Inhibition of HIF-1α Attenuates Silica-Induced Pulmonary Fibrosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Culture
2.3. Modeling of Transdifferentiation
2.4. Small-Molecule Intervention and Treatment of NIH-3T3 Cells
2.5. Transfection of Small-Interfering RNA
2.6. Animals
2.7. Animal Model of Silicosis
2.8. RT-qPCR
2.9. Western Blot Analysis
2.10. Histology
2.11. Immunohistochemistr (IHC)
2.12. Statistical Analysis
3. Result
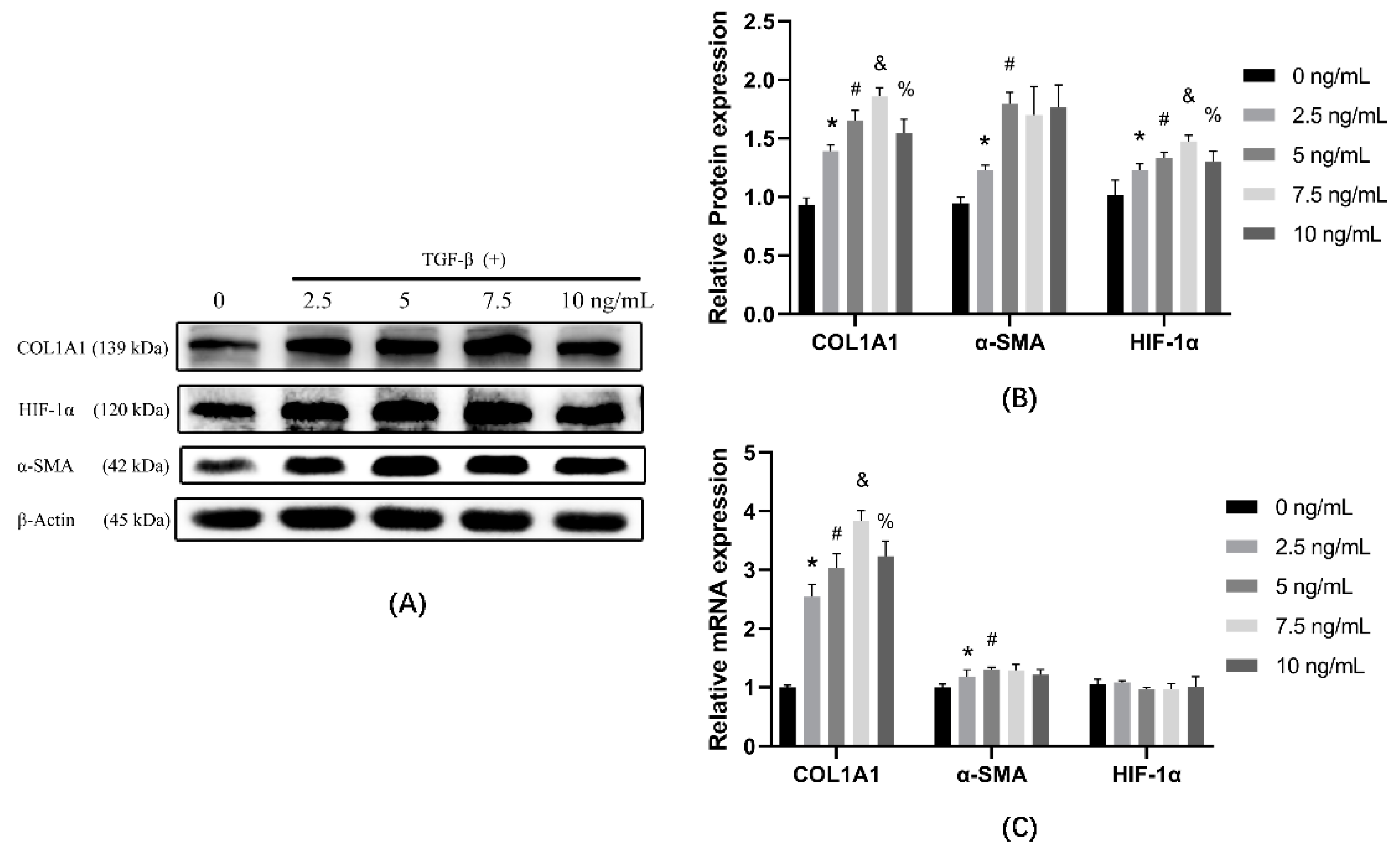
3.1. TGF-β1 Induces Cells Transdifferentiation in a Dose-Dependent Manner
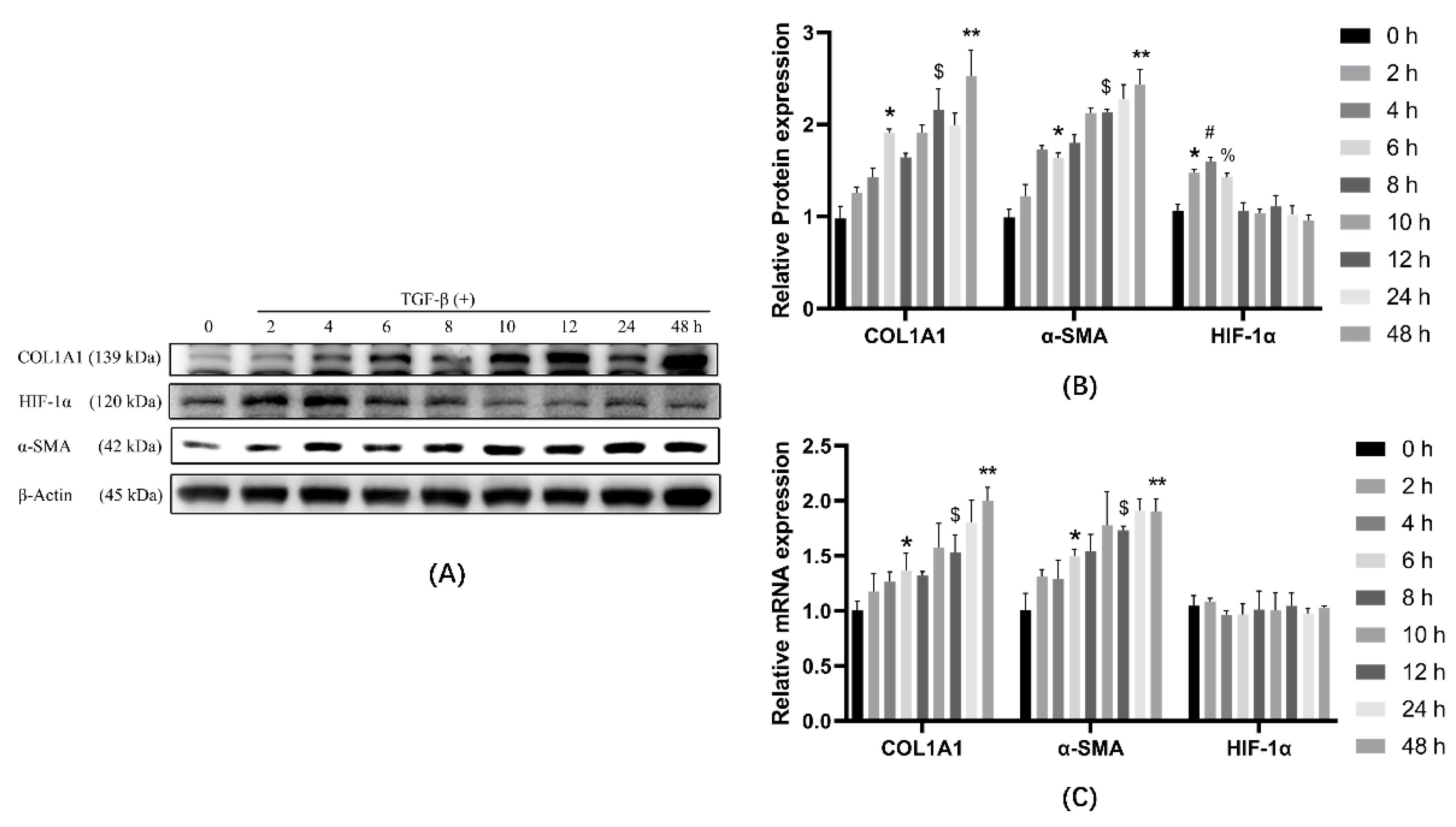
3.2. TGF-β1 Causes Cellular Transdifferentiation in a Time-Dependent Manner
3.3. HIF-1α and PSMAD3 Reached Maximum Expression at the Same Time
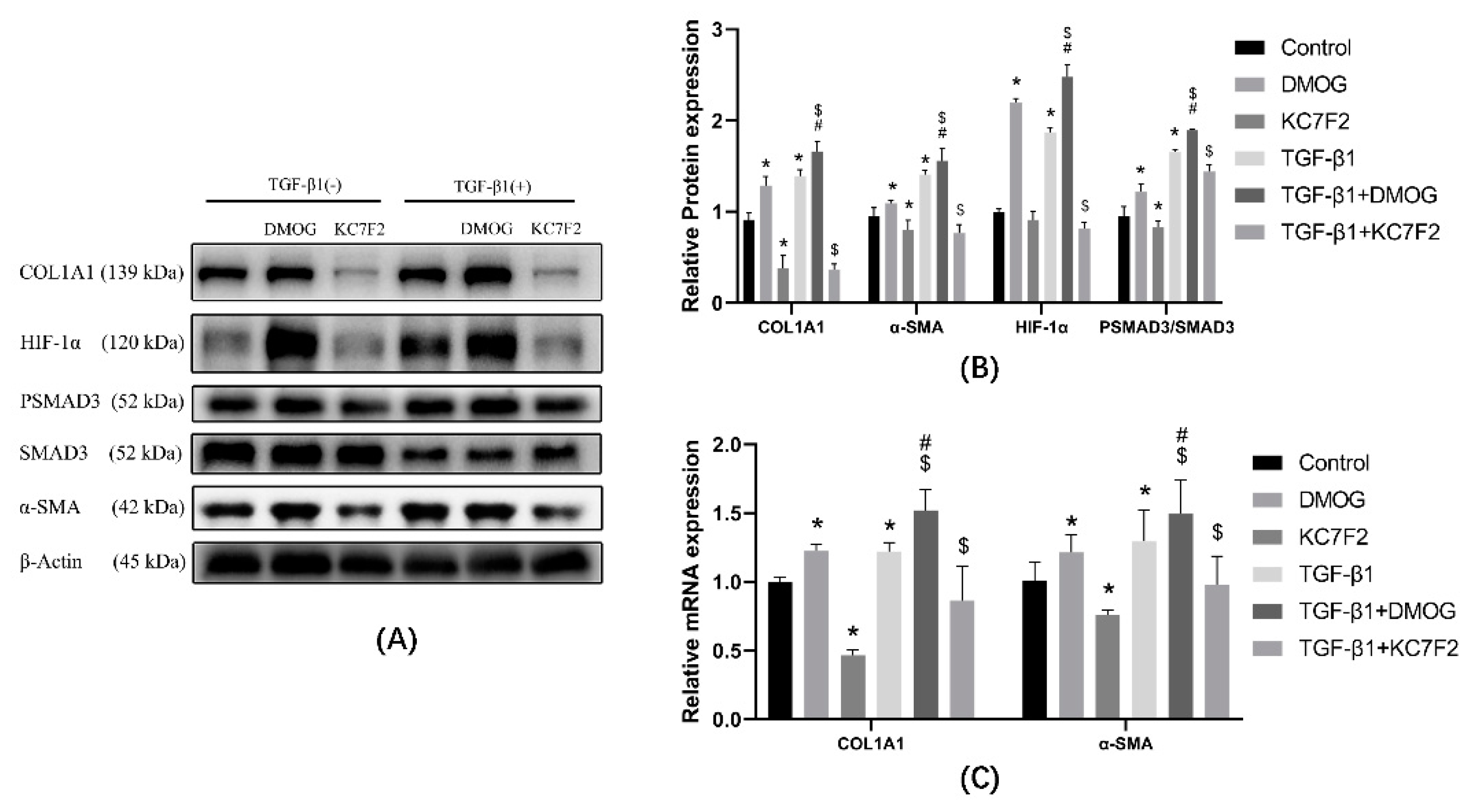
3.4. HIF-1α Affects the Phosphorylation of SMAD3 and the Expression of COL1A1
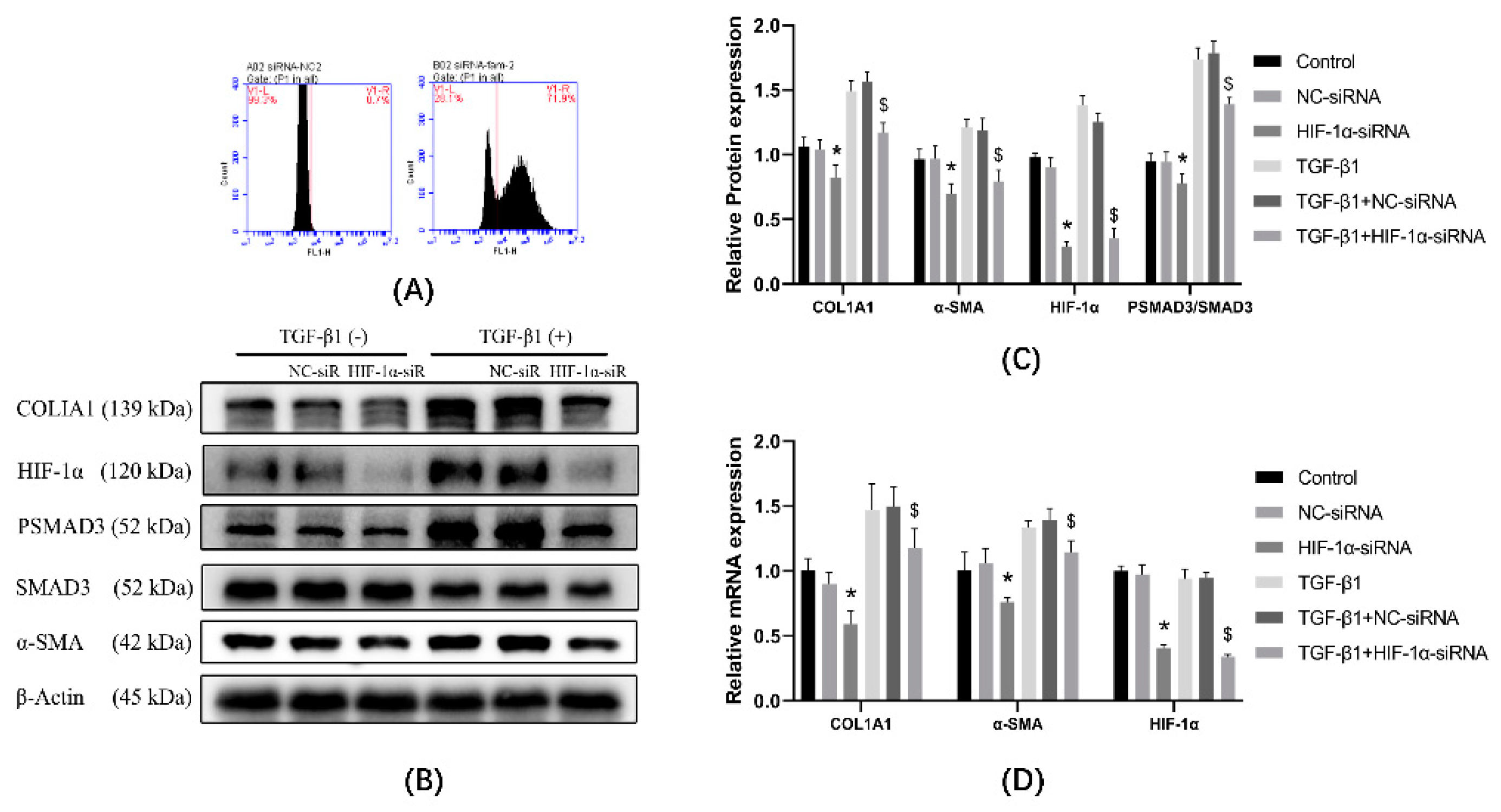
3.5. Expression of α-SMA and Col1a Was Affected by Knockdown of HIF-1α Using siRNA
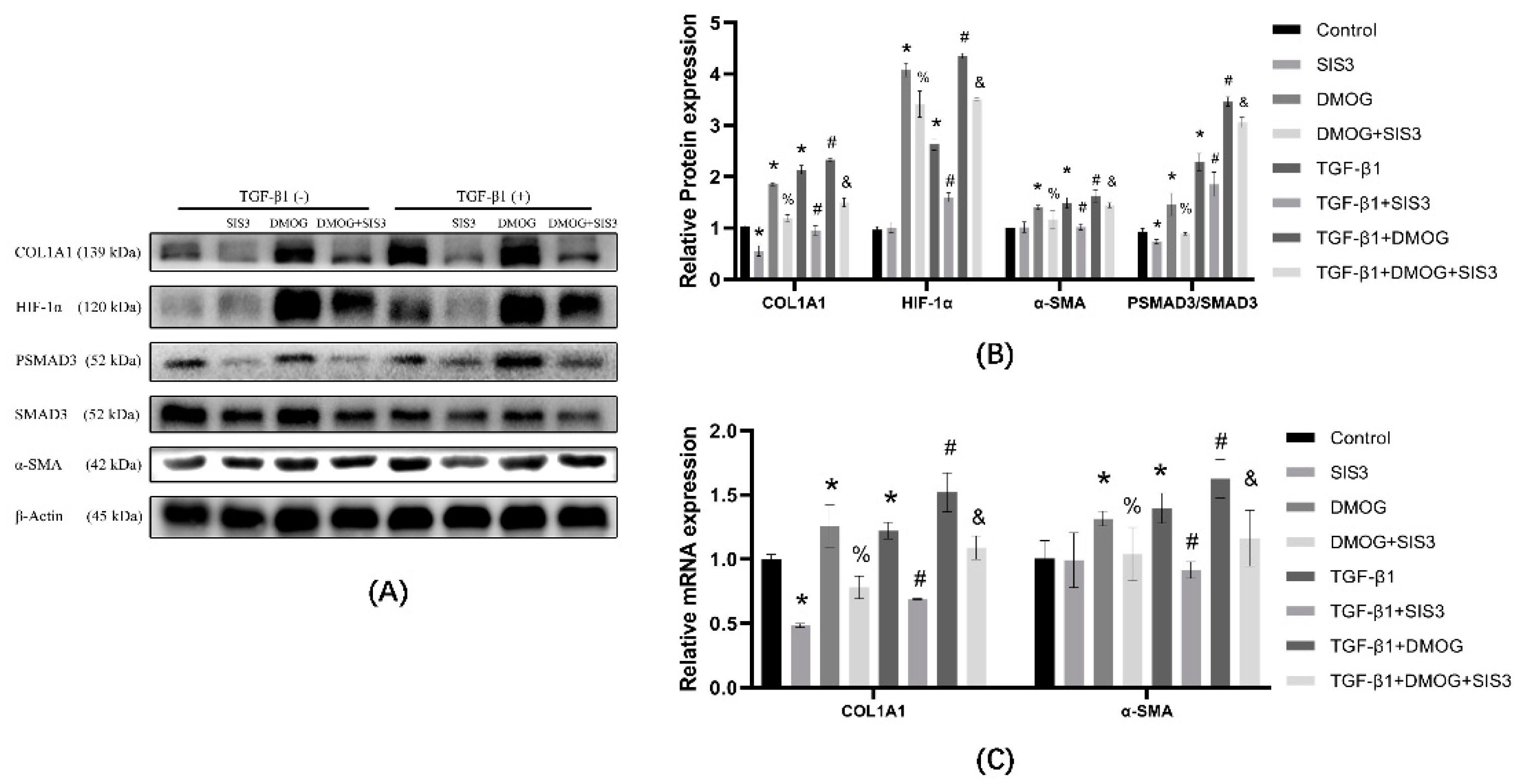
3.6. SIS3 Inhibits DMOG Induced Elevation of COL1A1
3.7. KC7F2 Alleviates Silica Particles Induced PF in Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Higgins, D.F.; Biju, M.P.; Akai, Y.; Wutz, A.; Johnson, R.S.; Haase, V.H. Hypoxic induction of Ctgf is directly mediated by Hif-1. Am. J. Physiol. Renal. Physiol. 2004, 287, F1223–F1232. [Google Scholar] [CrossRef]
- Li, J.; Yao, W.; Hou, J.Y.; Zhang, L.; Bao, L.; Chen, H.T.; Wang, D.; Yue, Z.Z.; Li, Y.P.; Zhang, M.; et al. The Role of Fibrocyte in the Pathogenesis of Silicosis. Biomed. Environ. Sci. 2018, 31, 311–316. [Google Scholar] [CrossRef]
- Ikonen, E. Molecular mechanisms of intracellular cholesterol transport. Curr. Opin. Lipidol. 1997, 8, 60–64. [Google Scholar] [CrossRef]
- Kage, H.; Borok, Z. EMT and interstitial lung disease: A mysterious relationship. Curr. Opin. Pulm. Med. 2012, 18, 517–523. [Google Scholar] [CrossRef]
- Miao, R.M.; Fang, Z.H. Therapeutic efficacy of Jinshuibao capsules in treatment of silicosis. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi Zhonghua Laodong Weisheng Zhiyebing Zazhi Chin. J. Ind. Hyg. Occup. Dis. 2012, 30, 781–782. [Google Scholar]
- Sun, J.; Song, P.; Wang, Y.; Chen, Y. Clinical efficacy of acetylcysteine combined with tetrandrine tablets in the treatment of silicosis and the effect on serum IL-6 and TNF-α. Exp. Ther. Med. 2019, 18, 3383–3388. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.Y.; Liang, J.T.; Wu, J.; Liu, L.; Lu, M.Z.; He, X.Y.; Wu, L.J.; Jiang, H.Y.; Wang, F.; Meng, X.; et al. Efficacy and Safety of Dahuang Zhechong Pill in Silicosis: A Randomized Controlled Trial. Evid. Based Complementary Altern. Med. eCAM 2021, 2021, 4354054. [Google Scholar] [CrossRef]
- Ma, T.T.; Meng, X.M. TGF-β/Smad and Renal Fibrosis. Adv. Exp. Med. Biol. 2019, 1165, 347–364. [Google Scholar] [CrossRef]
- Margadant, C.; Sonnenberg, A. Integrin-TGF-beta crosstalk in fibrosis, cancer and wound healing. EMBO Rep. 2010, 11, 97–105. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Liu, C.; Zhou, D.; Zhang, L. TGF-β/SMAD Pathway and Its Regulation in Hepatic Fibrosis. J. Histochem. Cytochem. 2016, 64, 157–167. [Google Scholar] [CrossRef]
- Das, F.; Bera, A.; Ghosh-Choudhury, N.; Abboud, H.E.; Kasinath, B.S.; Choudhury, G.G. TGFβ-induced deptor suppression recruits mTORC1 and not mTORC2 to enhance collagen I (α2) gene expression. PLoS ONE 2014, 9, e109608. [Google Scholar] [CrossRef]
- Rozen-Zvi, B.; Hayashida, T.; Hubchak, S.C.; Hanna, C.; Platanias, L.C.; Schnaper, H.W. TGF-β/Smad3 activates mammalian target of rapamycin complex-1 to promote collagen production by increasing HIF-1α expression. Am. J. Physiol. Renal. Physiol. 2013, 305, F485–F494. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Lv, F.L.; Wang, G.H. Effects of HIF-1α on diabetic retinopathy angiogenesis and VEGF expression. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5071–5076. [Google Scholar] [CrossRef]
- Lin, C.J.; Lan, Y.M.; Ou, M.Q.; Ji, L.Q.; Lin, S.D. Expression of miR-217 and HIF-1α/VEGF pathway in patients with diabetic foot ulcer and its effect on angiogenesis of diabetic foot ulcer rats. J. Endocrinol. Investig. 2019, 42, 1307–1317. [Google Scholar] [CrossRef]
- Elson, D.A.; Ryan, H.E.; Snow, J.W.; Johnson, R.; Arbeit, J.M. Coordinate up-regulation of hypoxia inducible factor (HIF)-1alpha and HIF-1 target genes during multi-stage epidermal carcinogenesis and wound healing. Cancer Res. 2000, 60, 6189–6195. [Google Scholar]
- Bruick, R.K.; McKnight, S.L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 2001, 294, 1337–1340. [Google Scholar] [CrossRef] [Green Version]
- Iwai, K.; Yamanaka, K.; Kamura, T.; Minato, N.; Conaway, R.C.; Conaway, J.W.; Klausner, R.D.; Pause, A. Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA 1999, 96, 12436–12441. [Google Scholar] [CrossRef] [Green Version]
- Kallio, P.J.; Okamoto, K.; O’Brien, S.; Carrero, P.; Makino, Y.; Tanaka, H.; Poellinger, L. Signal transduction in hypoxic cells: Inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. EMBO J. 1998, 17, 6573–6586. [Google Scholar] [CrossRef]
- Jaakkola, P.; Mole, D.R.; Tian, Y.M.; Wilson, M.I.; Gielbert, J.; Gaskell, S.J.; von Kriegsheim, A.; Hebestreit, H.F.; Mukherji, M.; Schofield, C.J.; et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001, 292, 468–472. [Google Scholar] [CrossRef]
- Otali, D.; Fredenburgh, J.; Oelschlager, D.K.; Grizzle, W.E. A standard tissue as a control for histochemical and immunohistochemical staining. Biotech. Histochem. 2016, 91, 309–326. [Google Scholar] [CrossRef] [Green Version]
- Milito, A.; Brancaccio, M.; D’Argenio, G.; Castellano, I. Natural Sulfur-Containing Compounds: An Alternative Therapeutic Strategy against Liver Fibrosis. Cells 2019, 8, 1356. [Google Scholar] [CrossRef] [Green Version]
- Brancaccio, M.; D’Argenio, G.; Lembo, V.; Palumbo, A.; Castellano, I. Antifibrotic Effect of Marine Ovothiol in an In Vivo Model of Liver Fibrosis. Oxid. Med. Cell. Longev. 2018, 2018, 5045734. [Google Scholar] [CrossRef] [Green Version]
- Ryu, D.R.; Yu, M.R.; Kong, K.H.; Kim, H.; Kwon, S.H.; Jeon, J.S.; Han, D.C.; Noh, H. Sirt1-hypoxia-inducible factor-1α interaction is a key mediator of tubulointerstitial damage in the aged kidney. Aging Cell 2019, 18, e12904. [Google Scholar] [CrossRef]
- Zhang, H.X.; Li, Y.N.; Wang, X.L.; Ye, C.L.; Zhu, X.Y.; Li, H.P.; Yang, T.; Liu, Y.J. Probucol ameliorates EMT and lung fibrosis through restoration of SIRT3 expression. Pulm. Pharmacol. Ther. 2019, 57, 101803. [Google Scholar] [CrossRef]
- Janbandhu, V.; Tallapragada, V.; Patrick, R.; Li, Y.; Abeygunawardena, D.; Humphreys, D.T.; Martin, E.; Ward, A.O.; Contreras, O.; Farbehi, N.; et al. Hif-1a suppresses ROS-induced proliferation of cardiac fibroblasts following myocardial infarction. Cell Stem Cell 2022, 29, 281–297.e212. [Google Scholar] [CrossRef]
- Li, A.; Peng, R.; Sun, Y.; Liu, H.; Peng, H.; Zhang, Z. LincRNA 1700020I14Rik alleviates cell proliferation and fibrosis in diabetic nephropathy via miR-34a-5p/Sirt1/HIF-1α signaling. Cell Death Dis. 2018, 9, 461. [Google Scholar] [CrossRef]
- Zhu, Y.; Tan, J.; Xie, H.; Wang, J.; Meng, X.; Wang, R. HIF-1alpha regulates EMT via the Snail and beta-catenin pathways in paraquat poisoning-induced early pulmonary fibrosis. J. Cell. Mol. Med. 2016, 20, 688–697. [Google Scholar] [CrossRef]
- Ichihara, S.; Li, P.; Mise, N.; Suzuki, Y.; Izuoka, K.; Nakajima, T.; Gonzalez, F.; Ichihara, G. Ablation of aryl hydrocarbon receptor promotes angiotensin II-induced cardiac fibrosis through enhanced c-Jun/HIF-1α signaling. Arch. Toxicol. 2019, 93, 1543–1553. [Google Scholar] [CrossRef]
- Miranda da Costa, N.M.; Saab Abe, C.T.; Mitre, G.P.; Mesquita, R.A.; Silva Kataoka, M.S.D.; Ribeiro Ribeiro, A.L.; Jaeger, R.G.; Alves-Júnior, S.M.; Smith, A.M.; Pinheiro, J.J.V. HIF-1α is Overexpressed in Odontogenic Keratocyst Suggesting Activation of HIF-1α and NOTCH1 Signaling Pathways. Cells 2019, 8, 731. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; He, Y.; Zhao, H.; Xu, X. Hypoxia inducible factor-1 promotes liver fibrosis in nonalcoholic fatty liver disease by activating PTEN/p65 signaling pathway. J. Cell Biochem. 2019, 120, 14735–14744. [Google Scholar] [CrossRef]
- Zhang, Y.; Bian, Y.; Wang, Y.; Wang, Y.; Duan, X.; Han, Y.; Zhang, L.; Wang, F.; Gu, Z.; Qin, Z. HIF-1α is necessary for activation and tumour-promotion effect of cancer-associated fibroblasts in lung cancer. J. Cell. Mol. Med. 2021, 25, 5457–5469. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, X.M.; Huang, X.R.; Lan, H.Y. The preventive and therapeutic implication for renal fibrosis by targetting TGF-β/Smad3 signaling. Clin. Sci. 2018, 132, 1403–1415. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Dalgard, C.L.; Mohyeldin, A.; McFate, T.; Tait, A.S.; Verma, A. Reversible inactivation of HIF-1 prolyl hydroxylases allows cell metabolism to control basal HIF-1. J. Biol. Chem. 2005, 280, 41928–41939. [Google Scholar] [CrossRef] [Green Version]
- Lv, X.M.; Li, M.D.; Cheng, S.; Liu, B.L.; Liu, K.; Zhang, C.F.; Xu, X.H.; Zhang, M. Neotuberostemonine inhibits the differentiation of lung fibroblasts into myofibroblasts in mice by regulating HIF-1α signaling. Acta Pharmacol. Sin. 2018, 39, 1501–1512. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, X.; Wang, J.; Zhu, Y.; Li, R.; Wang, J.; He, X.; Shan, L.; Wang, R.; Wang, L.; et al. Paeoniflorin alleviates liver fibrosis by inhibiting HIF-1α through mTOR-dependent pathway. Fitoterapia 2014, 99, 318–327. [Google Scholar] [CrossRef]
- Mabjeesh, N.J.; Escuin, D.; LaVallee, T.M.; Pribluda, V.S.; Swartz, G.M.; Johnson, M.S.; Willard, M.T.; Zhong, H.; Simons, J.W.; Giannakakou, P. 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell 2003, 3, 363–375. [Google Scholar] [CrossRef] [Green Version]
- Mabjeesh, N.J.; Post, D.E.; Willard, M.T.; Kaur, B.; Van Meir, E.G.; Simons, J.W.; Zhong, H. Geldanamycin induces degradation of hypoxia-inducible factor 1alpha protein via the proteosome pathway in prostate cancer cells. Cancer Res. 2002, 62, 2478–2482. [Google Scholar]
- Shi, J.; Xiao, P.; Liu, X.; Chen, Y.; Xu, Y.; Fan, J.; Yin, Y. Notch3 Modulates Cardiac Fibroblast Proliferation, Apoptosis, and Fibroblast to Myofibroblast Transition via Negative Regulation of the RhoA/ROCK/Hif1alpha Axis. Front. Physiol. 2020, 11, 669. [Google Scholar] [CrossRef]
- Narita, T.; Yin, S.; Gelin, C.F.; Moreno, C.S.; Yepes, M.; Nicolaou, K.C.; Van Meir, E.G. Identification of a novel small molecule HIF-1alpha translation inhibitor. Clin. Cancer Res. 2009, 15, 6128–6136. [Google Scholar] [CrossRef] [Green Version]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Seagroves, T.N.; Ryan, H.E.; Lu, H.; Wouters, B.G.; Knapp, M.; Thibault, P.; Laderoute, K.; Johnson, R.S. Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol. Cell. Biol. 2001, 21, 3436–3444. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.Z.; Zhu, W.; Han, J.; Yang, X.; Zhou, R.; Lu, H.C.; Yu, H.; Yuan, W.B.; Li, P.C.; Tao, J.; et al. The role of the HIF-1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer. Cancer Commun. 2021, 41, 560–575. [Google Scholar] [CrossRef]


| Gene | Forward, 5′-3′ | Reverse, 5′-3′ |
|---|---|---|
| COL1A1 | TAAGGGTCCCCAATGGTGAGA | GGGTCCCTCGACTCCTACAT |
| α-SMA | GGCACCACTGAACCCTAAGG | ACAATACCAGTTGTACGTCCAGA |
| SMAD3 | AGACGCCAGTTCTACCTCCAGTG | GCCAGCAGGGAAGTTAGTGTTCTC |
| HIF-1α | CTGCCACTGCCACCACAACTG | TGCCACTGTATGCTGATGCCTTAG |
| β-Actin | GTGCTATGTTGCTCTAGACTTCG | ATGCCACAGGATTCCATACC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Li, Y.; Niu, Z.; Xia, J.; Dai, K.; Wang, C.; Yao, W.; Guo, Y.; Deng, X.; He, J.; et al. Inhibition of HIF-1α Attenuates Silica-Induced Pulmonary Fibrosis. Int. J. Environ. Res. Public Health 2022, 19, 6775. https://doi.org/10.3390/ijerph19116775
Xu X, Li Y, Niu Z, Xia J, Dai K, Wang C, Yao W, Guo Y, Deng X, He J, et al. Inhibition of HIF-1α Attenuates Silica-Induced Pulmonary Fibrosis. International Journal of Environmental Research and Public Health. 2022; 19(11):6775. https://doi.org/10.3390/ijerph19116775
Chicago/Turabian StyleXu, Xiao, Yiping Li, Zhuoya Niu, Jiarui Xia, Kai Dai, Chen Wang, Wu Yao, Yonghua Guo, Xuedan Deng, Jing He, and et al. 2022. "Inhibition of HIF-1α Attenuates Silica-Induced Pulmonary Fibrosis" International Journal of Environmental Research and Public Health 19, no. 11: 6775. https://doi.org/10.3390/ijerph19116775
APA StyleXu, X., Li, Y., Niu, Z., Xia, J., Dai, K., Wang, C., Yao, W., Guo, Y., Deng, X., He, J., Deng, M., Si, H., & Hao, C. (2022). Inhibition of HIF-1α Attenuates Silica-Induced Pulmonary Fibrosis. International Journal of Environmental Research and Public Health, 19(11), 6775. https://doi.org/10.3390/ijerph19116775







