Degradation of Three Microcystin Variants in the Presence of the Macrophyte Spirodela polyrhiza and the Associated Microbial Communities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Organisms
2.2. Biodegradation Assays
2.3. Parameters Measured
- C is the concentration of particular MCs
- Cs is the concentration of particular MCs at the beginning of the experiments
- Cd is the concentration of particular MCs after four and nine days of incubation
2.4. Microorganism Identification and Counting
2.5. Plant Extraction for MC Analysis
2.6. HPLC-PDA Analysis of Microcystins and Their Biodegradation Products in Water and Plants
2.7. Data Analysis
3. Results
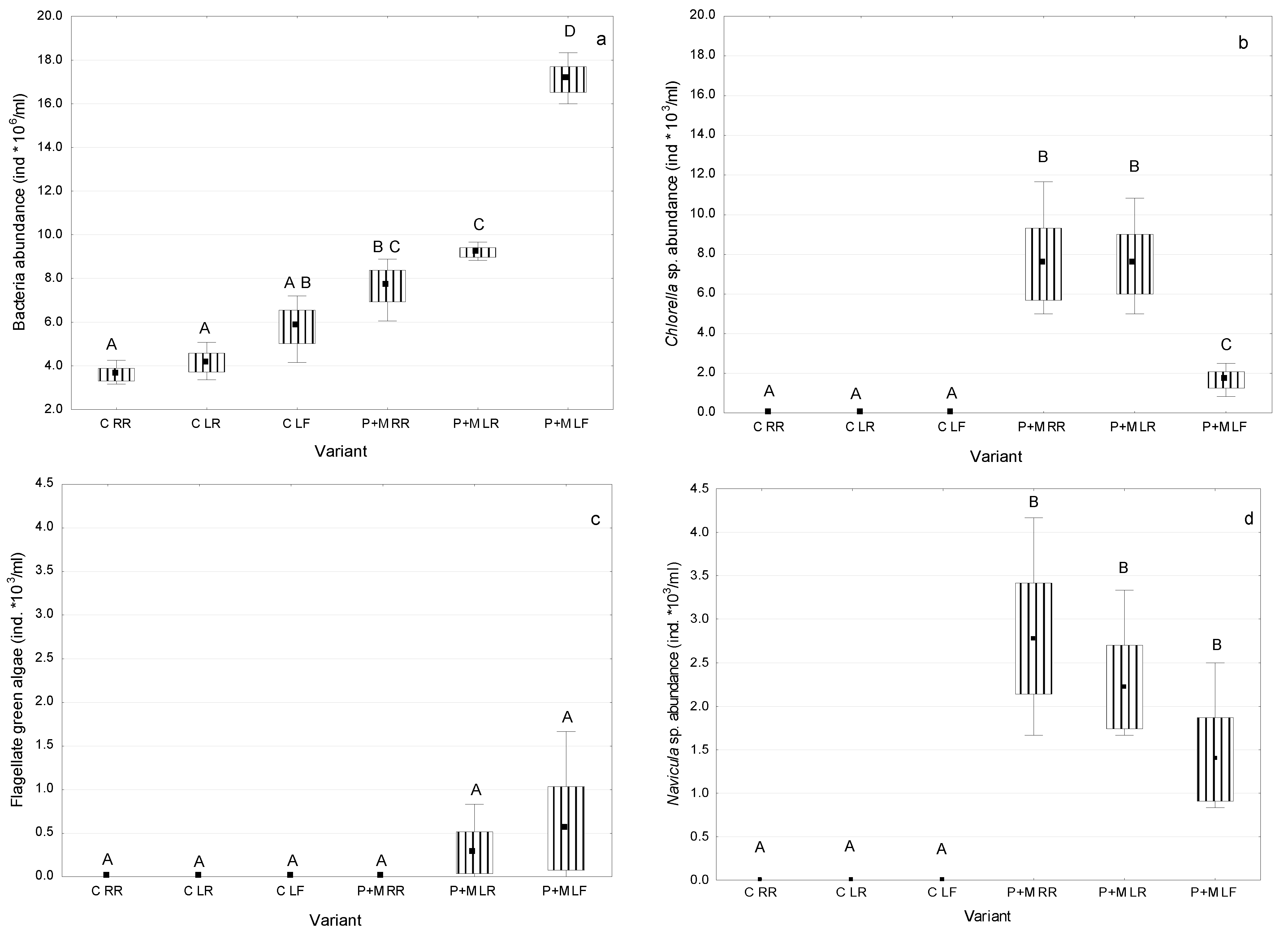
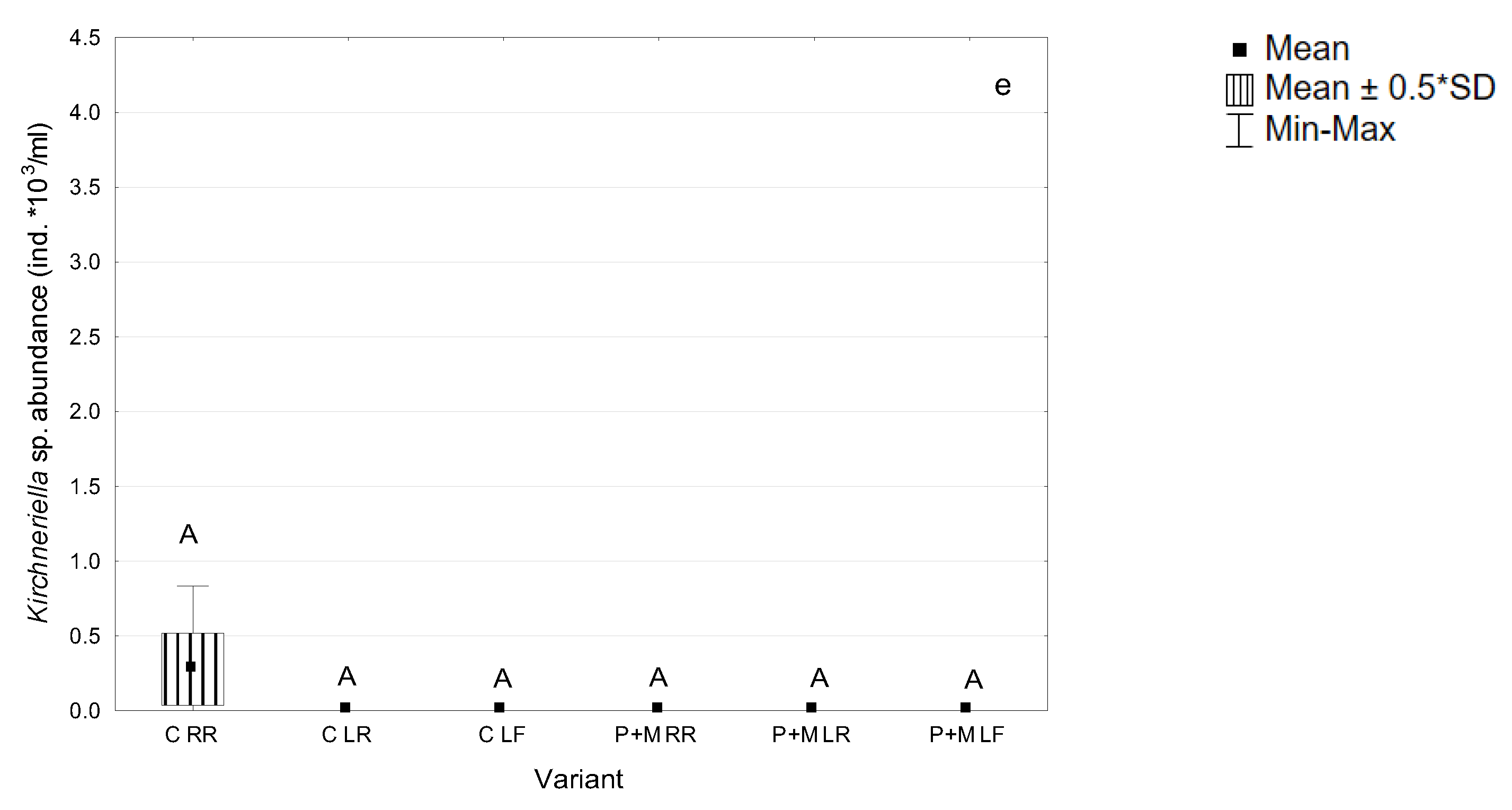
3.1. Microorganisms Assocciated with S. polyrhiza
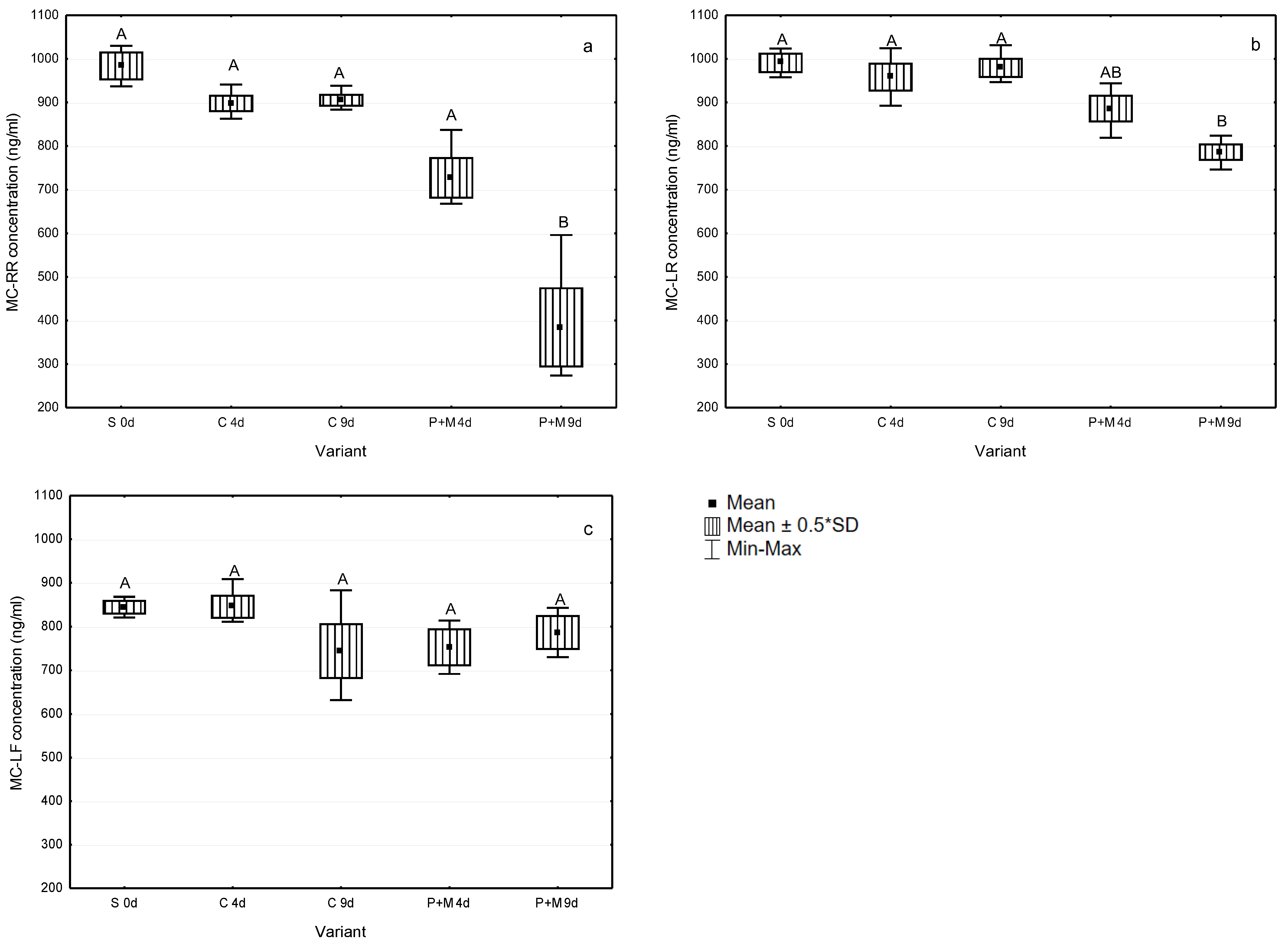
3.2. Biodegradation of MCs after the Incubation with S. polyrhiza and Associated Microorganisms
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paerl, H.W.; Huisman, J. Blooms like it hot. Science 2008, 320, 57–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toporowska, M.; Pawlik-Skowrońska, B.; Kalinowska, R. Mass development of diazotrophic cyanobacteria (Nostocales) and production of neurotoxic anatoxin-a in a Planktothrix (Oscillatoriales) dominated temperate lake. Water Air Soil Pollut. 2016, 227, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantzouki, E.; Lürling, M.; Fastner, J.; de Senerpont Domis, L.; Wilk-Woźniak, E.; Koreiviene, J.; Seelen, L.; Teurlincx, S.; Verstijnen, Y.; Krztoń, W.; et al. Temperature effects explain continental scale distribution of cyanobacterial toxins. Toxins 2018, 10, 156. [Google Scholar] [CrossRef] [Green Version]
- Van Hassel, W.H.R.; Andjelkovic, M.; Durieu, B.; Marroquin, V.A.; Masquelier, J.; Huybrechts, B.; Wilmotte, A.A. Summer of cyanobacterial blooms in Belgian Waterbodies: Microcystin quantification and molecular characterizations. Toxins 2022, 14, 61. [Google Scholar] [CrossRef] [PubMed]
- Sivonen, K.; Börner, T. Bioactive compounds produced by cyanobacteria. In The Cyanobacteria; Herrero, A., Flores, E., Eds.; Caister Academic Press: Norfolk, UK, 2008; pp. 159–197. [Google Scholar]
- Spoof, L.; Catherine, A. Table of microcystins and nodularins. In Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis; Meriluoto, J., Spoof, L., Codd, G.A., Eds.; John Wiley & Sons: Chichester, UK, 2017; pp. 526–537. [Google Scholar]
- Pawlik-Skowronska, B.; Toporowska, M. How to mitigate cyanobacterial blooms and cyanotoxin production in eutrophic water reservoirs? Hydrobiologia 2016, 778, 45–59. [Google Scholar] [CrossRef] [Green Version]
- Massey, I.Y.; Yang, F. A mini review on microcystins and bacterial degradation. Toxins 2020, 12, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codd, G.; Bell, S.; Kaya, K.; Ward, C.; Beattie, K.; Metcalf, J. Cyanobacterial toxins, exposure routes and human health. Eur. J. Phycol. 1999, 34, 405–415. [Google Scholar] [CrossRef]
- Gilroy, D.J.; Kauffman, K.W.; Hall, R.A.; Huang, X.; Chu, F.S. Assessing potential health risks from microcystin toxins in blue-green algae dietary supplements. Environ. Health Perspect. 2000, 108, 435–439. [Google Scholar] [CrossRef]
- Martins, J.C.; Vasconcelos, V.M. Microcystin dynamics in aquatic organisms. J. Toxicol. Environ. Health Part B 2009, 12, 65–82. [Google Scholar] [CrossRef]
- Pawlik-Skowrońska, B.; Toporowska, M.; Mazur-Marzec, H. Toxic oligopeptides in the cyanobacterium Planktothrix agardhii-dominated blooms and their effects on duckweed (Lemnaceae) development. KMAE 2018, 419, 41. [Google Scholar] [CrossRef] [Green Version]
- Sivonen, K.; Jones, G. Cyanobacterial toxins. In Toxic Cyanobacteria in Water. A Guide to Their Public Health Consequences, Monitoring and Management; Chorus, I., Bartram, J., Eds.; E and FN Spon: London, UK, 1999; pp. 41–111. [Google Scholar]
- Foss, A.J.; Aubel, M.T.; Gallagher, B.; Mettee, N.; Miller, A.; Fogelson, S.B. Diagnosing microcystin intoxication of canines: Clinicopathological indications, pathological characteristics, and analytical detection in postmortem and antemortem samples. Toxins 2019, 11, 456. [Google Scholar] [CrossRef] [Green Version]
- Grabowska, M.; Kobos, J.; Toruńska-Sitarz, A.; Mazur-Marzec, H. Non-ribosomal peptides produced by Planktothrix agardhii from Siemianówka Dam Reservoir SDR (northeast Poland). Arch. Microbiol. 2014, 19610, 697–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svircev, Z.; Lalic, D.; Bojadzija, S.G.; Tokodi, N.; Drobac, B.D.; Chen, L.; Meriluoto, J.; Codd, G.A. Global geographical and historical overview of cyanotoxin distribution and cyanobacterial poisonings. Arch. Toxicol. 2019, 93, 2429–2481. [Google Scholar] [CrossRef] [PubMed]
- Toporowska, M.; Mazur-Marzec, H.; Pawlik-Skowrońska, B. The effects of cyanobacterial bloom extracts on the biomass, Chl-a, MC and other oligopeptides contents in a natural Planktothrix agardhii population. Int. J. Environ. Res. Public Health 2020, 17, 2881. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.; Gaudieux, A.L.; Fanok, S.; Newcombe, G.; Humpage, A.R. Bacterial degradation of microcystin toxins in drinking water eliminates their toxicity. Toxicon 2007, 50, 438–441. [Google Scholar] [CrossRef]
- Dziga, D.; Maksylewicz, A.; Maroszek, M.; Budzyńska, A.; Napiorkowska-Krzebietke, A.; Toporowska, M.; Grabowska, M.; Kozak, A.; Rosińska, J.; Meriluoto, J. The biodegradation of microcystins in temperate freshwater bodies with previous cyanobacterial history. Ecotoxicol. Environ. Saf. 2017, 145, 420–430. [Google Scholar] [CrossRef] [PubMed]
- León, C.; Boix, C.; Beltrán, E.; Peñuela, G.; López, F.; Sancho, J.V.; Hernández, F. Study of cyanotoxin degradation and evaluation of their transformation products in surface waters by LC-QTOF MS. Chemosphere 2019, 229, 538–548. [Google Scholar] [CrossRef]
- Harada, K.; Tsuji, K.; Watanabe, M.F.; Kondo, F. Stability of microcystins from cyanobacteria—III. Effect of pH and temperature. Phycologia 1996, 35, 83–88. [Google Scholar] [CrossRef]
- Li, J.; Li, R.; Li, J. Current research scenario for microcystins biodegradation–a review on fundamental knowledge, application prospects and challenges. STOTEN 2017, 595, 615–632. [Google Scholar] [CrossRef]
- Jones, G.J.; Bourne, D.G.; Blakeley, R.L.; Doelle, H. Degradation of the cyanobacterial hepatotoxin microcystin by aquatic bacteria. Nat. Toxins 1994, 2, 228–235. [Google Scholar] [CrossRef]
- Manage, P.M.; Edwards, C.; Singh, B.K.; Lawton, L.A. Isolation and identification of novel microcystin-degrading bacteria. Appl. Environ. Microbiol. 2009, 75, 6924–6928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alamri, S.A. Biodegradation of microcystin by a new Bacillus sp. isolated from a Saudi freshwater lake. Afr. J. Biotechnol. 2010, 9, 6552–6559. [Google Scholar]
- Dziga, D.; Wasylewski, M.; Wladyka, B.; Nybom, S.; Meriluoto, J. Microbial degradation of microcystins. Chem. Res. Toxicol. 2013, 26, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Mou, X.; Lu, X.; Jacob, J.; Sun, S.; Heath, R. Metagenomic Identification of bacterioplankton taxa and pathways involved in microcystin degradation in Lake Erie. PLoS ONE 2013, 8, e61890. [Google Scholar] [CrossRef] [Green Version]
- Ding, Q.; Liu, K.Y.; Xu, K.; Sun, R.L.; Zhang, J.; Yin, L.H.; Pu, Y.P. Further understanding of degradation pathways of Microcystin-LR by an indigenous Sphingopyxis sp. in environmentally relevant pollution concentrations. Toxins 2018, 10, 536. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wu, P.; Chen, J.; Yan, H. Biodegradation of Microcystin-RR by a new isolated Sphingopyxis sp. USTB-05. Chin. J. Chem. Eng. 2010, 18, 108–112. [Google Scholar] [CrossRef]
- Zhang, L.; Gu, L.; Wei, Q.; Zhu, X.X.; Wang, J.; Wang, X.J.; Yang, Z. High temperature favors elimination of toxin-producing Microcystis and degradation of microcystins by mixotrophic Ochromonas. Chemosphere 2017, 172, 96–102. [Google Scholar] [CrossRef]
- Xiao, W.; Yan, W.; Jian, O.; Wei, N.; Pan, G. Microcystin-LR biodegradation by Sphingopyxis sp. USTB-05. Front. Environ. Sci. Eng. China 2011, 5, 526–532. [Google Scholar] [CrossRef]
- Xu, H.; Wang, H.; Xu, Q.; Lv, L.; Yin, C.; Liu, X.; Du, H.; Yan, H. Pathway for biodegrading microcystin-YR by Sphingopyxis sp. USTB-05. PLoS ONE 2015, 10, e0124425. [Google Scholar] [CrossRef]
- Briand, E.; Humbert, J.F.; Tambosco, K.; Bormans, M.; Gerwick, W.H. Role of bacteria in the production and degradation of Microcystis cyanopeptides. Microbiologyopen 2016, 5, 469–478. [Google Scholar] [CrossRef]
- Romero-Oliva, C.S.; Contardo-Jara, V.; Block, T.; Pflugmacher, S. Accumulation of microcystin congeners in different aquatic plants and crops—A case study from lake Amatitlan, Guatemala. Ecotoxicol. Environ. Saf. 2014, 102, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Pflugmacher, S.; Wiegand, C.; Beattie, K.A.; Krause, E.; Steinberg, C.E.; Codd, G.A. Uptake, effects, and metabolism of cyanobacterial toxins in the emergent reed plant Phragmites australis (cav.) trin. ex steud. Environ. Toxicol. Chem. Int. J. 2001, 20, 846–852. [Google Scholar] [CrossRef]
- de Morais Calado, S.L.; Santos, G.S.; Leite, T.P.B.; Wojciechowski, J.; Junior, M.N.; Bozza, D.C.; de Magalhaes, V.F.; Cestari, M.M.; Prodocimo, V.; de Assis, H.C.S. Depuration time and sublethal effects of microcystins in a freshwater fish from water supply reservoir. Chemosphere 2018, 210, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Machado, J.; Campos, A.; Vasconcelos, V.; Freitas, M. Effects of microcystin-LR and cylindrospermopsin on plant-soil systems: A review of their relevance for agricultural plant quality and public health. Environ. Res. 2017, 153, 191–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landolt, E. Lemnaceae. In Flowering Plants Monocotyledons; Kubitzki, K., Ed.; Springer: Berlin/Heidelberg, Germany, 1998; pp. 264–270. [Google Scholar]
- Kim, I.S. Microbial colonization of the aquatic duckweed, Spirodela polyrhiza, during development. Appl. Microsc. 2004, 34, 103–111. [Google Scholar]
- Matsuzawa, H.; Tanaka, Y.; Tamaki, H.; Kamagata, Y.; Mori, K. Culture-dependent and independent analyses of the microbial communities inhabiting the giant duckweed (Spirodela polyrrhiza) rhizoplane and isolation of a variety of rarely cultivated organisms within the phylum Verrucomicrobia. Microbes Environ. 2009, 25, 302–308. [Google Scholar] [CrossRef] [Green Version]
- Baudo, R.; Foudoulakis, M.; Arapis, G.; Perdaen, K.; Lanneau, W.; Paxinou, A.C.; Kouvdou, S.; Persoone, G. History and sensitivity comparison of the Spirodela polyrhiza microbiotest and Lemna toxicity tests. KMAE 2015, 416, 23. [Google Scholar] [CrossRef] [Green Version]
- ISO 20079:2005; Water Quality—Determination of the Toxic Effect of Water Constituents and Wastewater on Duckweed (Lemna Minor)—Duckweed Growth Inhibition Test. ISO: Geneva, Switzerland, 2005.
- Starmach, K. Freshwater Phytoplankton. Study Methods, Key to Freshwater Species of Central Europe; PWN: Warszawa/Kraków, Poland, 1989; pp. 1–496. (In Polish) [Google Scholar]
- Cox, E.J. Identification of Freshwater Diatoms from Live Material; Chapman and Hall: London, UK, 1996. [Google Scholar]
- Ziegler, P.; Adelmann, K.; Zimmer, S.; Schmidt, C.; Appenroth, K.J. Relative in vitro growth rates of duckweeds (Lemnaceae)–the most rapidly growing higher plants. Plant Biol. 2015, 17, 33–41. [Google Scholar] [CrossRef]
- Ishii, H.; Nishijima, M.; Abe, T. Characterization of degradation process of cyanobacterial hepatotoxins by a gram-negative aerobic bacterium. Water Res. 2004, 38, 2667–2676. [Google Scholar] [CrossRef]
- Iwashita, T.; Tanaka, Y.; Tamaki, H.; Yoneda, Y.; Makino, A.; Tateno, Y.; Li, Y.; Toyama, T.; Kamagata, Y.; Mori, K. Comparative analysis of microbial communities in fronds and roots of three duckweed species: Spirodela polyrhiza, Lemna minor, and Lemna aequinoctialis. Microbes Environ. 2020, 35, ME20081. [Google Scholar] [CrossRef]
- Phujomjai, Y.; Somdee, A.; Somdee, T. Biodegradation of microcystin Dha(7) MC-LR by a novel microcystin-degrading bacterium in an internal airlift loop bioreactor. Water Sci. Technol. 2016, 73, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Huang, F.; Feng, H.; Wei, J.; Massey, I.Y.; Liang, G.; Zhang, F.; Yin, L.; Kacew, S.; Zhang, X.; et al. A complete route for biodegradation of potentially carcinogenic cyanotoxin microcystin-LR in a novel indigenous bacterium. Water Res. 2020, 174, 115638. [Google Scholar] [CrossRef]
- Lawton, L.A.; Welgamage, A.; Manage, P.M.; Edwards, C. Novel bacterial strains for the removal of microcystins from drinking water. Water Sci. Technol. 2011, 63, 1137–1142. [Google Scholar] [CrossRef] [PubMed]
- Idroos, E.S.; De Silva, B.; Manage, P.M. Biodegradation of microcystin analogues by Stenotrophomonas maltophilia isolated from Beira Lake Sri Lanka. J. Natl. Sci. Found. 2017, 45, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Nájera, A.F.; Serwecińska, L.E.; Gągała-Borowska, I.; Jurczak, T.E.; Mankiewicz-Boczek, J.D. The characterization of a novel bacterial strain capable of microcystin degradation from the Jeziorsko reservoir, Poland: A preliminary study. Biologia 2017, 72, 1394–1402. [Google Scholar] [CrossRef]
- Tsao, S.; Wei, D.J.; Chang, Y.T.; Lee, J.F. Aerobic biodegradation of microcystin-LR by an indigenous bacterial mixed culture isolated in Taiwan. Int. Biodeterior. Biodegrad. 2017, 124, 101–108. [Google Scholar] [CrossRef]
- Manheim, D.; Cheung, Y.M.; Jiang, S. The effect of organic carbon addition on the community structure and kinetics of microcystin-degrading bacterial consortia. Water 2018, 10, 1523. [Google Scholar] [CrossRef] [Green Version]
- Christoffersen, K.; Lyck, S.; Winding, A. Microbial activity and bacterial community structure during degradation of microcystins. Aquat. Microb. Ecol. 2002, 27, 125–136. [Google Scholar] [CrossRef] [Green Version]
- Edwards, C.; Graham, D.; Fowler, N.; Lawton, L.A. Biodegradation of microcystins and nodularin in freshwaters. Chemosphere 2008, 73, 1315–1321. [Google Scholar] [CrossRef]
- Mohamed, Z.A.; Al-Shehri, A.M. Grazing on Microcystis aeruginosa and degradation of microcystins by the heterotrophic flagellate Diphylleia rotans. Ecotoxicol. Environ. Saf. 2013, 96, 48–52. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, H.Y.; Hong, Y.; Yang, J. Isolation of a Poterioochromonas capable of feeding on Microcystis aeruginosa and degrading microcystin-LR. FEMS Microbiol. Lett. 2008, 288, 241–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Hu, H.Y.; Men, Y.J.; Christoffersen, K.S. The effect of Poterioochromonas abundance on production of intra- and extracellular microcystin-LR concentration. Hydrobiologia 2010, 652, 237–246. [Google Scholar] [CrossRef]
- Maruyama, T.; Kato, K.; Yokoyama, A.; Tanaka, T.; Hiraishi, A.; Park, H.D. Dynamics of microcystin-degrading bacteria in mucilage of Microcystis. Microb. Ecol. 2003, 46, 279–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Shimizu, K.; Zhou, Y.; Utsumi, M.; Sakharkar, M.K.; Zhang, Z.; Sun, H.; Sugiura, N. Biodegradation of microcystins by bacterial communities co-existing with the flagellate Monas guttula and concurrent succession of community structures. J. Water Supply Res. Technol.—AQUA 2011, 60, 352–363. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Pan, G.; Yan, H. Microbial biodegradation of microcystin-RR by bacterium Sphingopyxis sp. USTB-05. J. Environ. Sci. 2010, 22, 168–175. [Google Scholar] [CrossRef]
- Pham, T.L.; Utsumi, M. An overview of the accumulation of microcystins in aquatic ecosystems. J. Environ. Manag. 2018, 213, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Mitrovic, S.M.; Allis, O.; Furey, A.; James, K.J. Bioaccumulation and harmful effects of microcystin-LR in the aquatic plants Lemna minor and Wolffia arrhiza and the filamentous alga Chladophora fracta. Ecotoxicol. Environ. Saf. 2005, 61, 345–352. [Google Scholar] [CrossRef]
- Isobe, T.; Okuhata, H.; Miyasaka, H.; Jeon, B.S.; Park, H.D. Detoxification of microcystin-LR in water by Portulaca oleracea cv. J. Biosci. Bioeng. 2014, 117, 330–332. [Google Scholar] [CrossRef]
- Omidi, A.; Pflugmacher, S.; Kaplan, A.; Kim, Y.J.; Esterhuizen, M. Reviewing Interspecies Interactions as a Driving Force Affecting the community structure in lakes via cyanotoxins. Microorganisms 2021, 9, 1583. [Google Scholar] [CrossRef]
- Fischer, A.; Hoeger, S.J.; Stemmer, K.; Feurstein, D.J.; Knobeloch, D.; Nussler, A.; Dietrich, D.R. The role of organic anion transporting polypeptides (OATPs/SLCOs) in the toxicity of different microcystin congeners in vitro: A comparison of primary human hepatocytes and OATP-transfected HEK293 cells. Toxicol. Appl. Pharmacol. 2010, 245, 9–20. [Google Scholar] [CrossRef] [Green Version]
- Vesterkvist, P.S.M.; Misiorek, J.O.; Spoof, L.E.M.; Toivola, D.M.; Meriluoto, J.A.O. Comparative cellular toxicity of hydrophilic and hydrophobic microcystins on Caco-2 cells. Toxins 2012, 4, 1008–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, C.J.; Codd, G.A. Comparative toxicity of four microcystins of different hydrophobicities to the protozoan, Tetrahymena pyriformis. J. Appl. Microb. 1999, 86, 874–882. [Google Scholar] [CrossRef] [PubMed]
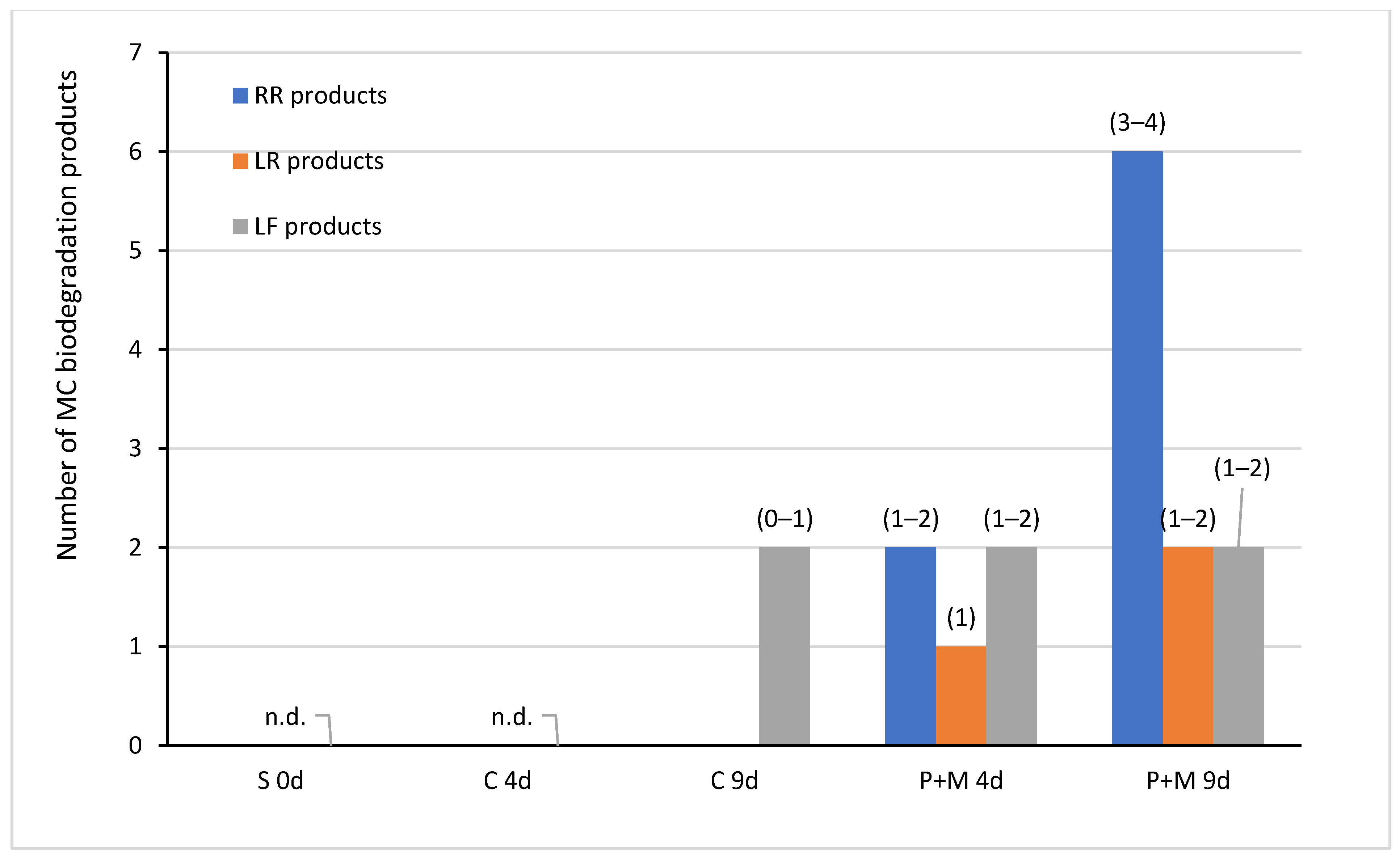
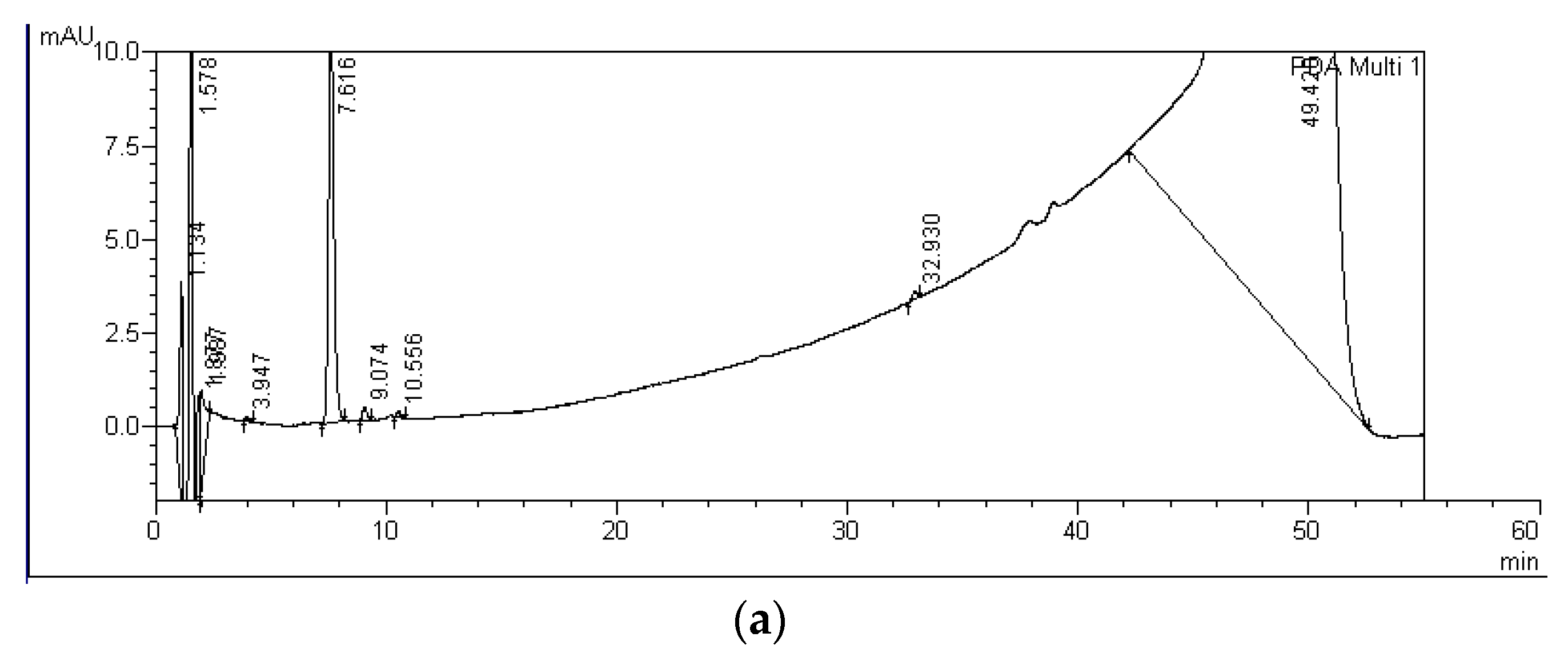
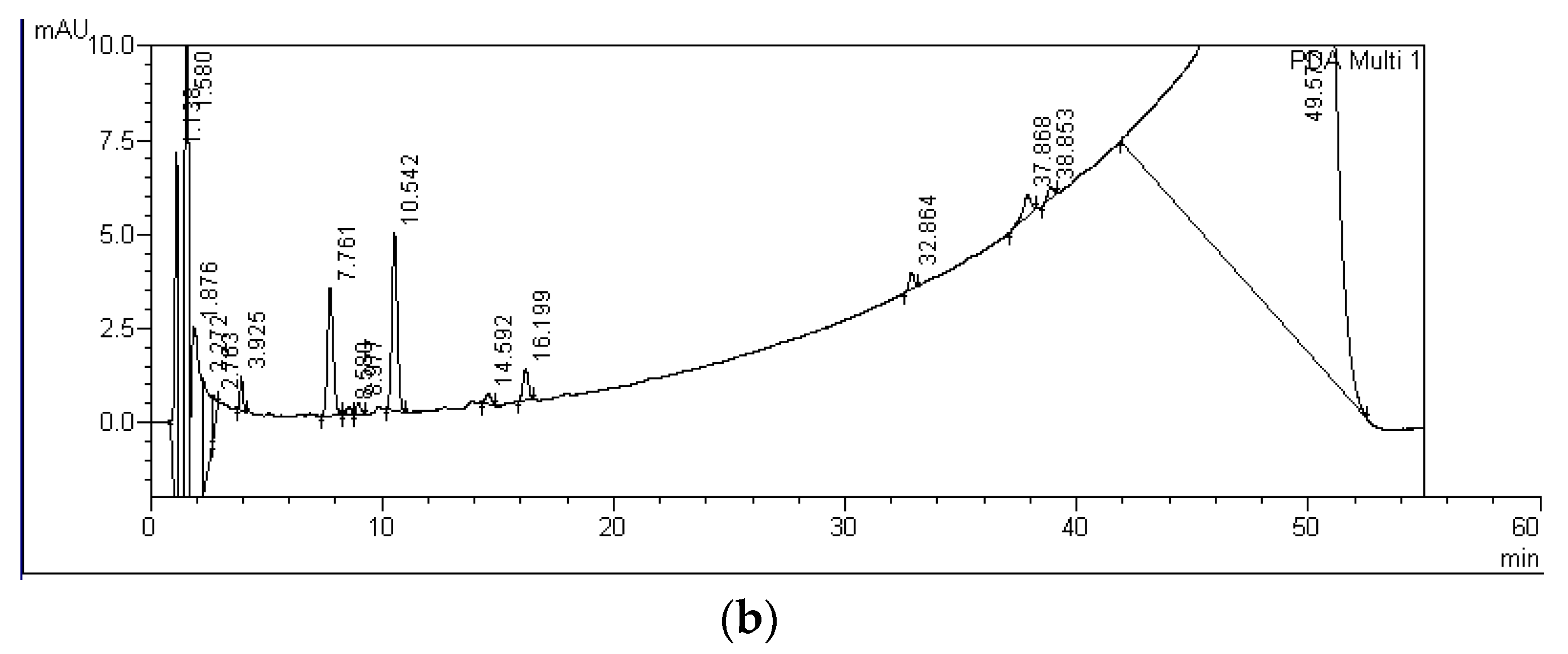
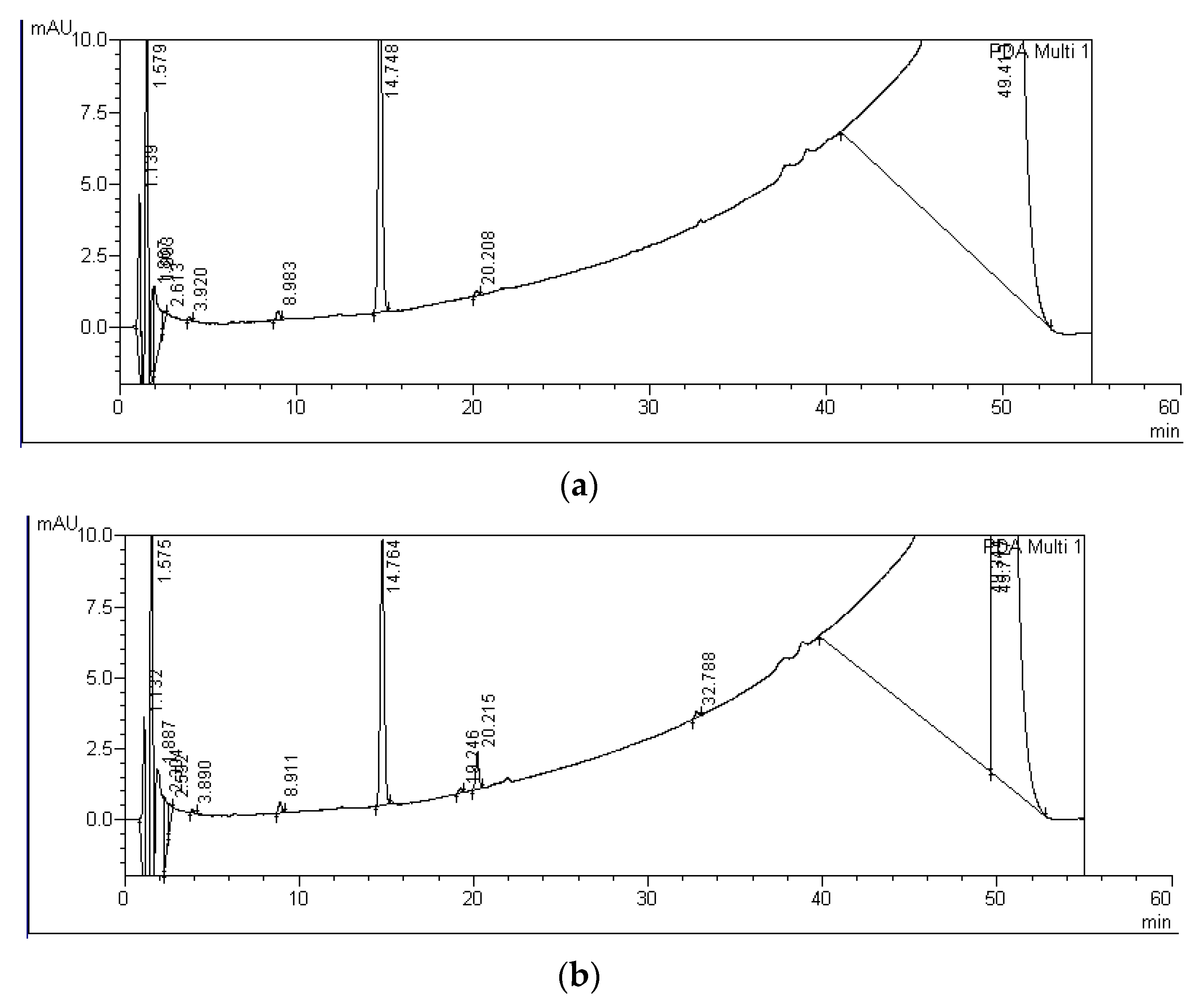
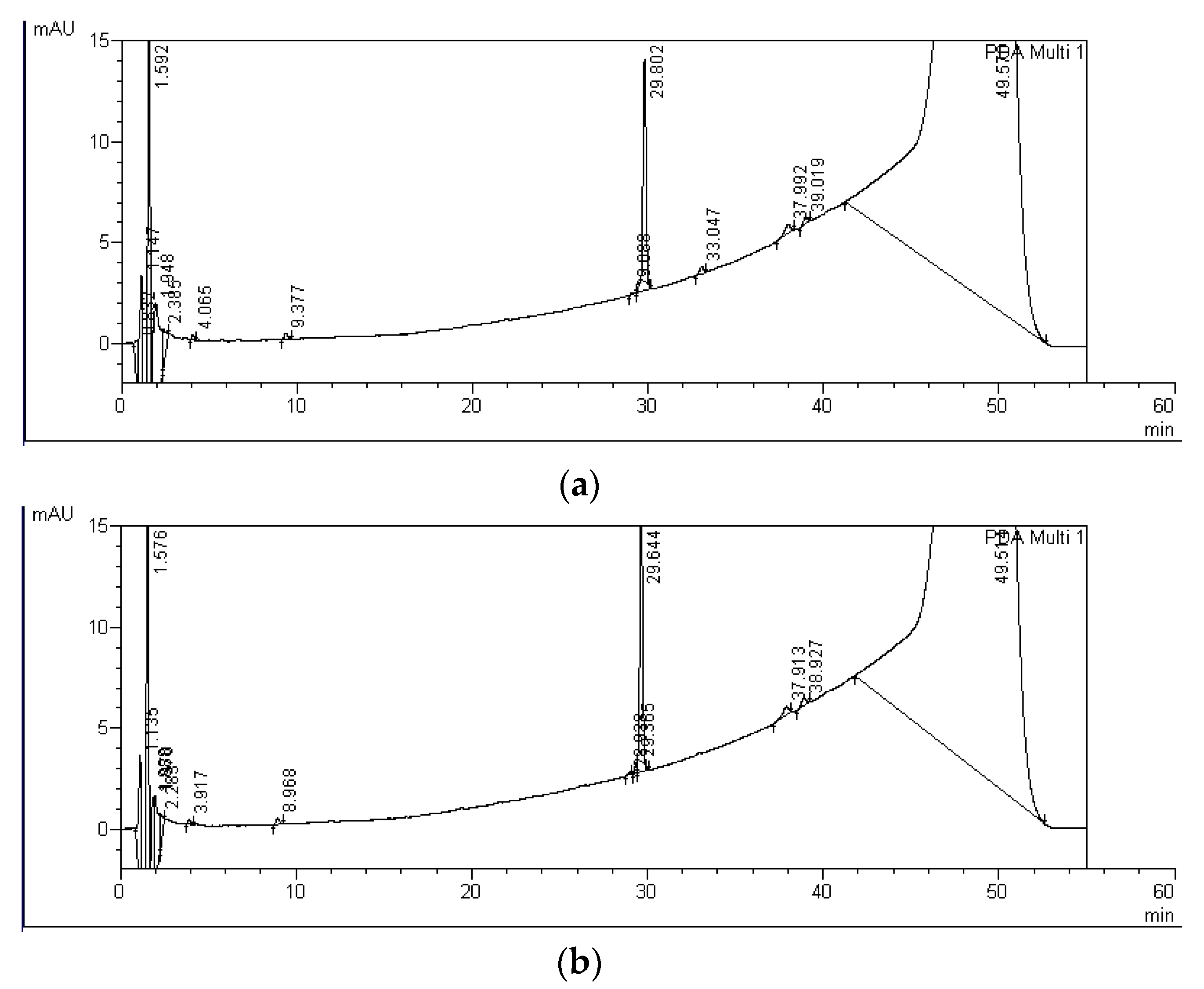
| MC Variants | MCs Degradation Rates (%) | HPLC Detection of Degradation Products and Their Retention Times (Minutes) | ||||||
|---|---|---|---|---|---|---|---|---|
| 4 d | 9 d | A-* | B-* | C-* | D-* | E- | F- | |
| MC-RR C | 9 ± 4 | 8 ± 3 | - | - | - | - | - | - |
| MC-LR C | 3 ± 7 | 1 ± 5 | - | - | - | - | - | - |
| MC-LF C | 0 ± 6 | 11 ± 15 | - | C-LF: 30.0 | D-LF: 33.0 | - | - | |
| MC-RR P+M | 26 ± 10 | 61 ± 19 | A-RR: 5.6 | B-RR: 6.5 | C-RR: 8.6 | D-RR: 10.6 | E-RR: 14.6 | F-RR: 16.2 |
| MC-LR P+M | 11 ± 6 | 21 ± 4 | A-LR: 19.3 | B-LR: 20.2 | - | - | - | - |
| MC-LF P+M | 11 ± 10 | 7 ± 9 | A-LF: 28.9 | B-LF: 29.4 | C-LF: 30.0 | D-LF: 33.0 | - | - |
| MCs and Their Degradation Products | Units | Experimental Variants | ||||
|---|---|---|---|---|---|---|
| S 0d | C 4d | C 9d | P + M 4d | P + M 9d | ||
| A-RR | ng equiv. MC-RR/mL | 0–7.9 | ||||
| B-RR | 0–5.5 | |||||
| MC-RR | ng MC-RR/mL | 936.8–1030.2 | 862.9–941.06 | 883.6–938.2 | 667.9–836.8 | 273.4–595.7 |
| C-RR | ng equiv. MC-RR/mL | 0–16.0 | ||||
| D-RR | 11.3–35.9 | 331.1–379.5 | ||||
| E-RR | 0–20.0 | |||||
| F-RR | 0–45.7 | 16.5–58.3 | ||||
| Total concentration | ng equiv. MC-RR/mL | 936.8–1030.2 | 862.9–941.06 | 883.6–938.2 | 712.9–848.0 * | 729.5–851.3 ** |
| MC-LR | ng MC-LR/mL | 958.5–1023.0 | 892.4–1024.7 | 947.0–1031.2 | 818.8–943.9 | 745.8–824.2 |
| A-LR | ng equiv. MC-LR/mL | 0–10.0 | ||||
| B-LR | 0–27.8 | 37.0–80.4 | ||||
| Total concentration | ng equiv. MC-LR/mL | 958.5–1030.2 | 892.4–1024.7 | 947.0–1031.2 | 846.6–950.17 | 825.3–862.4 |
| A-LF | ng equiv. MC-LF/mL | 0–5.2 | 0–7.0 | 5.1–5.9 | ||
| B-LF | 0–23.0 | |||||
| MC-LF | ng MC-LF/mL | 820.9–868.5 | 811.6–908.4 | 631.4–882.6 | 691.7–813.9 | 730.1–843.4 |
| C-LF | ng equiv. MC-LF/mL | 0–6.6 | ||||
| D-LF | 0–30.6 | |||||
| Total concentration | ng equiv. MC-LF/mL | 820.9–868.5 | 811.6–908.4 | 636.6–882.6 | 697.0–851.6 | 735.1–872.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toporowska, M. Degradation of Three Microcystin Variants in the Presence of the Macrophyte Spirodela polyrhiza and the Associated Microbial Communities. Int. J. Environ. Res. Public Health 2022, 19, 6086. https://doi.org/10.3390/ijerph19106086
Toporowska M. Degradation of Three Microcystin Variants in the Presence of the Macrophyte Spirodela polyrhiza and the Associated Microbial Communities. International Journal of Environmental Research and Public Health. 2022; 19(10):6086. https://doi.org/10.3390/ijerph19106086
Chicago/Turabian StyleToporowska, Magdalena. 2022. "Degradation of Three Microcystin Variants in the Presence of the Macrophyte Spirodela polyrhiza and the Associated Microbial Communities" International Journal of Environmental Research and Public Health 19, no. 10: 6086. https://doi.org/10.3390/ijerph19106086
APA StyleToporowska, M. (2022). Degradation of Three Microcystin Variants in the Presence of the Macrophyte Spirodela polyrhiza and the Associated Microbial Communities. International Journal of Environmental Research and Public Health, 19(10), 6086. https://doi.org/10.3390/ijerph19106086






