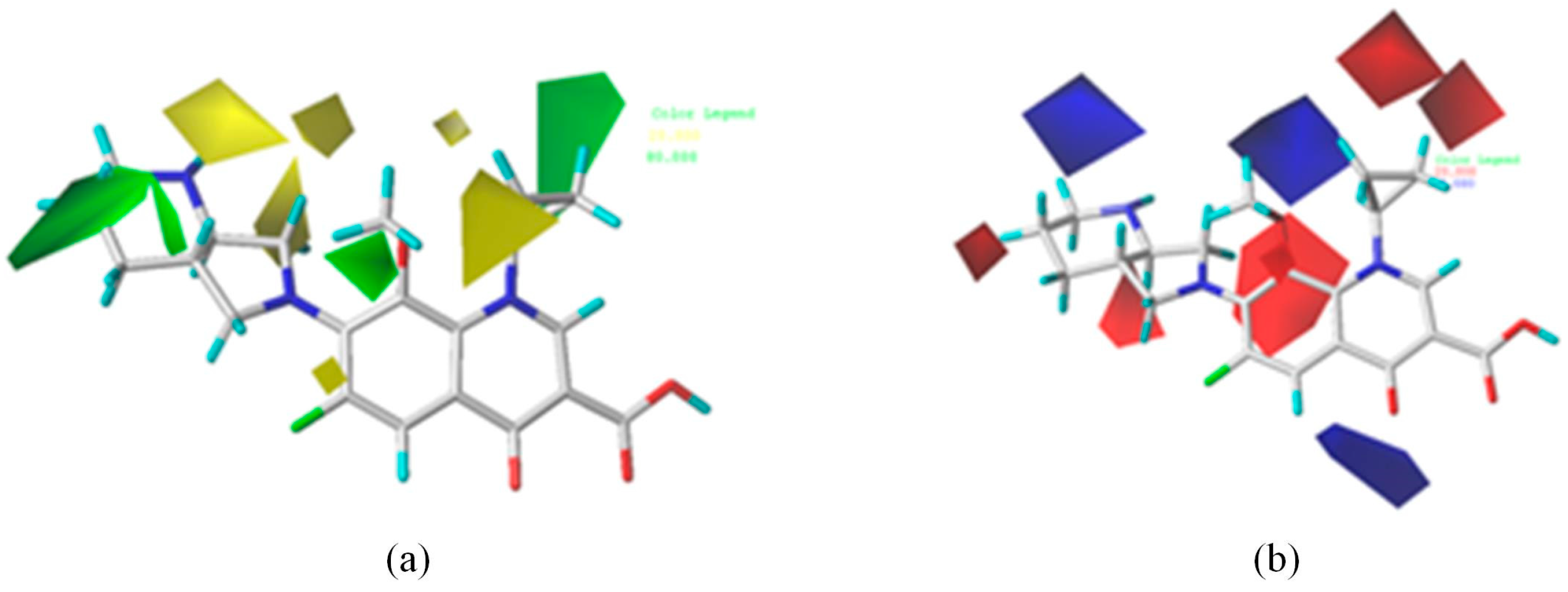
3.3.1. Determination of Modified Sites and Groups for FQs with High Combined Biodegradability Based on the Contour Maps
3.3.2. Evaluation of Biodegradability, Environmental Friendliness, and Functionality of MOX Derivatives
3.4. Regulation of External Environmental Conditions to Enhance the Combined Biodegradation of FQs by the Thermophilic Group in Sewage Sludge Composting Stage
3.4.1. Effectiveness Validation of External Environmental Conditions to Enhance the Combined Biodegradation of FQs by Thermophilic Group
3.4.2. Identification of Interactions between the External Environmental Conditions for Enhancing the Combined Biodegradation of FQs by Thermophilic Group
3.5. Regulatory Measures to Enhance the Ability of FQs Antibiotics to Inhibit Expression of Bacterial Resistance in Agricultural Soils
3.5.1. Characterization of the Ability of FQs Antibiotics to Inhibit the Expression of Bacterial Resistance
3.5.2. A Survey on Background Values of Nutrients in Agricultural Soils Based on the Sampling Method
3.5.3. Validation of Field Measures to Strengthen FQs Antibiotics to Inhibit Bacterial Resistance Expression
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone antibiotics. MedChemComm 2019, 10, 1719–1739. [Google Scholar] [CrossRef] [PubMed]
- Li, X.A.; Hou, Y.L.; Li, Q.; Gu, W.W.; Li, Y. Molecular design of high-efficacy and high drug safety Fluoroquinolones suitable for a variety of aerobic biodegradation bacteria. J. Environ. Manag. 2021, 299, 113628. [Google Scholar] [CrossRef] [PubMed]
- Pu, Q.K.; Han, Z.Z.; Li, X.A.; Li, Q.; Li, Y. Designing and screening of fluoroquinolone substitutes using combined in silico approaches: Biological metabolism–bioconcentration bilateral selection and their mechanism analyses. Green Chem. 2022, 24, 3778–3793. [Google Scholar] [CrossRef]
- Liu, X.; Li, M.; Chen, G.L. Fluoroquinolone Antibacterial Agent Contaminants in Soil/Groundwater: A Literature Review of Sources, Fate, and Occurrence. Water Air Soil Pollut. 2015, 226, 418. [Google Scholar]
- Hendricks, R.; Pool, E.J. The effectiveness of sewage treatment processes to remove faecal pathogens and antibiotic residues. J. Environ. Health Sci. 2012, 47, 289–297. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Liu, Y.; Dong, W.; Li, M.; Lu, S.; Wang, W.; Fan, Y. Occurrence and fate of ten sulfonamide antibiotics in typical wastewater treatment plants in the City of Jinan of Northeastern China. Desalin. Water Treat. 2020, 206, 340–348. [Google Scholar] [CrossRef]
- Paiva, M.C.; Reis, M.P.; Costa, P.S.; Dias, M.F.; Bleicher, L.; Scholte, L.L.S.; Nardi, R.M.D.; Nascimento, A.M.A. Identification of new bacteria harboring qnrS and aac(6′)-Ib/cr and mutations possibly involved in fluoroquinolone resistance in raw sewage and activated sludge samples from a full-scale WWTP. Water Res. 2017, 110, 27–37. [Google Scholar] [CrossRef]
- Jiang, Y. Removal of Fluoroquinolones Antibiotics in Sewage Sludge during Composting Fprocess: Characteristics and Mechanism; Guilin University of Technology: Guilin, China, 2017. (In Chinese) [Google Scholar]
- Selvam, A.; Zhao, Z.; Wong, J.W.C. Composting of swine manure spiked with sulfadiazine, chlortetracycline and ciprofloxacin. Bioresour. Technol. 2012, 126, 412–417. [Google Scholar] [CrossRef]
- Meng, L. Effect of Inoculation of a High Temperature-Resistant Bacterium and Thermophilic Composting on the Removal of Fluoroquinolones in Broiler Manures; Hunan Agricultural University: Changsha, China, 2015. (In Chinese) [Google Scholar]
- Pan, L.J.; Li, J.; Li, C.X.; Wang, Y. Biodegradation of fluoroquinolones by Thermus thermophilus. Chin. J. Environ. Eng. 2018, 14, 1092–1102. (In Chinese) [Google Scholar]
- Tumini, M.; Nagel, O.; Molina, M.P.; Althaus, R. Microbiological assay with Bacillus licheniformis for the easy detection of quinolones in milk. Int. Dairy J. 2017, 64, 9–13. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Ouyang, W.Y.; Wu, N.; Su, J.; Qiao, M. Antibiotic resistance: Sources and mitigation. Bull. Chin. Acad. Sci. 2015, 30, 509–516. [Google Scholar]
- National Health Commission of the People’s Republic of China (NHC). Antimicrobial Management and Bacterial Resistance in China; Peking Union Medical College Press: Beijing, China, 2018. [Google Scholar]
- Lewin, C.S.; Morrissey, I.; Smith, J.T. The mode of action of quinolones: The paradox in activity of low and high concentrations and activity in the anaerobic environment. Eur. J. Clin. Microbiol. 1991, 10, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.X.; Wang, Y.W.; Xu, H.H.; Li, Y.F.; Han, S. Fuzzy Comprehensive Evaluation Assistant 3D-QSAR of Environmentally Friendly FQs to Reduce ADRs. Int. J. Environ. Res. Public Health 2019, 16, 3161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, Z.X.; Xu, H.H.; Li, Y.F.; Wang, Y.W.; Han, S.; Ren, J.B. Combined toxicity characteristics and regulation of residual quinolone antibiotics in water environment. Chemosphere 2021, 263, 128301. [Google Scholar] [CrossRef]
- Mutiyar, P.K.; Mittal, A.K. Occurrences and fate of selected human antibiotics in influents and effluents of sewage treatment plant and effluent-receiving river Yamuna in Delhi (India). Environ. Monit. Assess. 2014, 186, 541–557. [Google Scholar] [CrossRef]
- Barancheshme, F.; Munir, M. Strategies to combat antibiotic resistance in the wastewater treatment plants. Front. Microbiol. 2018, 8, 2603. [Google Scholar] [CrossRef] [Green Version]
- Ju, F.; Beck, K.; Yin, X.; Maccagnan, A.; McArdell, C.S.; Singer, H.P.; Johnson, D.R.; Zhang, T.; Bürgmann, H. Wastewater treatment plant resistomes are shaped by bacterial composition, genetic exchange, and upregulated expression in the effluent microbiomes. ISME J. 2019, 13, 346–360. [Google Scholar] [CrossRef] [Green Version]
- Lopsik, K. Life cycle assessment of small-scale constructed wetland and extended aeration activated sludge wastewater treatment system. Int. J. Environ. Sci. Technol. 2013, 10, 1295–1308. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Chen, H.; Liu, S.; Xiao, L. Removal of pathogen and antibiotic resistance genes from waste activated sludge by different pre-treatment approaches. Sci. Total Environ. 2021, 763, 143014. [Google Scholar] [CrossRef]
- Migliore, L.; Cozzolino, S.; Fiori, M. Phytotoxicity to and uptake of enrofloxacin in crop plants. Chemosphere 2003, 52, 1233–1244. [Google Scholar] [CrossRef]
- Li, X.X.; Zhao, Y.Y.; Chen, B.; Zhu, Z.W.; Kang, Q.; Husain, T.; Zhang, B.Y. Inhalation and ingestion of Synthetic musks in pregnant women: In silico spontaneous abortion risk evaluation and control. Environ. Int. 2022, 158, 106911. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.X.; Wang, S.; Liu, D.; Yu, J.; Zhang, X.Y.; Zhao, P.N.; Sun, Y.X.; Han, S. Control strategies for the vertical gene transfer of quinolone ARGs in Escherichia coli through molecular modification and molecular dynamics. J. Hazard. Mater. 2021, 420, 126667. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.W.; Zhao, Y.Y.; Li, Q.; Li, Y. Environmentally friendly polychlorinated naphthalenes (PCNs) derivatives designed using 3D-QSAR and screened using molecular docking, density functional theory and health-based risk assessment. J. Hazard. Mater. 2019, 363, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Qu, R.J.; Li, C.G.; Liu, J.Q.; Xiao, R.Y.; Pan, X.X.; Zeng, X.L.; Wang, Z.Y.; Wu, J.C. Hydroxyl radical based photocatalytic degradation of halogenated organic contaminants and paraffin on silica gel. Environ. Sci. Technol. 2018, 52, 7220–7229. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Hou, Y.L.; Li, Y. Multi-directional selective toxicity effects on farmland ecosystems: A novel design of green substitutes for neonicotinoid insecticides. J. Clean. Prod. 2020, 272, 122715. [Google Scholar] [CrossRef]
- Wang, Q.; Awasthi, M.K.; Zhao, J.; Ren, X.; Li, R.; Wang, Z.; Wang, M.; Zhang, Z. Improvement of pig manure compost lignocellulose degradation, organic matter humification and compost quality with medical stone. Bioresour. Technol. 2017, 243, 771–777. [Google Scholar] [CrossRef]
- Guo, R.; Li, G.; Jiang, T.; Schuchardt, F.; Chen, T.; Zhao, Y.; Shen, Y. Effect of aeration rate, C/N ratio and moisture content on the stability and maturity of compost. Bioresour. Technol. 2012, 112, 171–178. [Google Scholar] [CrossRef]
- Bernal, M.P.; Alburquerque, J.A.; Moral, R. Composting of animal manures and chemical criteria for compost maturity assessment. A review. Bioresour. Technol. 2009, 100, 5444–5453. [Google Scholar] [CrossRef]
- Iqbal, M.K.; Shafiq, T.; Ahmed, K. Characterization of bulking agents and its effects on physical properties of compost. Bioresour. Technol. 2010, 101, 1913–1919. [Google Scholar] [CrossRef]
- Wang, M.J.; Awasthi, M.K.; Wang, Q.; Chen, H.Y.; Ren, X.N.; Zhao, J.C.; Li, R.H.; Zhang, Z.Q. Comparison of additives amendment for mitigation of greenhouse gases and ammonia emission during sewage sludge co-composting based on correlation analysis. Bioresour. Technol. 2017, 243, 520–527. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Li, Y. Design of environmentally friendly neonicotinoid insecticides with bioconcentration tuning and Bi-directional selective toxic effects. J. Clean. Prod. 2019, 221, 113–121. [Google Scholar] [CrossRef]
- Zhao, X.H.; Chu, Z.H.; Li, Y. Molecular Design of Lower Photodegradation Fluoroquinolone Antibiotics and Their Photolysis Paths Inference. Chem. J. Chin. Univ. 2018, 12, 2707–2718. (In Chinese) [Google Scholar]
- Zhao, X.H.; Zhao, Y.Y.; Ren, Z.X.; Li, Y. Combined QSAR/QSPR and molecular docking study on fluoroquinolones to reduce biological enrichment. Comput. Biol. Chem. 2019, 79, 177–184. [Google Scholar] [CrossRef]
- Zhao, X.H.; Wang, X.L.; Li, Y. Combined HQSAR method and molecular docking study on genotoxicity mechanism of quinolones with higher genotoxicity. Environ. Sci. Pollut. Res. 2019, 26, 34830–34853. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Li, G.X.; Yang, Q.Y.; Luo, W.H. Effect of bulking agents on maturity and gaseous emissions during kitchen waste composting. Chemosphere 2013, 93, 1393–1399. [Google Scholar] [CrossRef]
- Yang, J.W.; Gu, W.W.; Chu, Z.H.; Li, Y. Adsorption Mechanism of Metalaxyl Pesticide in Pesticide/Heavy Metal Sediment Using Fractional Factorial Design/Fixed Effects Models. Pol. J. Environ. Stud. 2019, 29, 1435–1449. [Google Scholar] [CrossRef]
- Gu, W.J.; Zhang, F.B.; Xu, P.Z.; Xie, K.Z.; Tang, S.H. Nitrogen conservation by adding sulfur to dairy manure in compost bioreactors. Plant Nutr. Fertil. Sci. 2017, 17, 224–230. (In Chinese) [Google Scholar]
- Tao, Y.; Zhan, H.H.; Tang, L.S. Effect of adding apple residue on physicochemical properties of swine feces aerobic compost. China Soil Fertil. 2019, 5, 135–140. (In Chinese) [Google Scholar]
- Seleiman, M.F.; Santanen, A.; Mäkelä, P.S. Recycling sludge on cropland as fertilizer—Advantages and risks. Resour. Conserv. Recycl. 2020, 155, 104647. [Google Scholar] [CrossRef]
- Tang, X.H.; Zhao, L. The development of sludge disposal strategy. Environ. Sci. Manag. 2005, 30, 68–70. [Google Scholar]
- Acharya, A.S.; Prakash, A.; Saxena, P.; Nigam, A. Sampling: Why and how of it. Indian J. Med. Spec. 2013, 4, 330–333. [Google Scholar] [CrossRef]
- Bandick, A.K.; Dick, R.P. Field management effects on soil enzyme activities. Soil Biol. Biochem. 1999, 31, 1471–1479. [Google Scholar] [CrossRef]
- Yang, H.S. Resource management, soil fertility and sustainable crop production: Experiences of China. Agric. Ecosyst. Environ. 2006, 116, 27–33. [Google Scholar] [CrossRef]
- Xie, J.; Sun, X.; Ren, L.; Zhang, Y.Z.; Yi, Z. Studies on Lignocellulolytic Enzymes Production and Biomass Degradation of Pleurotus sp2 and Trametes gallica in Wheat Straw Cultures. Chin. J. Biotechnol. 2001, 17, 575–578. [Google Scholar]
- Saar, R.A.; Weber, J.H. Fulvic acid: Modifier of metal-ion chemistry. Environ. Sci. Technol. 1982, 16, 510A–517A. [Google Scholar] [CrossRef]

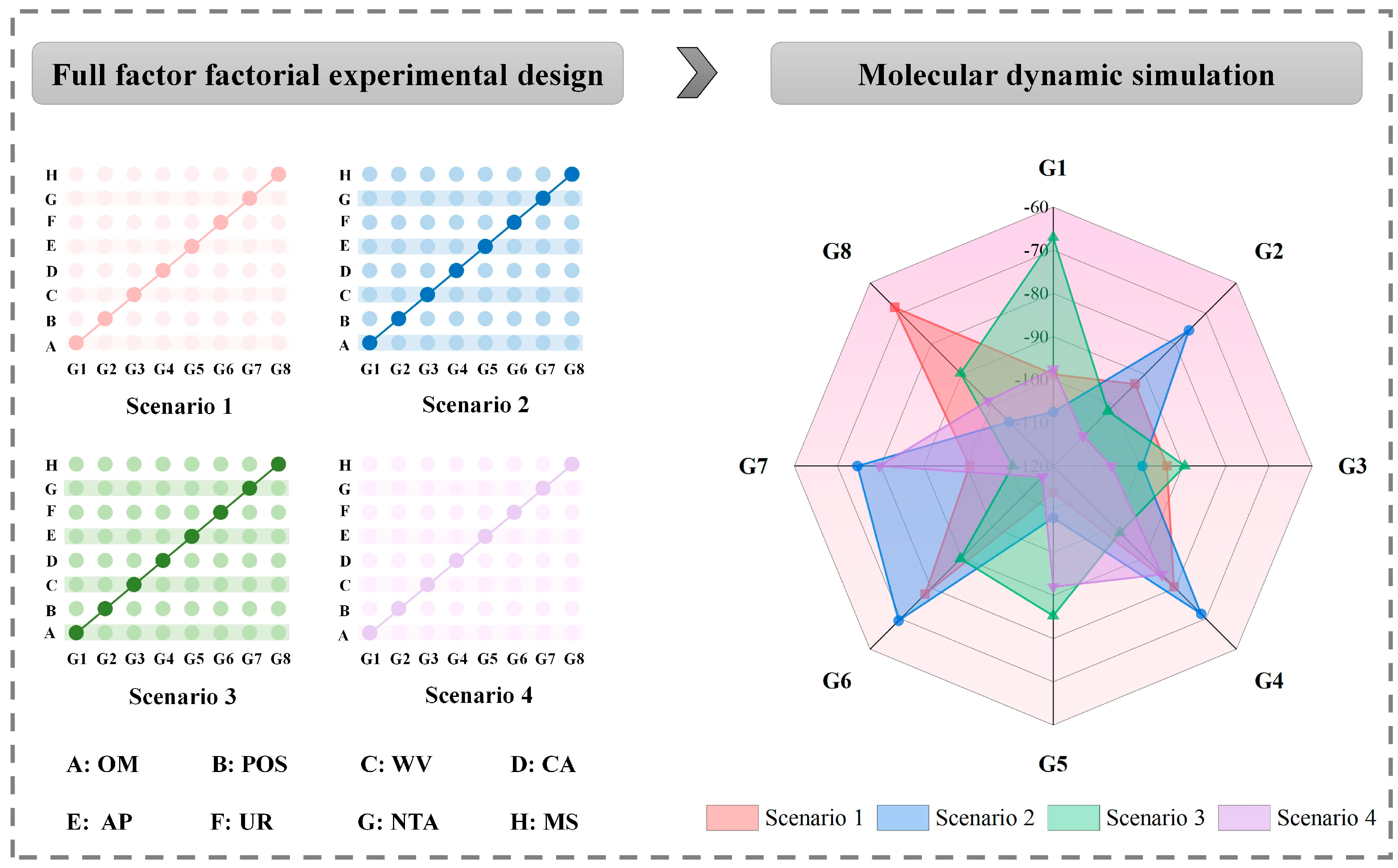

| Scenarios | Group | OM | POS | WV | CA | AP | UR | NTA | MS |
|---|---|---|---|---|---|---|---|---|---|
| Scenarios 1–4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| 5 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| 6 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| No. | LibDock Scores (Å) | No. | LibDock Scores (Å) | ||||
|---|---|---|---|---|---|---|---|
| 1GKQ | 1OB0 | 5M0K | 1GKQ | 1OB0 | 5M0K | ||
| DIF | 84.12 | 75.48 | 140.10 | GRE | 112.04 | 96.52 | 131.73 |
| ENR | 100.78 | 86.76 | 134.64 | ORB | 109.02 | 73.32 | 123.93 |
| NOR | 104.68 | 72.91 | 112.09 | SIT | 113.51 | 44.62 | 116.03 |
| LOM | 101.81 | 77.37 | 102.15 | TEM | 101.77 | 66.88 | 128.92 |
| OFL | 98.41 | 78.28 | 116.92 | D1 | 97.29 | 65.18 | 117.57 |
| PEF | 99.23 | 79.78 | 116.54 | D12 | 103.11 | 62.64 | 121.15 |
| FLE | 99.72 | 79.14 | 119.69 | D13 | 110.35 | 72.08 | 140.83 |
| CIP | 104.38 | 70.57 | 121.27 | D14 | 108.14 | 72.45 | 126.07 |
| BAL | 93.22 | 70.74 | 112.77 | D16 | 111.53 | 70.42 | 118.88 |
| MAR | 95.34 | 71.23 | 112.43 | D28 | 105.93 | 68.43 | 119.98 |
| PIP | 98.32 | 80.33 | 115.43 | D29 | 106.31 | 72.58 | 93.84 |
| CIN | 82.01 | 77.73 | 106.75 | D32 | 109.95 | 66.48 | 123.98 |
| ENO | 105.20 | 69.22 | 116.45 | D36 | 108.31 | 71.45 | 111.83 |
| DAN | 113.74 | 74.73 | 117.32 | D37 | 109.31 | 77.10 | 124.29 |
| GAT | 93.02 | 54.82 | 104.21 | F1 | 101.62 | 83.50 | 118.65 |
| LEV | 98.41 | 78.28 | 116.92 | F2 | 111.46 | 77.43 | 111.20 |
| RUF | 82.48 | 77.59 | 102.54 | F3 | 61.76 | 72.02 | 56.16 |
| PAZ | 95.01 | 79.25 | 123.49 | F4 | 91.93 | 76.03 | 91.83 |
| NAD | 108.43 | 81.41 | 112.09 | F5 | 99.71 | 65.73 | 113.26 |
| MOX | 83.16 | 69.91 | 83.47 | F6 | 113.41 | 81.72 | 105.99 |
| SPA | 114.87 | 72.45 | 121.10 | Gat-29 | 116.29 | 72.32 | 130.15 |
| SAR | 88.81 | 75.82 | 135.60 | Gat-30 | 117.05 | 77.81 | 129.55 |
| AMI | 97.24 | 73.04 | 105.85 | Gat-31 | 91.29 | 63.80 | 117.09 |
| BES | 82.54 | 59.00 | 119.14 | Gat-33 | 106.93 | 80.69 | 125.73 |
| CLI | 109.08 | 69.81 | 115.70 | ||||
| No. | CV | No. | CV | No. | CV | No. | CV |
|---|---|---|---|---|---|---|---|
| 1 | 5.857 | 14 | 6.205 | 27 | 6.185 | F1 | 6.154 |
| 2 | 6.792 | 15 | 4.129 | 28 | 4.882 | F2 | 6.027 |
| 3 | 5.605 | 16 | 5.752 | 29 | 5.800 | F3 | 2.050 |
| 4 | 5.351 | 17 | 4.630 | D1 | 5.177 | F4 | 4.571 |
| 5 | 5.752 | 18 | 5.881 | D12 | 5.413 | F5 | 5.149 |
| 6 | 5.834 | 19 | 6.108 | D13 | 6.747 | F6 | 6.110 |
| 7 | 5.931 | 20 | 3.696 | D14 | 6.185 | Gat-29 | 6.629 |
| 8 | 5.801 | 21 | 6.278 | D16 | 5.990 | Gat-30 | 6.871 |
| 9 | 5.095 | 22 | 5.902 | D28 | 5.727 | Gat-31 | 4.872 |
| 10 | 5.186 | 23 | 5.116 | D29 | 5.045 | Gat-33 | 6.475 |
| 11 | 5.785 | 24 | 4.401 | D32 | 5.933 | ||
| 12 | 4.758 | 25 | 5.764 | D36 | 5.675 | ||
| 13 | 5.615 | 26 | 7.541 | D37 | 6.367 |
| Compounds | Substituent Groups | Predicted CV | Change Range (%) | Predicted 1GKQ | Change Range (%) | Predicted 1OB0 | Change Range (%) | Predicted 5M0K | Change Range (%) |
|---|---|---|---|---|---|---|---|---|---|
| Moxifloxacin | - | 3.696 | - | 1.920 | - | 1.845 | - | 1.922 | - |
| Derivative-1 | 1-NO | 3.823 | 3.44 | 2.049 | 6.72 | 1.873 | 1.52 | 2.062 | 7.28 |
| Derivative-2 | 1-COOH | 3.712 | 0.43 | 2.048 | 6.67 | 1.880 | 1.90 | 2.066 | 7.49 |
| Derivative-3 | 2-CH3 | 4.079 | 10.36 | 2.051 | 6.82 | 1.891 | 2.49 | 2.058 | 7.08 |
| Derivative-4 | 2-C2H5 | 4.315 | 16.75 | 2.049 | 6.72 | 1.889 | 2.38 | 2.073 | 7.86 |
| Derivative-5 | 2-C3H7 | 4.592 | 24.24 | 2.049 | 6.72 | 1.887 | 2.28 | 2.093 | 8.90 |
| Derivative-6 | 2-C4H9 | 4.592 | 24.24 | 2.049 | 6.72 | 1.885 | 2.17 | 2.100 | 9.26 |
| Derivative-7 | 2-C5H11 | 4.623 | 25.08 | 2.049 | 6.72 | 1.883 | 2.06 | 2.106 | 9.57 |
| Derivative-8 | 2-C=C | 4.144 | 12.12 | 2.050 | 6.77 | 1.885 | 2.17 | 2.070 | 7.70 |
| Derivative-9 | 2-CH2NH2 | 4.088 | 10.61 | 2.050 | 6.77 | 1.890 | 2.44 | 2.064 | 7.39 |
| Derivative-10 | 2-NH2 | 4.454 | 20.51 | 2.047 | 6.61 | 1.896 | 2.76 | 2.048 | 6.56 |
| No. | Genotoxicity | Bioaccumulation | Photodegradability | |||
|---|---|---|---|---|---|---|
| Predicted | Change Range (%) | Predicted | Change Range (%) | Predicted | Change Range (%) | |
| MOX | 8.869 | 0.950 | 1.975 | |||
| Derivative-1 | 7.978 | −10.05 | 0.211 | −77.79 | 0.965 | −51.14 |
| Derivative-2 | 8.163 | −7.96 | 0.541 | −43.05 | 0.921 | −53.37 |
| Derivative-3 | 8.389 | −5.41 | 1.147 | 20.74 | 1.07 | −45.82 |
| Derivative-4 | 8.343 | −5.93 | 1.258 | 32.42 | 1.094 | −44.61 |
| Derivative-5 | 8.437 | −4.87 | 1.497 | 57.58 | 1.117 | −43.44 |
| Derivative-6 | 8.537 | −3.74 | 1.547 | 62.84 | 1.153 | −41.62 |
| Derivative-7 | 8.467 | −4.53 | 1.643 | 72.95 | 1.156 | −41.47 |
| Derivative-8 | 8.373 | −5.59 | 1.102 | 16.00 | 1.068 | −45.92 |
| Derivative-9 | 8.549 | −3.61 | 0.912 | −4.00 | −0.582 | −129.47 |
| Derivative-10 | 8.808 | −0.69 | 0.453 | −52.32 | −0.219 | −111.09 |
| Area | No. | Coordinate | Nutrient Content (g/kg) | |||
|---|---|---|---|---|---|---|
| E (°) | N (°) | C | N | P | ||
| I | 1 | 125.60 | 50.15 | 1.33 | 0.85 | 2.17 |
| 2 | 125.62 | 48.46 | 0.87 | 0.56 | 1.27 | |
| 3 | 125.56 | 46.64 | 0.43 | 0.24 | 0.84 | |
| 4 | 125.45 | 45.23 | 0.68 | 0.58 | 0.99 | |
| II | 5 | 120.20 | 40.41 | 0.96 | 0.53 | 1.23 |
| 6 | 122.95 | 42.00 | 0.76 | 0.49 | 1.14 | |
| 7 | 124.79 | 41.33 | 0.37 | 0.24 | 0.57 | |
| 8 | 121.44 | 38.91 | 0.96 | 0.53 | 1.23 | |
| III | 9 | 125.97 | 41.94 | 2.13 | 0.49 | 1.14 |
| 10 | 120.07 | 43.63 | 1.42 | 0.88 | 1.36 | |
| 11 | 127.55 | 42.82 | 1.23 | 0.72 | 0.99 | |
| 12 | 130.71 | 44.47 | 2.13 | 1.15 | 2.17 | |
| Average value | 1.11 | 0.61 | 1.26 | |||
| Addition of Nutrient Elements | 2 | 1 | 2 | |||
| FQs | Nutrient Elements | Field Measures | Binding Energy | Change Range | ||||
|---|---|---|---|---|---|---|---|---|
| C | N | P | Organic Fertilizer Application | Straw Returning | Plowing | |||
| MOX | 2 | 1 | 2 | - | - | - | −49.445 | - |
| Derivative-10 | - | - | - | −73.608 | 48.87% | |||
| 0 | 0 | 0 | −73.608 | - | ||||
| 1 | 0 | 0 | −85.565 | 16.24% | ||||
| 0 | 1 | 0 | −78.515 | 6.67% | ||||
| 0 | 0 | 1 | −93.523 | 27.06% | ||||
| 1 | 1 | 0 | −95.529 | 29.78% | ||||
| 1 | 0 | 1 | −87.956 | 19.49% | ||||
| 0 | 1 | 1 | −85.619 | 16.32% | ||||
| 1 | 1 | 1 | −104.013 | 41.31% | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, X.; Zhao, Y.; Ren, Z.; Wang, P.; Li, Y. Bio-Enhanced Degradation Strategies for Fluoroquinolones in the Sewage Sludge Composting Stage: Molecular Modification and Resistance Gene Regulation. Int. J. Environ. Res. Public Health 2022, 19, 7766. https://doi.org/10.3390/ijerph19137766
Jin X, Zhao Y, Ren Z, Wang P, Li Y. Bio-Enhanced Degradation Strategies for Fluoroquinolones in the Sewage Sludge Composting Stage: Molecular Modification and Resistance Gene Regulation. International Journal of Environmental Research and Public Health. 2022; 19(13):7766. https://doi.org/10.3390/ijerph19137766
Chicago/Turabian StyleJin, Xingyan, Yuanyuan Zhao, Zhixing Ren, Panpan Wang, and Yu Li. 2022. "Bio-Enhanced Degradation Strategies for Fluoroquinolones in the Sewage Sludge Composting Stage: Molecular Modification and Resistance Gene Regulation" International Journal of Environmental Research and Public Health 19, no. 13: 7766. https://doi.org/10.3390/ijerph19137766
APA StyleJin, X., Zhao, Y., Ren, Z., Wang, P., & Li, Y. (2022). Bio-Enhanced Degradation Strategies for Fluoroquinolones in the Sewage Sludge Composting Stage: Molecular Modification and Resistance Gene Regulation. International Journal of Environmental Research and Public Health, 19(13), 7766. https://doi.org/10.3390/ijerph19137766







