Mercury and Antibiotic Resistance Co-Selection in Bacillus sp. Isolates from the Almadén Mining District
Abstract
:1. Introduction
2. Materials and Methods
2.1. Obtaining Mercury-Tolerant Strains
2.2. Molecular Identification of Microorganisms
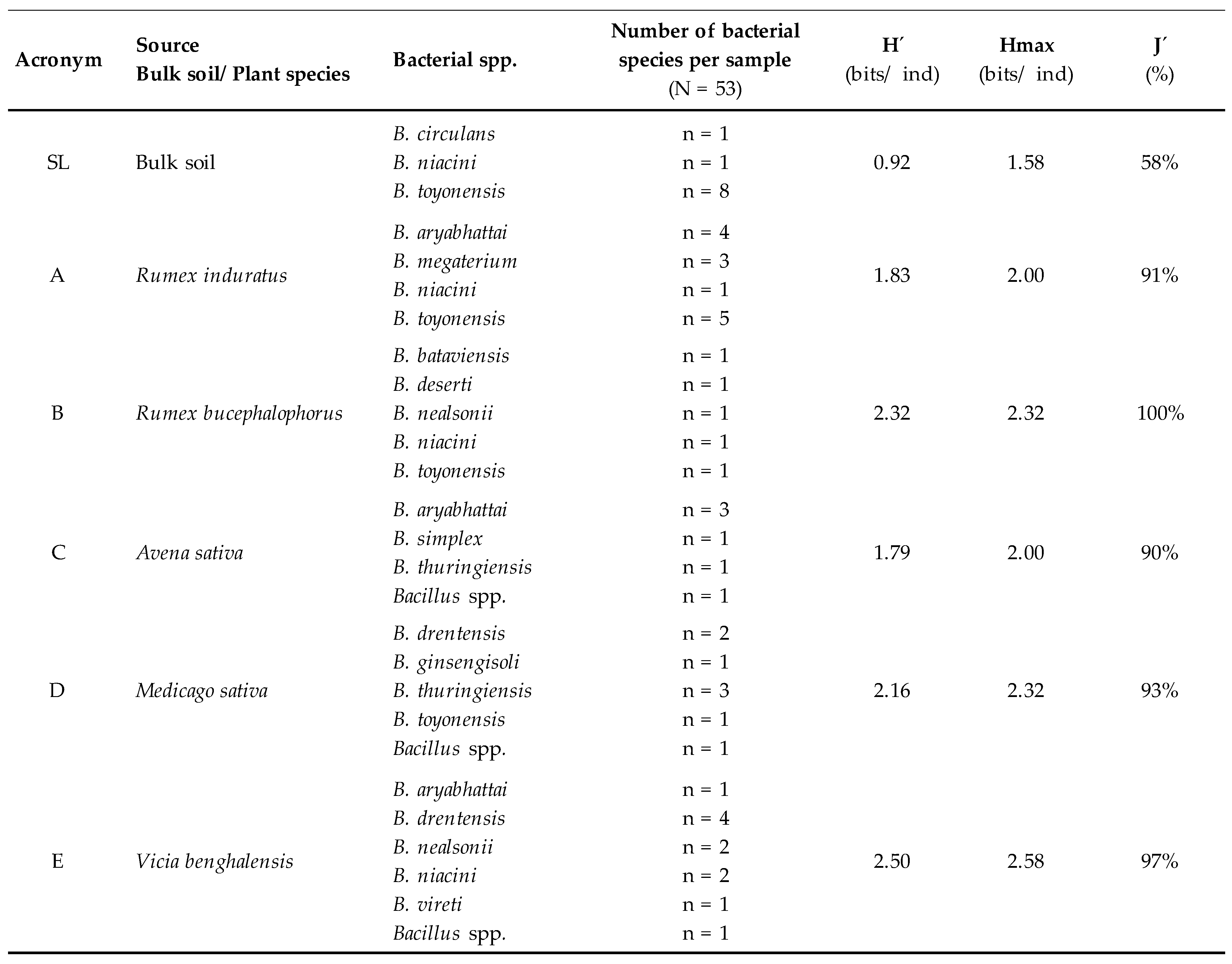
2.3. Microbial Diversity
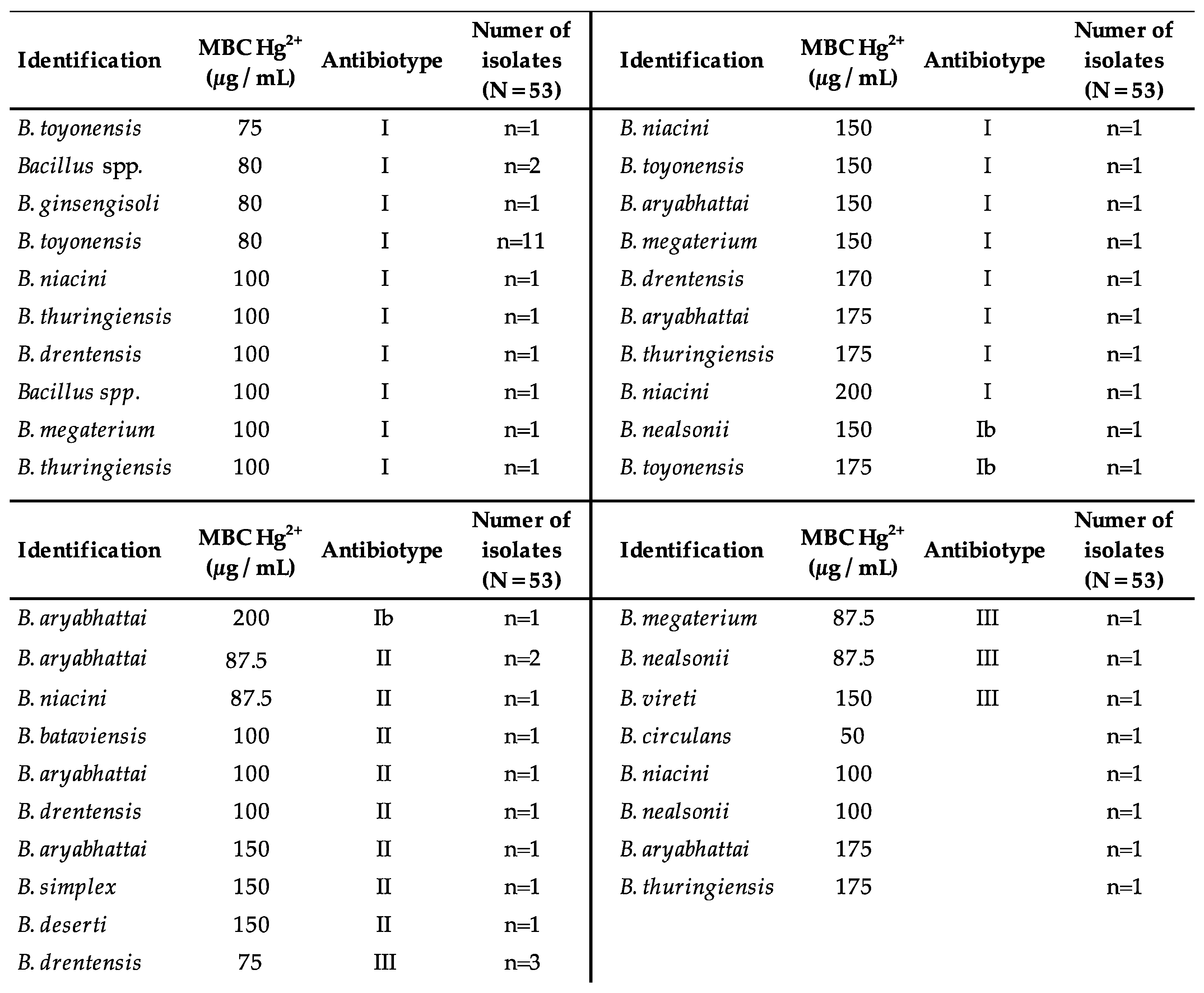
2.4. Hg Minimum Bactericidal Concentration (MBC)
2.5. Antibiogram and Antibiotic Minimum Inhibitory Concentration (MIC)
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
- Most of the mercurotolerant isolates of the Bacillus spp. genus are multi-resistant. Only antibiotypes II (n = 9) and III (n = 6), poorly represented, presented a resistance profile similar to the wild-type phenotype.
- Of the 53 strains tested, 38 expressed a phenotype of resistance to cephalosporins. This finding suggests that the incidence of bacterial resistance to these antibiotics in Hg-contaminated soils is higher than estimated in previous works.
- The correlation between the Hg minimum bactericidal concentration (CMB) and the CMI of cefotaxime, ceftazidime, and tetracycline MIC was significant (p-value ≤ 0.05). This finding statistically demonstrates the co-selection of bacterial resistance to cephalosporins and tetracyclines under Hg-selective environmental pressure.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Canteón, R. Antibiotic resistance genes from the environment: A perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clin. Microbiol. Infect. 2009, 15, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Martinez, J.L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef]
- Wright, G.D. Antibiotic resistance in the environment: A link to the clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Buergmann, H.; Sørum, H.; Norström, M.; Pons, M.-N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Martinez, J.L. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. R. Soc. B Boil. Sci. 2009, 276, 2521–2530. [Google Scholar] [CrossRef] [Green Version]
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the Antibiotic Resistome. Science 2006, 311, 374–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization (WHO). Global Action Plan on Antimicrobial Resistance. 2015. Available online: https://apps.who.int/iris/handle/10665/193736 (accessed on 10 July 2021).
- Chen, J.; Li, J.; Zhang, H.; Shi, W.; Liu, Y. Bacterial Heavy-Metal and Antibiotic Resistance Genes in a Copper Tailing Dam Area in Northern China. Front. Microbiol. 2019, 10, 1916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldi, F.; Marchetto, D.; Gallo, M.; Fani, R.; Maida, I.; Covelli, S.; Fajon, V.; Žižek, S.; Hines, M.; Horvat, M. Chlor-alkali plant contamination of Aussa River sediments induced a large Hg-resistant bacterial community. Estuar. Coast. Shelf Sci. 2012, 113, 96–104. [Google Scholar] [CrossRef]
- Ballabio, C.; Jiskra, M.; Osterwalder, S.; Borrelli, P.; Montanarella, L.; Panagos, P. A spatial assessment of mercury content in the European Union topsoil. Sci. Total Environ. 2021, 769, 144755. [Google Scholar] [CrossRef] [PubMed]
- Wohlgemuth, L.; Osterwalder, S.; Joseph, C.; Kahmen, A.; Hoch, G.; Alewell, C.; Jiskra, M. A bottom-up quantification of foliar mercury uptake fluxes across Europe. Biogeosciences 2020, 17, 6441–6456. [Google Scholar] [CrossRef]
- Gil-Hernández, F.; Gómez-Fernández, A.R.; La Torre-Aguilar, M.J.; Pérez-Navero, J.L.; Flores-Rojas, K.; Martín-Borreguero, P.; Gil-Campos, M. Neurotoxicity by mercury is not associated with autism spectrum disorders in Spanish children. Ital. J. Pediatr. 2020, 46, 1–7. [Google Scholar] [CrossRef]
- Das, S.; Dash, H.R.; Chakraborty, J. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl. Microbiol. Biotechnol. 2016, 100, 2967–2984. [Google Scholar] [CrossRef]
- United Nations Environment Assembly. Minamata Convention on Mercury. Collection, UN Treaty. 2013. Available online: http://www.mercuryconvention.org/Convention/Text (accessed on 10 July 2021).
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef]
- Huerta, B.; Marti, E.; Gros, M.; López, P.; Pompêo, M.; Armengol, J.; Barceló, D.; Balcazar, J.L.; Rodriguez-Mozaz, S.; Marcé, R. Exploring the links between antibiotic occurrence, antibiotic resistance, and bacterial communities in water supply reservoirs. Sci. Total Environ. 2013, 456, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Knapp, C.W.; Callan, A.; Aitken, B.; Shearn, R.; Koenders, A.; Hinwood, A. Relationship between antibiotic resistance genes and metals in residential soil samples from Western Australia. Environ. Sci. Pollut. Res. 2017, 24, 2484–2494. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Li, L.; Ashbolt, N.; Wang, X.; Cui, Y.; Zhu, X.; Xu, Y.; Yang, Y.; Mao, D.; Luo, Y. Arctic antibiotic resistance gene contamination, a result of anthropogenic activities and natural origin. Sci. Total Environ. 2018, 621, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Timoney, J.F.; Port, J.; Giles, J.; Spanier, J. Heavy-Metal and Antibiotic Resistance in the Bacterial Flora of Sediments of New York Bight. Appl. Environ. Microbiol. 1978, 36, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 2012, 3, 399. [Google Scholar] [CrossRef] [Green Version]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Nemergut, D.R.; Martin, A.P.; Schmidt, S.K. Integron Diversity in Heavy-Metal-Contaminated Mine Tailings and Inferences about Integron Evolution. Appl. Environ. Microbiol. 2004, 70, 1160–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanauskas, R.; Glenn, T.; Jagoe, C.H.; Tuckfield, R.C.; Lindell, A.H.; King, C.J.; McArthur, J.V. Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ. Microbiol. 2006, 8, 1510–1514. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wang, J.; Li, J.; Li, J.; Ma, Y.; Chen, D.; He, J. Field-based evidence for copper contamination induced changes of antibiotic resistance in agricultural soils. Environ. Microbiol. 2016, 18, 3896–3909. [Google Scholar] [CrossRef]
- Di Cesare, A.; Eckert, E.; D’Urso, S.; Bertoni, R.; Gillan, D.C.; Wattiez, R.; Corno, G. Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Res. 2016, 94, 208–214. [Google Scholar] [CrossRef]
- Wright, M.S.; Peltier, G.L.; Stepanauskas, R.; McArthur, J.V. Bacterial tolerances to metals and antibiotics in metal-contaminated and reference streams. FEMS Microbiol. Ecol. 2006, 58, 293–302. [Google Scholar] [CrossRef]
- Monier, J.-M.; Demaneche, S.; Delmont, T.; Mathieu, A.; Vogel, T.; Simonet, P. Metagenomic exploration of antibiotic resistance in soil. Curr. Opin. Microbiol. 2011, 14, 229–235. [Google Scholar] [CrossRef]
- Nesme, J.; Simonet, P. The soil resistome: A critical review on antibiotic resistance origins, ecology and dissemination potential in telluric bacteria. Environ. Microbiol. 2015, 17, 913–930. [Google Scholar] [CrossRef]
- Wright, G.D. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat. Rev. Genet. 2007, 5, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.; Reyes, A.; Wang, B.; Selleck, E.; Sommer, M.O.A.; Dantas, G. The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Baquerizo, M.; Guerra, C.A.; Cano-Díaz, C.; Egidi, E.; Wang, J.-T.; Eisenhauer, N.; Singh, B.K.; Maestre, F.T. The proportion of soil-borne pathogens increases with warming at the global scale. Nat. Clim. Chang. 2020, 10, 550–554. [Google Scholar] [CrossRef]
- Domínguez-Begines, J.; Ávila, J.M.; García, L.V.; Gómez-Aparicio, L. Soil-borne pathogens as determinants of regeneration patterns at community level in Mediterranean forests. New Phytol. 2020, 227, 588–600. [Google Scholar] [CrossRef]
- Millán, R.; Carpena, R.O.; Schmid, T.; Sierra, M.J.; Moreno, E.; Peñalosa, J.; Gamarra, R.; Esteban, E. Rehabilitación de suelos contaminados con mercurio: Estrategias aplicables en el área de Almadén. Ecosistemas 2007, 16, 56–66. [Google Scholar]
- Velasco, A.G.-V.; Probanza, A.; Gutierrez-Mañero, F.J.; Ramos, B.; Garcia, J.A.L. Functional diversity of rhizosphere microorganisms from different genotypes of Arabidopsis thaliana. Community Ecol. 2009, 10, 111–119. [Google Scholar] [CrossRef]
- Mathema, V.B.; Thakuri, B.K.C.; Sillanpää, M.; Shrestha, R.A. Study of mercury (II) tolerant bacterial isolates from Baghmati River with estimation of plasmid size and growth variation for the high mercury (II) resistant Enterobacter spp. J. Biotechnol. Res. 2011, 3, 72–77. [Google Scholar]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; The University of Illinois Press: Urbana, IL, USA, 1949. [Google Scholar]
- Pielou, E.C. An Introduction to Mathematical Ecology; Wiley: Hoboken, NJ, USA, 1969; 285p. [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria: Proposed Guideline M45-P, 3rd ed.; CLSI: Wayne, PA, USA, 2016. [Google Scholar]
- Schlegelová, J.; Brychta, J.; Klimova, E.; Nápravníková, E.; Babak, V. The prevalence of and resistance to antimicrobial agents of Bacillus cereus isolates from foodstuffs. Veterinární Med. 2012, 48, 331–338. [Google Scholar] [CrossRef] [Green Version]
- Narita, M.; Chiba, K.; Nishizawa, H.; Ishii, H.; Huang, C.-C.; Kawabata, Z.; Silver, S.; Endo, G. Diversity of mercury resistance determinants among Bacilluss trains isolated from sediment of Minamata Bay. FEMS Microbiol. Lett. 2003, 223, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Garbeva, P.; Van Veen, J.; Van Elsas, J. Predominant Bacillus spp. in Agricultural Soil under Different Management Regimes Detected via PCR-DGGE. Microb. Ecol. 2003, 45, 302–316. [Google Scholar] [CrossRef]
- Bartelt-Hunt, S.; Snow, D.; Damon-Powell, T.; Miesbach, D. Occurrence of steroid hormones and antibiotics in shallow groundwater impacted by livestock waste control facilities. J. Contam. Hydrol. 2011, 123, 94–103. [Google Scholar] [CrossRef]
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243. [Google Scholar] [CrossRef]
- Benveniste, R.; Davies, J. Aminoglycoside Antibiotic-Inactivating Enzymes in Actinomycetes Similar to Those Present in Clinical Isolates of Antibiotic-Resistant Bacteria. Proc. Natl. Acad. Sci. USA 1973, 70, 2276–2280. [Google Scholar] [CrossRef] [Green Version]
- D’Costa, V.M.; Griffiths, E.; Wright, G. Expanding the soil antibiotic resistome: Exploring environmental diversity. Curr. Opin. Microbiol. 2007, 10, 481–489. [Google Scholar] [CrossRef]
- Witte, W. Ecological impact of antibiotic use in animals on different complex microflora: Environment. Int. J. Antimicrob. Agents 2000, 14, 321–325. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing: Sixteenth Informational Supplement M100-S16; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters, version 11.0. 2021.
- Bucher, T.; Keren-Paz, A.; Hausser, J.; Olender, T.; Cytryn, E.; Kolodkin-Gal, I. An active β-lactamase is a part of an orchestrated cell wall stress resistance network of Bacillus subtilis and related rhizosphere species. Environ. Microbiol. 2019, 21, 1068–1085. [Google Scholar] [CrossRef]
- Kong, X.-X.; Jiang, J.-L.; Qiao, B.; Liu, H.; Cheng, J.-S.; Yuan, Y.-J. The biodegradation of cefuroxime, cefotaxime and cefpirome by the synthetic consortium with probiotic Bacillus clausii and investigation of their potential biodegradation pathways. Sci. Total Environ. 2019, 651, 271–280. [Google Scholar] [CrossRef]
- Abdu, N.; Abdullahi, A.A.; Abdulkadir, A. Heavy metals and soil microbes. Environ. Chem. Lett. 2017, 15, 65–84. [Google Scholar] [CrossRef]
- Borymski, S.; Cycoń, M.; Beckmann, M.; Mur, L.; Piotrowska-Seget, Z. Plant Species and Heavy Metals Affect Biodiversity of Microbial Communities Associated with Metal-Tolerant Plants in Metalliferous Soils. Front. Microbiol. 2018, 9, 1425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chelliah, R.; Wei, S.; Park, B.-J.; Park, J.-H.; Park, Y.-S.; Kim, S.-H.; Jin, Y.-G.; Oh, D.-H. New perspectives on Mega plasmid sequence (poh1) in Bacillus thuringiensis ATCC 10792 harbouring antimicrobial, insecticidal and antibiotic resistance genes. Microb. Pathog. 2019, 126, 14–18. [Google Scholar] [CrossRef]
- Roberts, M.C. Epidemiology of tetracycline-resistance determinants. Trends Microbiol. 1994, 2, 353–357. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michalova, E.; Novotna, P.; Schlegelová, J. Tetracyclines in veterinary medicine and bacterial resistance to them. Veterinární Med. 2012, 49, 79–100. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, L.D.; Sørensen, S. The Effect of Longterm Exposure to Mercury on the Bacterial Community in Marine Sediment. Curr. Microbiol. 1998, 36, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J. Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol. Rev. 1983, 47, 361–409. [Google Scholar] [CrossRef] [PubMed]
- Summers, A.; Wireman, J.; Vimy, M.J.; Lorscheider, F.L.; Marshall, B.; Levy, S.B.; Bennett, S.; Billard, L. Mercury released from dental “silver” fillings provokes an increase in mercury- and antibiotic-resistant bacteria in oral and intestinal floras of primates. Antimicrob. Agents Chemother. 1993, 37, 825–834. [Google Scholar] [CrossRef] [Green Version]
- Ji, L.; Xie, S.; Feng, J.; Li, Y.; Chen, L. Heavy metal uptake capacities by the common freshwater green alga Cladophora fracta. Environ. Boil. Fishes 2011, 24, 979–983. [Google Scholar] [CrossRef]

| Family | Antibiotic | p-Value |
|---|---|---|
| Betalactams | benzylpenicillin | 0.597 |
| ampicillin | 0.992 | |
| cefotaxime | 0.035 * | |
| ceftriaxone | 0.038 * | |
| Lincosamines | clindamycin | 0.051 |
| Glycopeptides | vancomycin | 0.988 |
| Tetracyclines | tetracycline | 0.001 * |
| Sulfamides | cotrimoxazole | 0.322 |
| Macrolides | erythromycin | 0.289 |
| Quinolones | levofloxacin | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robas, M.; Probanza, A.; González, D.; Jiménez, P.A. Mercury and Antibiotic Resistance Co-Selection in Bacillus sp. Isolates from the Almadén Mining District. Int. J. Environ. Res. Public Health 2021, 18, 8304. https://doi.org/10.3390/ijerph18168304
Robas M, Probanza A, González D, Jiménez PA. Mercury and Antibiotic Resistance Co-Selection in Bacillus sp. Isolates from the Almadén Mining District. International Journal of Environmental Research and Public Health. 2021; 18(16):8304. https://doi.org/10.3390/ijerph18168304
Chicago/Turabian StyleRobas, Marina, Agustín Probanza, Daniel González, and Pedro A. Jiménez. 2021. "Mercury and Antibiotic Resistance Co-Selection in Bacillus sp. Isolates from the Almadén Mining District" International Journal of Environmental Research and Public Health 18, no. 16: 8304. https://doi.org/10.3390/ijerph18168304
APA StyleRobas, M., Probanza, A., González, D., & Jiménez, P. A. (2021). Mercury and Antibiotic Resistance Co-Selection in Bacillus sp. Isolates from the Almadén Mining District. International Journal of Environmental Research and Public Health, 18(16), 8304. https://doi.org/10.3390/ijerph18168304









