Seasonal and Technological Shifts of the WHO Priority Multi-Resistant Pathogens in Municipal Wastewater Treatment Plant and Its Receiving Surface Water: A Case Study
Abstract
:1. Introduction
2. Materials and Methods
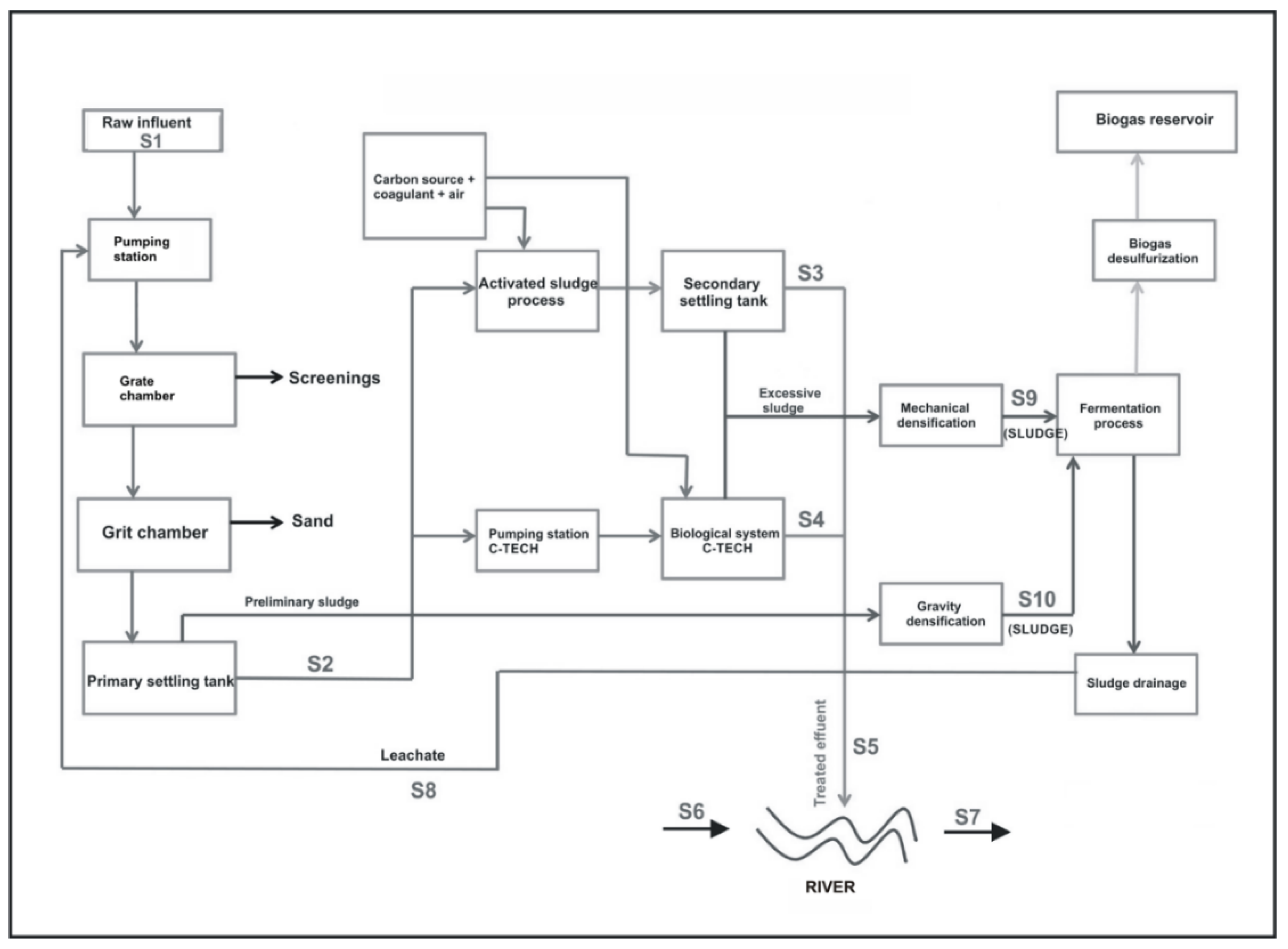
2.1. Description of WWTP and Sample Collection
2.2. DNA Extraction and Illumina Sequencing
2.3. Data Analysis
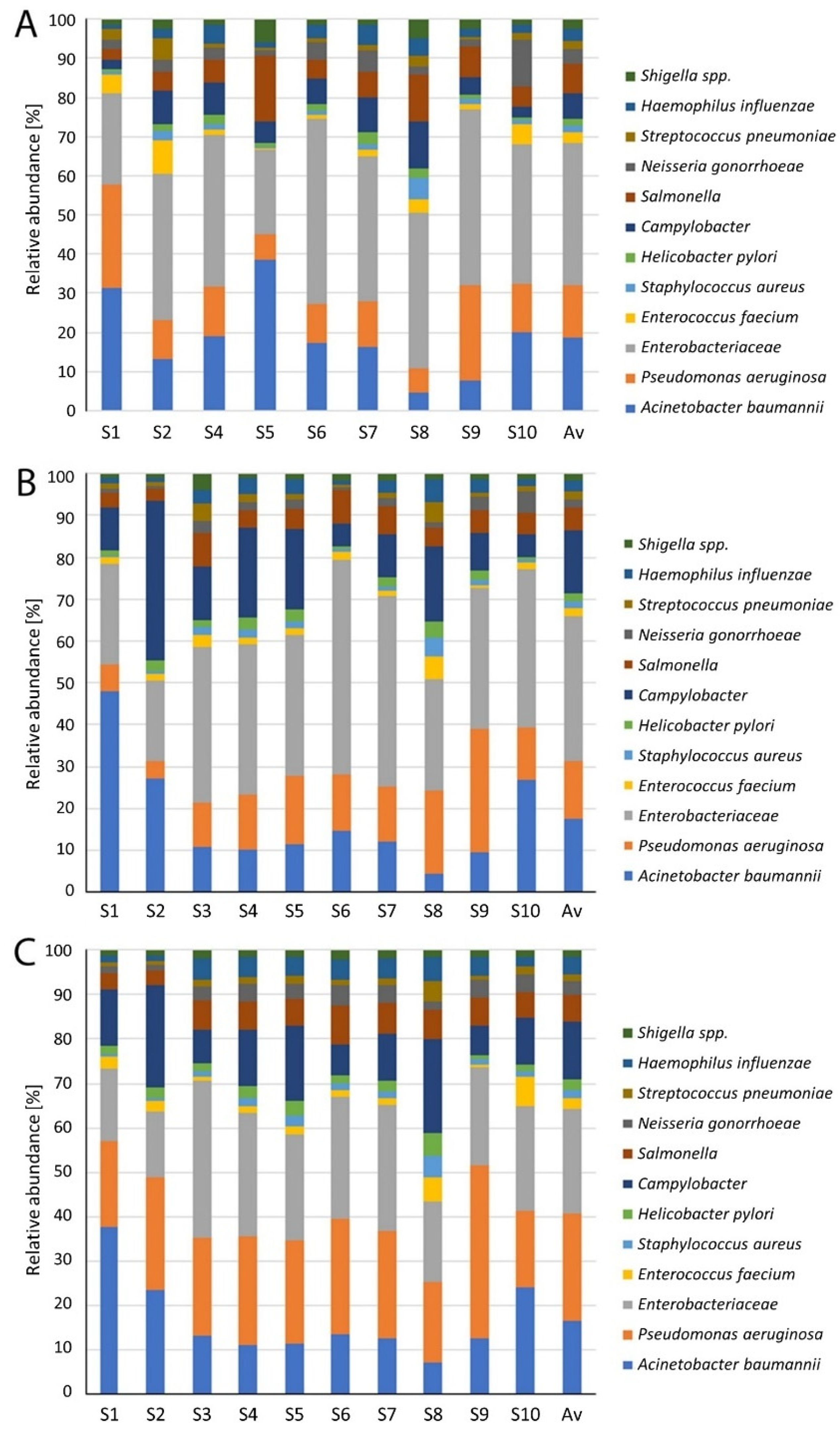
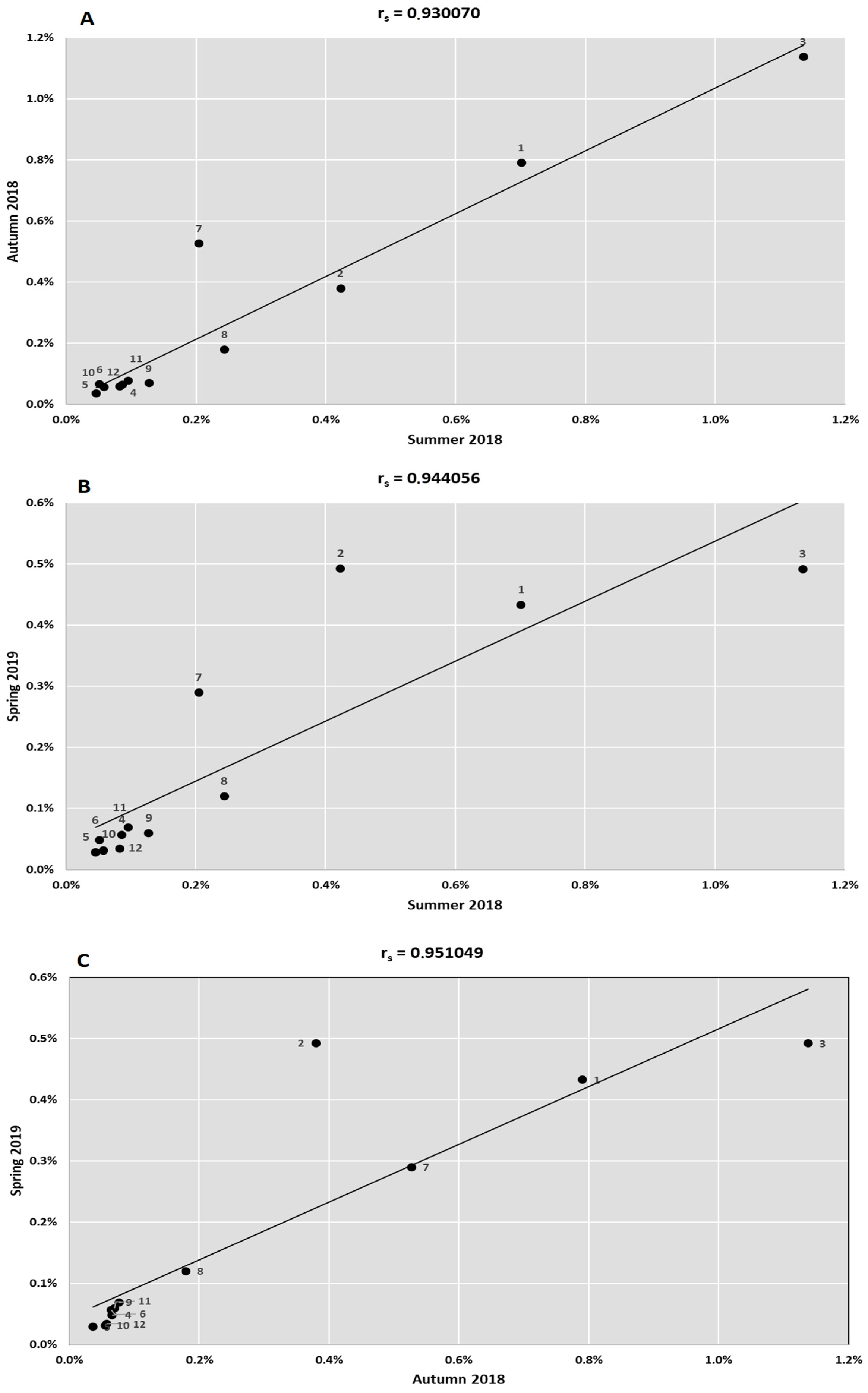
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dias, D.A.; Urban, S.; Roessner, U. A historical overview of natural products in drug discovery. Metabolites 2012, 2, 303–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rex, J.H. ND4BB: Addressing the antimicrobial resistance crisis. Nat. Rev. Microbiol. 2014, 12, 231–242. [Google Scholar] [CrossRef]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. Available online: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (accessed on 14 March 2021).
- WHO. Antimicrobial Resistance. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 16 March 2021).
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The antimicrobial resistance crisis: Causes, consequences, and management. Front. Public Health 2014, 2, 145. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Trotter, A.J.; Aydin, A.; Strinden, M.J.; O’Grady, J. Recent and emerging technologies for the rapid diagnosis of infection and antimicrobial resistance. Curr. Opin. Microbiol. 2019, 51, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, L.; Zhang, T. Detecting human bacterial pathogens in wastewater treatment plants by a high-throughput shotgun sequencing technique. Environ. Sci. Technol. 2013, 47, 5433–5441. [Google Scholar] [CrossRef]
- Kumaraswamy, R.; Amha, Y.M.; Anwar, M.Z.; Henschel, A.; Rodriguez, J.; Ahmad, F. Molecular analysis for screening human bacterial pathogens in municipal wastewater treatment and reuse. Environ. Sci. Technol. 2014, 48, 11610–11619. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.T.T.; Petrovich, M.L.; Chaudhary, A.; Wright, D.; Murphy, B.; Wells, G.; Poretsky, R. Metagenomics reveals the impact of wastewater treatment plants on the dispersal of microorganisms and genes in aquatic sediments. Appl. Environ. Microbiol. 2018, 84, e02168-17. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.; Zhao, F.; Zhang, X.-X.; Ye, L.; Ren, H.; Zhang, T.; Mao, Y.; Ju, F.; Wang, Y.; Li, B. Free-living bacteria and potential bacterial pathogens in sewage treatment plants. Appl. Microbiol. Biotechnol. 2018, 102, 2455–2464. [Google Scholar] [CrossRef]
- Osunmakinde, C.O.; Selvarajan, R.; Mamba, B.B.; Msagati, T.A.M. Profiling bacterial diversity and potential pathogens in wastewater treatment plants using high-throughput sequencing analysis. Microorganisms 2019, 7, 506. [Google Scholar] [CrossRef] [Green Version]
- Rajasulochana, P. Comparison on efficiency of various techniques in treatment of waste and sewage water—A comprehensive review. Resour. Technol. 2016, 2, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Okoh, A.I.; Odjadjare, E.E.; Igbinosa, E.O.; Osode, A.N. Wastewater treatment plants as a source of microbial pathogens in receiving watersheds. Afr. J. Biotechnol. 2007, 6, 2932–2944. [Google Scholar] [CrossRef]
- Naidoo, S.; Olaniran, A.O. Treated wastewater effluent as a source of microbial pollution of surface water resources. Int. J. Environ. Res. Public Heal. 2013, 11, 249–270. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Zhang, X.; Wang, Z.; Huang, K.; Wang, Y.; Liang, W. Bacterial pathogens and community composition in advanced sewage treatment systems revealed by metagenomics analysis based on high-throughput sequencing. PLoS ONE 2015, 5, e0125549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucena, F.; Duran, A.E.; Morón, A.; Calderón, E.; Campos, C.; Gantzer, C.; Skraber, S.; Jofre, J. Reduction of bacterial indicators and bacteriophages infecting fecal bacteria in primary and secondary wastewater treatments. J. Appl. Microbiol. 2004, 97, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Action Plan on Antimicrobial Resistance. 2015. Available online: http://www.who.int/antimicrobial-resistance/global-action-plan/en/ (accessed on 20 February 2021).
- WHO. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug Resistant Bacterial Infections, Including Tuberculosis; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Rolbiecki, D.; Harnisz, M.; Korzeniewska, E.; Jałowiecki, Ł.; Płaza, G. Occurrence of fluoroquinolones and sulfonamides resistance genes in wastewater and sludge at different stages of wastewater treatment: A preliminary case study. Appl. Sci. 2020, 10, 5816. [Google Scholar] [CrossRef]
- Płaza, G.; Jałowiecki, Ł.; Głowacka, D.; Hubeny, J.; Harnisz, M.; Korzeniewska, E. Insights into the microbial diversity and structure in a full-scale municipal wastewater treatment plant with particular regard to Archaea. PLoS ONE 2021, 16, e0250514. [Google Scholar] [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [Green Version]
- De Lima, I.R.; Dos Santos, L.U.; Tosetto, M.S.; Franco, R.M.; Guimarães, J.R. Urban water reuse: Microbial pathogens control by direct filtration and ultraviolet disinfection. J. Water Health 2014, 12, 465–473. [Google Scholar] [CrossRef]
- Ashbolt, N.J.; Grabow, W.O.K.; Snozzi, M. Indicators of microbial water quality. In Water Quality: Guidelines, Standards and Health; Fewtrell, L., Bartram, J., Eds.; IWA Publishing: London, UK, 2001; pp. 127–145. [Google Scholar]
- Wen, X.; Chen, F.; Lin, Y.; Zhu, H.; Yuan, F.; Kuang, D.; Jia, Z.; Yuan, Z. Microbial indicators and their use for monitoring drinking water quality—A review. Sustainability 2020, 12, 2249. [Google Scholar] [CrossRef] [Green Version]
- Jurzik, L.; Hamza, I.A.; Puchert, W.; Uberla, K.; Wilhelm, M. Chemical and microbiological parameters as possible indicators for human enteric viruses in surface water. Int. J. Hyg. Environ. Health 2010, 213, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Ajonina, C.; Buzie, C.; Rubiandini, R.H.; Otterpohl, R. Microbial pathogens in wastewater treatment plants (WWTP) in Hamburg. J. Toxicol. Environ. Health Part A 2015, 78, 381–387. [Google Scholar] [CrossRef]
- Marie, V.; Lin, J. Microbial indicators and environmental relationships in the Umhlangane River, Durban, South Africa. Open Life Sci. 2018, 13, 385–395. [Google Scholar] [CrossRef]
- Garrido-Perez, M.C.; Anfuso, E.; Acevedo, A.; Perales-Vargas-Machuca, J.A. Microbial indicators of faecal contamination in waters and sediments of beach bathing zones. Int. J. Hyg. Environ. Health 2008, 211, 510–517. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, F.; Taban, B.M. A State-of-Art Review on Multi-Drug Resistant Pathogens in Foods of Animal Origin: Risk Factors and Mitigation Strategies. Front. Microbiol. 2019, 10, 2091. [Google Scholar] [CrossRef] [Green Version]
- Kakoullis, L.; Papachristodoulou, E.; Chra, P.; Panos, G. Mechanisms of antibiotic resistance in important gram-positive and gram-negative pathogens and novel antibiotic solutions. Antibiotics 2021, 10, 415. [Google Scholar] [CrossRef] [PubMed]
- ECDC. Antimicrobial Resistance in the EU/EEA—AER for 2019; ECDC: Stockholm, Sweden, 2020. [Google Scholar]
- Banin, E.; Hughes, D.; Kuipers, O.P. Bacterial pathogens, antibiotics and antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 450–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaha, D.C.; Bungau, S.; Aleya, S.; Tit, D.M.; Vesa, C.M.; Popa, A.R.; Pantis, C.; Maghiar, O.A.; Bratu, O.G.; Furau, C.; et al. What antibiotics for what pathogens? The sensitivity spectrum of isolated strains in an intensive care unit. Sci. Total Environ. 2019, 687, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Handal, R.; Qunibi, L.; Sahouri, I.; Juhari, M.; Dawodi, R.; Marzouqa, H.; Hindiyeh, M. Characterization of carbapenem-resistant Acinetobacter baumannii strains isolated from hospitalized patients in Palestine. Int. J. Microbiol. 2017, 2017, 8012104. [Google Scholar] [CrossRef] [Green Version]
- Fischbach, M.A.; Walsh, C.T. Antibiotics for Emerging Pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Mhondoro, M.; Ndlovu, N.; Donewell, B.; Juru, T.; Tafara, G.N.; Gerald, S.; Peter, N.; Mufuta, T. Trends in antimicrobial resistance of bacterial pathogens in Harare, Zimbabwe, 2012–2017: A secondary dataset analysis. BMC Infect. Dis. 2019, 19, 746. [Google Scholar] [CrossRef]
- Karkman, A.; Pärnänen, K.; Larsson, D.G.J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.R.; Montoya, V.; Gardy, J.L.; Patrick, D.M.; Tang, P. Metagenomics for pathogen detection in public health. Genome Med. 2013, 5, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibekwe, A.M.; Leddy, M.; Murinda, S.E. Potential human pathogenic bacteria in a mixed urban watershed as revealed by pyrosequencing. PLoS ONE 2013, 8, e79490. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-Castillo, F.; Loera-Muro, A.; Jacques, M.; Garneau, P.; Avelar-González, F.; Harel, J.; Guerrero-Barrera, A. Waterborne pathogens: Detection methods and challenges. Pathogens 2015, 4, 307–334. [Google Scholar] [CrossRef] [PubMed]
- Ricchi, M.; Bertasio, C.; Boniotti, M.B.; Vicari, N.; Russo, S.; Tilola, M.; Bellotti, M.A.; Bertasi, B. Comparison among the quantification of bacterial pathogens by qPCR, dPCR, and cultural methods. Front. Microbiol. 2017, 8, 1174. [Google Scholar] [CrossRef]



| Categories | Bacteria | Antibiotic-Resistance |
|---|---|---|
| Priority 1: Critical | Acinetobacter baumannii | Carbapenem-resistant |
| Pseudomonas aeruginosa | Carbapenem-resistant | |
| Enterobacteriaceae | Carbapenem-resistant, ESBL-producing | |
| Priority 2: High | Enterococcus faecium | Vancomycin-resistant |
| Staphylococcus aureus | Methicillin-resistant, vancomycin-resistant | |
| Helicobacter pylori | Clarithromycin-resistant | |
| Campylobacter spp. | Fluoroquinolone-resistant | |
| Salmonella spp. | Fluoroquinolone-resistant | |
| Neisseria gonorrhoeae | Cephalosporin-resistant, fluoroquinolone-resistant | |
| Priority 3: Medium | Streptococcus pneumoniae | Penicillin-non-susceptible |
| Haemophilus influenzae | Ampicillin-resistant | |
| Shigella spp. | Fluoroquinolone-resistant |
| Technological Parameters * | Unit | Wastewater | June 2018 | Autumn 2018 | March 2019 |
|---|---|---|---|---|---|
| Flow | m3/month | 960,077 | 722,516 | 793,234 | |
| Temperature | °C | Influent | 19.5 | 10.5 | 14.5 |
| Effluent | 21 | 19 | 13.5 | ||
| pH | Effluent | 7.2 | 7.2 | 7.2 | |
| COD | mg/L | Influent | 963 | 672 | 970 |
| Effluent | 36.5 | 30.5 | 35.0 | ||
| BOD5 | mg/L | Influent | 435 | 290 | 340 |
| Effluent | 4.8 | 4.7 | 6.0 | ||
| Suspension | mg/L | Influent | 525 | 310 | 455 |
| Effluent | 5.8 | 6.3 | 7.2 | ||
| NTOT | mg/L | Influent | 106.1 | 78.1 | 84.2 |
| Effluent | 10.4 | 7.9 | 6.2 | ||
| NNH4+ | mg/L | Influent | 32.25 | 48.90 | 56.90 |
| Effluent | 0.36 | 0.22 | 0.31 | ||
| PTOT | mg/L | Influent | 8.88 | 14.9 | 11.4 |
| Effluent | 0.74 | 1.1 | 0.5 | ||
| SRT | d | 17 | 19 | 20 | |
| HRT | h | 9 | 9 | 9 | |
| SS | kg/m3 | 4.5 | 5.0 | 5.5 | |
| Meterological parameters * | |||||
| Temperature | °C | 20.4 | 4.5 | 6.1 | |
| Rainfall | mm | 71 | 14 | 59 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jałowiecki, Ł.; Hubeny, J.; Harnisz, M.; Płaza, G. Seasonal and Technological Shifts of the WHO Priority Multi-Resistant Pathogens in Municipal Wastewater Treatment Plant and Its Receiving Surface Water: A Case Study. Int. J. Environ. Res. Public Health 2022, 19, 336. https://doi.org/10.3390/ijerph19010336
Jałowiecki Ł, Hubeny J, Harnisz M, Płaza G. Seasonal and Technological Shifts of the WHO Priority Multi-Resistant Pathogens in Municipal Wastewater Treatment Plant and Its Receiving Surface Water: A Case Study. International Journal of Environmental Research and Public Health. 2022; 19(1):336. https://doi.org/10.3390/ijerph19010336
Chicago/Turabian StyleJałowiecki, Łukasz, Jakub Hubeny, Monika Harnisz, and Grażyna Płaza. 2022. "Seasonal and Technological Shifts of the WHO Priority Multi-Resistant Pathogens in Municipal Wastewater Treatment Plant and Its Receiving Surface Water: A Case Study" International Journal of Environmental Research and Public Health 19, no. 1: 336. https://doi.org/10.3390/ijerph19010336
APA StyleJałowiecki, Ł., Hubeny, J., Harnisz, M., & Płaza, G. (2022). Seasonal and Technological Shifts of the WHO Priority Multi-Resistant Pathogens in Municipal Wastewater Treatment Plant and Its Receiving Surface Water: A Case Study. International Journal of Environmental Research and Public Health, 19(1), 336. https://doi.org/10.3390/ijerph19010336






