Tracking Fecal Bacterial Dispersion from Municipal Wastewater to Peri-Urban Farms during Monsoon Rains in Hue City, Vietnam
Abstract
1. Introduction
2. Materials and Methods
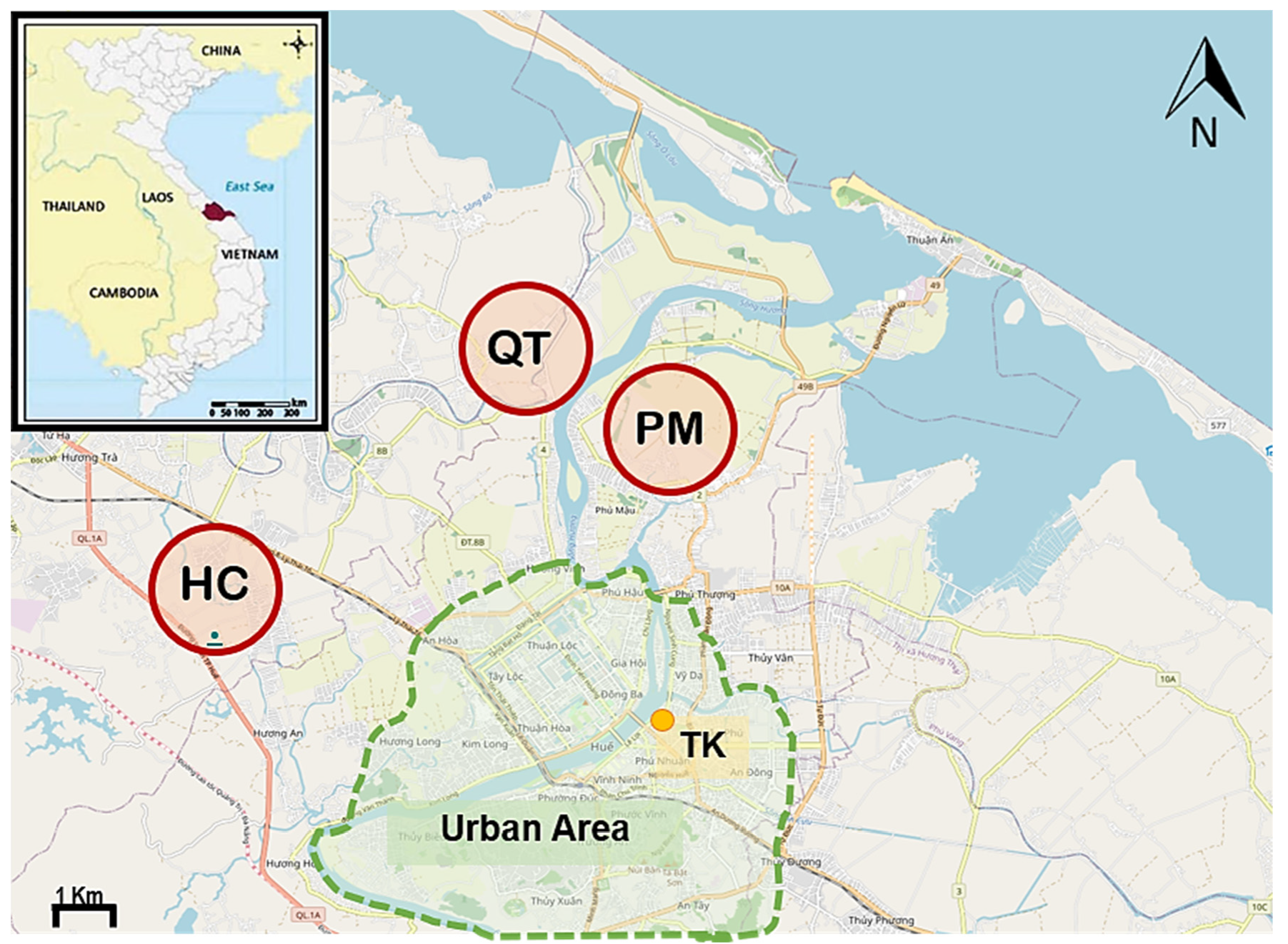
2.1. Sampling
2.2. Counting and Isolation of E. coli
2.3. Identification of E. coli Isolates
2.4. Multilocus Sequence Typing
2.5. GrapeTree Analysis
2.6. Phylogenetic and Diversity Analysis
2.7. Data Analysis
3. Results
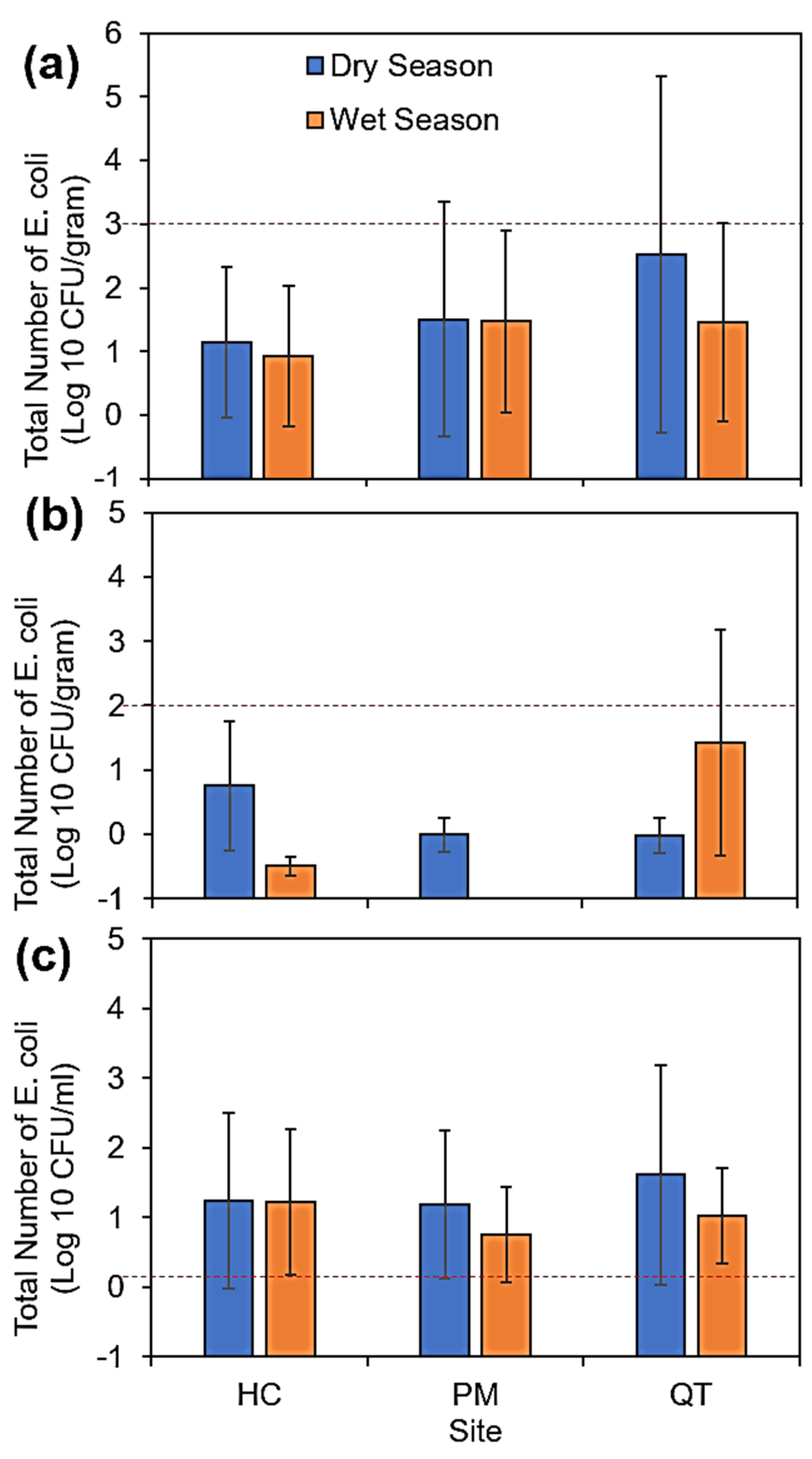
3.1. Concentration of E. coli
3.2. E. coli Isolates
3.3. Sequence Type Identities of E. coli Isolates
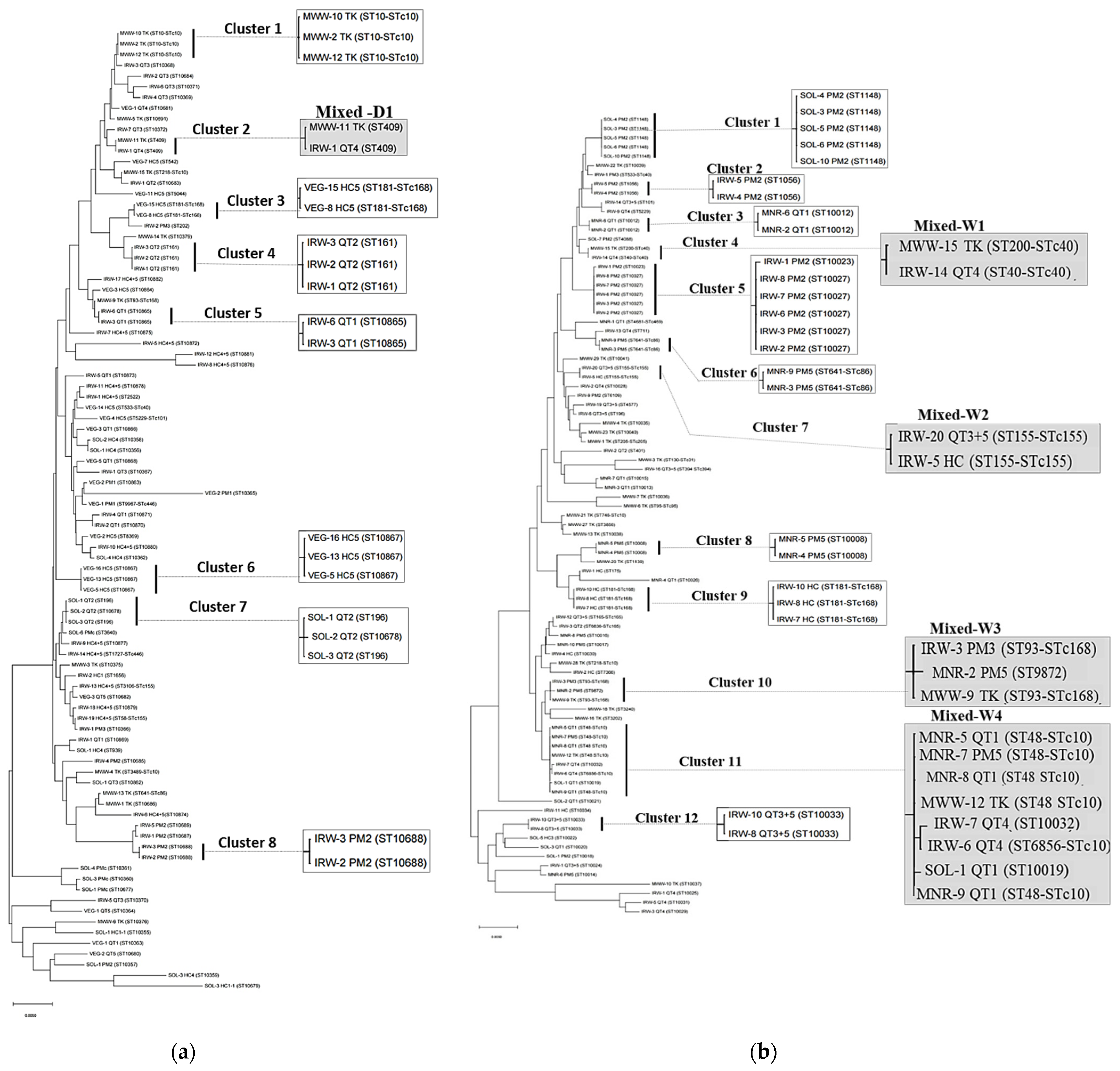
3.4. Relatedness of E. coli Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Talbot, C.J.; Bennett, E.M.; Cassell, K.; Hanes, D.M.; Minor, E.C.; Paerl, H.; Raymond, P.A.; Vargas, R.; Vidon, P.G.; Wollheim, W.; et al. The Impact of Flooding on Aquatic Ecosystem Services. Biogeochemistry 2018, 141, 439–461. [Google Scholar] [CrossRef] [PubMed]
- Alexander, K.A.; Heaney, A.K.; Shaman, J. Hydrometeorology and Flood Pulse Dynamics Drive Diarrheal Disease Outbreaks and Increase Vulnerability to Climate Change in Surface-Water-Dependent Populations: A Retrospective Analysis. PLoS Med. 2018, 15, e1002688. [Google Scholar] [CrossRef] [PubMed]
- Andrade, L.; O’Dwyer, J.; O’Neill, E.; Hynds, P. Surface Water Flooding, Groundwater Contamination, and Enteric Disease in Developed Countries: A Scoping Review of Connections and Consequences. Environ. Pollut. 2018, 236, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Lowe, D.; Ebi, K.; Forsberg, B. Factors Increasing Vulnerability to Health Effects before, during and after Floods. Int. J. Environ. Res. Public Health 2013, 10, 7015–7067. [Google Scholar] [CrossRef]
- Okaka, F.O.; Odhiambo, B.D.O. Relationship between Flooding and Out Break of Infectious Diseasesin Kenya: A Review of the Literature. J. Environ. Public Health 2018, 2018, 1–8. [Google Scholar] [CrossRef]
- Yu, P.; Zaleski, A.; Li, Q.; He, Y.; Mapili, K.; Pruden, A.; Alvarez, P.J.J.; Stadler, L.B. Elevated Levels of Pathogenic Indicator Bacteria and Antibiotic Resistance Genes after Hurricane Harvey’s Flooding in Houston. Environ. Sci. Technol. Lett. 2018, 5, 481–486. [Google Scholar] [CrossRef]
- De Man, H.; van den Berg, H.H.J.L.; Leenen, E.J.T.M.; Schijven, J.F.; Schets, F.M.; van der Vliet, J.C.; van Knapen, F.; de Roda Husman, A.M. Quantitative Assessment of Infection Risk from Exposure to Waterborne Pathogens in Urban Floodwater. Water Res. 2014, 48, 90–99. [Google Scholar] [CrossRef]
- Ten Veldhuis, J.A.E.; Clemens, F.H.L.R.; Sterk, G.; Berends, B.R. Microbial Risks Associated with Exposure to Pathogens in Contaminated Urban Flood Water. Water Res. 2010, 44, 2910–2918. [Google Scholar] [CrossRef]
- Yard, E.E.; Murphy, M.W.; Schneeberger, C.; Narayanan, J.; Hoo, E.; Freiman, A.; Lewis, L.S.; Hill, V.R. Microbial and Chemical Contamination during and after Flooding in the Ohio River—Kentucky, 2011. J. Environ. Sci. Health Part A 2014, 49, 1236–1243. [Google Scholar] [CrossRef]
- Carlton, E.J.; Eisenberg, J.N.S.; Goldstick, J.; Cevallos, W.; Trostle, J.; Levy, K. Heavy Rainfall Events and Diarrhea Incidence: The Role of Social and Environmental Factors. Am. J. Epidemiol. 2014, 179, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, K.; Yoshino, H.; Yanagi, U.; Azuma, K.; Osawa, H.; Kagi, N.; Shinohara, N.; Hasegawa, A. Indoor Environmental Problems and Health Status in Water-Damaged Homes Due to Tsunami Disaster in Japan. Build. Environ. 2015, 93, 24–34. [Google Scholar] [CrossRef]
- Masciopinto, C.; De Giglio, O.; Scrascia, M.; Fortunato, F.; La Rosa, G.; Suffredini, E.; Pazzani, C.; Prato, R.; Montagna, M.T. Human Health Risk Assessment for the Occurrence of Enteric Viruses in Drinking Water from Wells: Role of Flood Runoff Injections. Sci. Total Environ. 2019, 666, 559–571. [Google Scholar] [CrossRef]
- Taylor, J.; Davies, M.; Canales, M.; Lai, K.-M. The Persistence of Flood-Borne Pathogens on Building Surfaces under Drying Conditions. Int. J. Hyg. Environ. Health 2013, 216, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Bergholz, P.W.; Strawn, L.K.; Ryan, G.T.; Warchocki, S.; Wiedmann, M. Spatiotemporal Analysis of Microbiological Contamination in New York State Produce Fields Following Extensive Flooding from Hurricane Irene, August 2011. J. Food Prot. 2016, 79, 384–391. [Google Scholar] [CrossRef]
- Cann, K.F.; Thomas, D.R.; Salmon, R.L.; Wyn-Jones, A.P.; Kay, D. Extreme Water-Related Weather Events and Waterborne Disease. Epidemiol. Infect. 2013, 141, 671–686. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Wright, H.; Harris, P.N.A. Health Risks of Flood Disasters. Clin. Infect. Dis. 2018, 67, 1450–1454. [Google Scholar] [CrossRef] [PubMed]
- Anh, Tran Nguyen Quynh. ‘Characterization of Domestic Wastewater Discharge and Its Impact on Material Flows in Urban Hue, Vietnam’, 191. 2016. Available online: https://repository.kulib.kyoto-u.ac.jp/dspace/bitstream/2433/217214/2/dtikk00155.pdf (accessed on 27 May 2021).
- Tran, P.; Marincioni, F.; Shaw, R.; Sarti, M.; Van An, L. Flood Risk Management in Central Viet Nam: Challenges and Potentials. Nat. Hazards 2008, 46, 119–138. [Google Scholar] [CrossRef]
- Pandey, P.K.; Kass, P.H.; Soupir, M.L.; Biswas, S.; Singh, V.P. Contamination of Water Resources by Pathogenic Bacteria. AMB Expr. 2014, 4, 51. [Google Scholar] [CrossRef]
- Blyton, M.D.J.; Gordon, D.M. Genetic Attributes of E. coli Isolates from Chlorinated Drinking Water. PLoS ONE 2017, 12, e0169445. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Denamur, E.; Clermont, O.; Bonacorsi, S.; Gordon, D. The Population Genetics of Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2021, 19, 37–54. [Google Scholar] [CrossRef]
- Yu, F.; Chen, X.; Zheng, S.; Han, D.; Wang, Y.; Wang, R.; Wang, B.; Chen, Y. Prevalence and Genetic Diversity of Human Diarrheagenic Escherichia coli Isolates by Multilocus Sequence Typing. Int. J. Infect. Dis. 2018, 67, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Tram, N.T.; Dalsgaard, A. Water Used to Moisten Vegetables Is a Source of Escherichia Coli and Protozoan Parasite Contamination at Markets in Hanoi, Vietnam. J. Water Health 2014, 12, 896–900. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.Q.C.; Ho, T.T.; Nguyen, V.C.; Pham, H.S.H.; Vu, V.H.; Le, V.A.; Fujieda, A.; Ueru, T.; Akamatsu, M. Microbial and Parasitic Contamination on Fresh Vegetables Sold in Traditional Markets in Hue City, Vietnam. J. Food Nutr. 2014, 2, 959–964. [Google Scholar] [CrossRef][Green Version]
- Ngo, H.M.; Liu, R.; Moritaka, M.; Fukuda, S. Urban Consumer Trust in Safe Vegetables in Vietnam: The Role of Brand Trust and the Impact of Consumer Worry about Vegetable Safety. Food Control. 2020, 108, 106856. [Google Scholar] [CrossRef]
- Nguyen, T.K.; Bui, H.T.; Truong, T.A.; Lam, D.N.; Ikeuchi, S.; Ly, L.K.T.; Hara-Kudo, Y.; Taniguchi, T.; Hayashidani, H. Retail Fresh Vegetables as a Potential Source of Salmonella Infection in the Mekong Delta, Vietnam. Int. J. Food Microbiol. 2021, 341, 109049. [Google Scholar] [CrossRef]
- Statistics Office of Hue City (SOH). Statistical Yearbook; Statistical Publishing House: Hue City, Vietnam, 2019. [Google Scholar]
- Antony, A.; Paul, M.; Silvester, R.; Aneesa, P.A.; Suresh, K.; Divya, P.S.; Paul, S.; Fathima, P.A.; Abdulla, M. Comparative Evaluation of EMB Agar and Hicrome, E. coli Agar for Differentiation of Green Metallic Sheen Producing Non, E. coli and Typical, E. Coli Colonies from Food and Environmental Samples. J. Pure. Appl. Microbio. 2016, 10, 2863–2870. [Google Scholar] [CrossRef]
- Molina, F.; López-Acedo, E.; Tabla, R.; Roa, I.; Gómez, A.; Rebollo, J.E. Improved detection of Escherichia coli and coliform bacteria by multiplex PCR. BMC Biotechnol. 2015, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and Virulence in Escherichia coli: An Evolutionary Perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Maiden, M.C. Multilocus Sequence Typing of Bacteria. Annu. Rev. Microbiol. 2006, 60, 561–588. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355. [Google Scholar] [CrossRef] [PubMed]
- Enright, M.C.; Spratt, B.G. Multilocus Sequence Typing. Trends Microbiol. 1999, 7, 6. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Mohamed, K.; Fan, Y.; the Agama Study Group; Achtman, M. The EnteroBase User’s Guide, with Case Studies on Salmonella Transmissions, Yersinia Pestis Phylogeny, and Escherichia Core Genomic Diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of Core Genomic Relationships among 100,000 Bacterial Pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Ministry of Natural Resources and Environment. National technical regulation on surface water quality 2015 (QCVN 08-MT:2015/BTNMT). Available online: http://cem.gov.vn/storage/documents/5d6f3ecb26484qcvn-08-mt2015btnmt.pdf (accessed on 22 August 2019).
- Ministry of Health. National technical Regulation on Microbiological Contamination in Food 2012 (QCVN 8-3: 2012/BYT). Available online: http://www.fsi.org.vn/pic/files/qcvn-8-3_2011-byt-to-many-micro-vat-intp_bia_merged.pdf (accessed on 22 August 2019). (In Vietnamese).
- US-FDA. Code of Federal Regulations Title 21, 2020, Volume 2 (21CFR112.55). Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfCFR/CFRSearch.cfm?fr=112.55 (accessed on 10 January 2021).
- Lyautey, E.; Lu, Z.; Lapen, D.R.; Wilkes, G.; Scott, A.; Berkers, T.; Edge, T.A.; Topp, E. Distribution and Diversity of Escherichia coli Populations in the South Nation River Drainage Basin, Eastern Ontario, Canada. Appl. Environ. Microbiol. 2010, 76, 1486–1496. [Google Scholar] [CrossRef]
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the Environment. Microbiol. Mol. Biol. Rev. 2012, 76, 685–706. [Google Scholar] [CrossRef] [PubMed]
- Iwu, C.D.; Okoh, A.I. Preharvest Transmission routes of Fresh Produce Associated Bacterial Pathogens with Outbreak Potentials: A review. Int. J. Environ. Res. Public Health 2019, 16, 4407. [Google Scholar] [CrossRef] [PubMed]
- Mu, D.; Luo, P.; Lyu, J.; Zhou, M.; Huo, A.; Duan, W.; Nover, D.; He, B.; Zhao, X. Impact of Temporal Rainfall Patterns on Flash Floods in Hue City, Vietnam. J. Flood Risk Manag. 2021, 14. [Google Scholar] [CrossRef]
- Allard, S.M.; Callahan, M.T.; Bui, A.; Ferelli, A.M.C.; Chopyk, J.; Chattopadhyay, S.; Mongodin, E.F.; Micallef, S.A.; Sapkota, A.R. Creek to Table: Tracking Fecal Indicator Bacteria, Bacterial Pathogens, and Total Bacterial Communities from Irrigation Water to Kale and Radish Crops. Sci. Total Environ. 2019, 666, 461–471. [Google Scholar] [CrossRef]
- Weller, D.; Wiedmann, M.; Strawn, L.K. Spatial and Temporal Factors Associated with an Increased Prevalence of Listeria monocytogenes in Spinach Fields in New York State. Appl. Environ. Microbiol. 2015, 81, 6059–6069. [Google Scholar] [CrossRef]
- Chandran, A.; Mazumder, A. Pathogenic Potential, Genetic Diversity, and Population Structure of Escherichia Coli Strains Isolated from a Forest-Dominated Watershed (Comox Lake) in British Columbia, Canada. Appl. Environ. Microbiol. 2015, 81, 1788–1798. [Google Scholar] [CrossRef]
- Aibinu, I.; Odugbemi, T.; Koenig, W.; Ghebremedhin, B. Sequence Type ST131 and ST10 Complex (ST617) Predominant among CTX-M-15-Producing Escherichia coli Isolates from Nigeria* *This Study Has Been Partially Presented during the 51st Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC) in Chicago, IL, September 2011. Clin. Microbiol. Infect. 2012, 18, E49–E51. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chattaway, M.A.; Jenkins, C.; Rajendram, D.; Cravioto, A.; Talukder, K.A.; Dallman, T.; Underwood, A.; Platt, S.; Okeke, I.N.; Wain, J. Enteroaggregative Escherichia coli Have Evolved Independently as Distinct Complexes within the E. coli Population with Varying Ability to Cause Disease. PLoS ONE 2014, 9, e112967. [Google Scholar] [CrossRef]
- Manges, A.R. Escherichia coli and Urinary Tract Infections: The Role of Poultry-Meat. Clinical Microbiology and Infection 2016, 22, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Okeke, I.N.; Wallace-Gadsden, F.; Simons, H.R.; Matthews, N.; Labar, A.S.; Hwang, J.; Wain, J. Multi-Locus Sequence Typing of Enteroaggregative Escherichia coli Isolates from Nigerian Children Uncovers Multiple Lineages. PLoS ONE 2010, 5, e14093. [Google Scholar] [CrossRef]
- Nadimpalli, M.L.; Marks, S.J.; Montealegre, M.C.; Gilman, R.H.; Pajuelo, M.J.; Saito, M.; Tsukayama, P.; Njenga, S.M.; Kiiru, J.; Swarthout, J.; et al. Urban Informal Settlements as Hotspots of Antimicrobial Resistance and the Need to Curb Environmental Transmission. Nat Microbiol 2020, 5, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Shiraz, S.; Djebbi-Simmons, D.; Alhejaili, M.; Danos, K.; Janes, M.; Fontenot, K.; Xu, W. Evaluation of the Microbial Safety and Quality of Louisiana Strawberries after Flooding. Food Control. 2020, 110, 106970. [Google Scholar] [CrossRef]
- Unger, I.M.; Kennedy, A.C.; Muzika, R.-M. Flooding Effects on Soil Microbial Communities. Appl. Soil Ecol. 2009, 42, 1–8. [Google Scholar] [CrossRef]

| Target Gene | Primer | Sequence | Product Size | Annealing Temperature |
|---|---|---|---|---|
| adk | adk-F | TCATCATCTGCACTTTCCGC | 766 bp | 54 °C |
| adk-R | CCAGATCAGCGCGAACTTCA | |||
| fumC | fumC-F | TCACAGGTCGCCAGCGCTTC | 806 bp | 65 °C |
| fumC-R | GTACGCAGCGAAAAAGATTC | |||
| gyrB | gyrB-F | TCGGCGACACGGATGACGGC | 815 bp | 60 °C |
| gyrB-R | GTCCATGTAGGCGTTCAGGG | |||
| icd | icd-F | ATGGAAAGTAAAGTAGTTGTTCCGGCACA | 878 bp | 62 °C |
| icd-R | GGACGCAGCAGGATCTGTT | |||
| mdh | mdh-F | AGCGCGTTCTGTTCAAATGC | 799 bp | 56 °C |
| mdh-R | CAGGTTCAGAACTCTCTCTGT | |||
| purA | purA-F | TCGGTAACGGTGTTGTGCTG | 845 bp | 58 °C |
| purA-R | CATACGGTAAGCCACGCAGA | |||
| recA | recA-F | CGCATTCGCTTTACCCTGACC | 734 bp | 58 °C |
| recA-R | TCGTCGAAATCTACGGACCGGA |
| Sample Type | Number of E. coli Isolates | |
|---|---|---|
| Dry Season | Wet Season | |
| Manure (MNR) | - | 18 |
| Soil (SOL) | 16 | 11 |
| Irrigation water (IRW) | 40 | 39 |
| Wastewater (MWW) | 13 | 19 |
| Vegetable (VEG) | 21 | ND |
| Total | 90 | 87 |
| Season | Cluster | ST | ST Complex | Pathotype a | Phylogroup a (Clermont) | Phylogroup a (EzClermont) | Sample |
|---|---|---|---|---|---|---|---|
| Dry | 1 | 10 | STc10 | UPEC, STEC, VTEC, NMEC, ExPEC, ETEC, EPEC, EAEC, APEC, Non-pathogen | E or clade I, C, A | U, cryptic, C, A | MWW |
| 2 | 409 | None | Pathogen (Unknown) | B1 and A | A | MWW, IRW | |
| 3 | 181 | STc168 | Unknown, Non-pathogen | A | A | VEG | |
| 4 | 161 | None | Pathogen | A | A | IRW | |
| 5 | 10,865 | None | NA | IRW | |||
| 6 | 10,867 | None | NA | VEG | |||
| 7 | 196 | None | UPEC, Non-pathogen | B1 | B1 and A | SOL | |
| 10,678 | None | NA | SOL | ||||
| 8 | 10,688 | None | NA | IRW | |||
| Wet | 1 | 1148 | None | Pathogen, Non-pathogen | B1 | B1 | SOL |
| 2 | 1056 | None | NA | B1 | B1 | IRW | |
| 3 | 10,012 | None | NA | MNR | |||
| 4 | 200 | STc40 | VTEC, EHEC, EAEC, Non-pathogen | B1 | B1 | MWW | |
| 40 | STEC, EPEC, EAEC, Non-pathogen | B1 | B1 | IRW | |||
| 5 | 10,023 | None | NA | IRW | |||
| 10,027 | None | NA | IRW | ||||
| 6 | 641 | STc86 | ExPEC, Non-pathogen | D, B1, A | U, D, B1, A | MNR | |
| 7 | 155 | STc155 | UPEC, STEC, ExPEC, ETEC, EAEC, APEC, Non-pathogen | B1, A | U, B1, A | IRW | |
| 8 | 10,008 | None | NA | MNR | |||
| 9 | 181 | STc168 | Non-pathogen | A | A | IRW | |
| 10 | 93 | 168 | UPEC, ExPEC, ETEC, DAEC, APEC, Non-pathogen | E, D, A | D, A | IRW, MWW, MNR | |
| 9872 | None | NA | |||||
| 11 | 48 | 10 | UPEC, ExPEC, EAEC, APEC, Non-pathogen | E, A | Cryptic, E, A | MNR, MWW, IRW, SOL | |
| 10,032 | None | NA | No Database | ||||
| 6856 | 10 | NA | A | A | |||
| 10,019 | None | NA | |||||
| 12 | 10,033 | None | NA | IRW | |||
| Phylogroup | |||||||
| Summary | Sample | A | B1 | B2 | E | ||
| MNR | 3 | 4 | |||||
| VEG | 5 | 3 | |||||
| SOL | 1 | 9 | |||||
| IRW | 18 | 15 | |||||
| MWW | 14 | 3 | 1 | 1 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prayoga, W.; Nishiyama, M.; Praise, S.; Pham, D.V.; Duong, H.V.; Pham, L.K.; Dang, L.T.T.; Watanabe, T. Tracking Fecal Bacterial Dispersion from Municipal Wastewater to Peri-Urban Farms during Monsoon Rains in Hue City, Vietnam. Int. J. Environ. Res. Public Health 2021, 18, 9580. https://doi.org/10.3390/ijerph18189580
Prayoga W, Nishiyama M, Praise S, Pham DV, Duong HV, Pham LK, Dang LTT, Watanabe T. Tracking Fecal Bacterial Dispersion from Municipal Wastewater to Peri-Urban Farms during Monsoon Rains in Hue City, Vietnam. International Journal of Environmental Research and Public Health. 2021; 18(18):9580. https://doi.org/10.3390/ijerph18189580
Chicago/Turabian StylePrayoga, Windra, Masateru Nishiyama, Susan Praise, Dung Viet Pham, Hieu Van Duong, Lieu Khac Pham, Loc Thi Thanh Dang, and Toru Watanabe. 2021. "Tracking Fecal Bacterial Dispersion from Municipal Wastewater to Peri-Urban Farms during Monsoon Rains in Hue City, Vietnam" International Journal of Environmental Research and Public Health 18, no. 18: 9580. https://doi.org/10.3390/ijerph18189580
APA StylePrayoga, W., Nishiyama, M., Praise, S., Pham, D. V., Duong, H. V., Pham, L. K., Dang, L. T. T., & Watanabe, T. (2021). Tracking Fecal Bacterial Dispersion from Municipal Wastewater to Peri-Urban Farms during Monsoon Rains in Hue City, Vietnam. International Journal of Environmental Research and Public Health, 18(18), 9580. https://doi.org/10.3390/ijerph18189580







