Fusion of Higher Order Spectra and Texture Extraction Methods for Automated Stroke Severity Classification with MRI Images
Abstract
1. Introduction
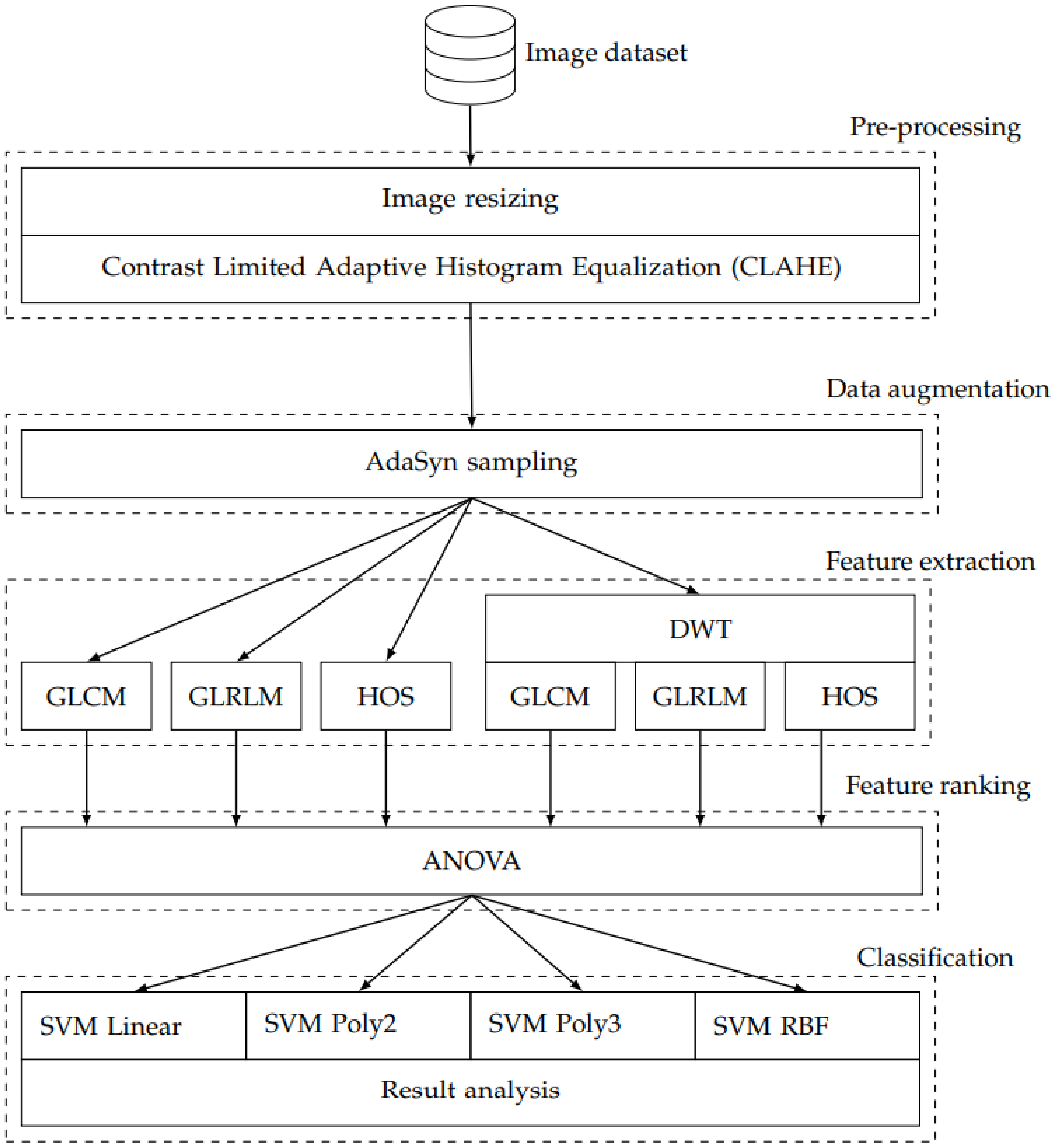
2. Methods
2.1. Image Dataset
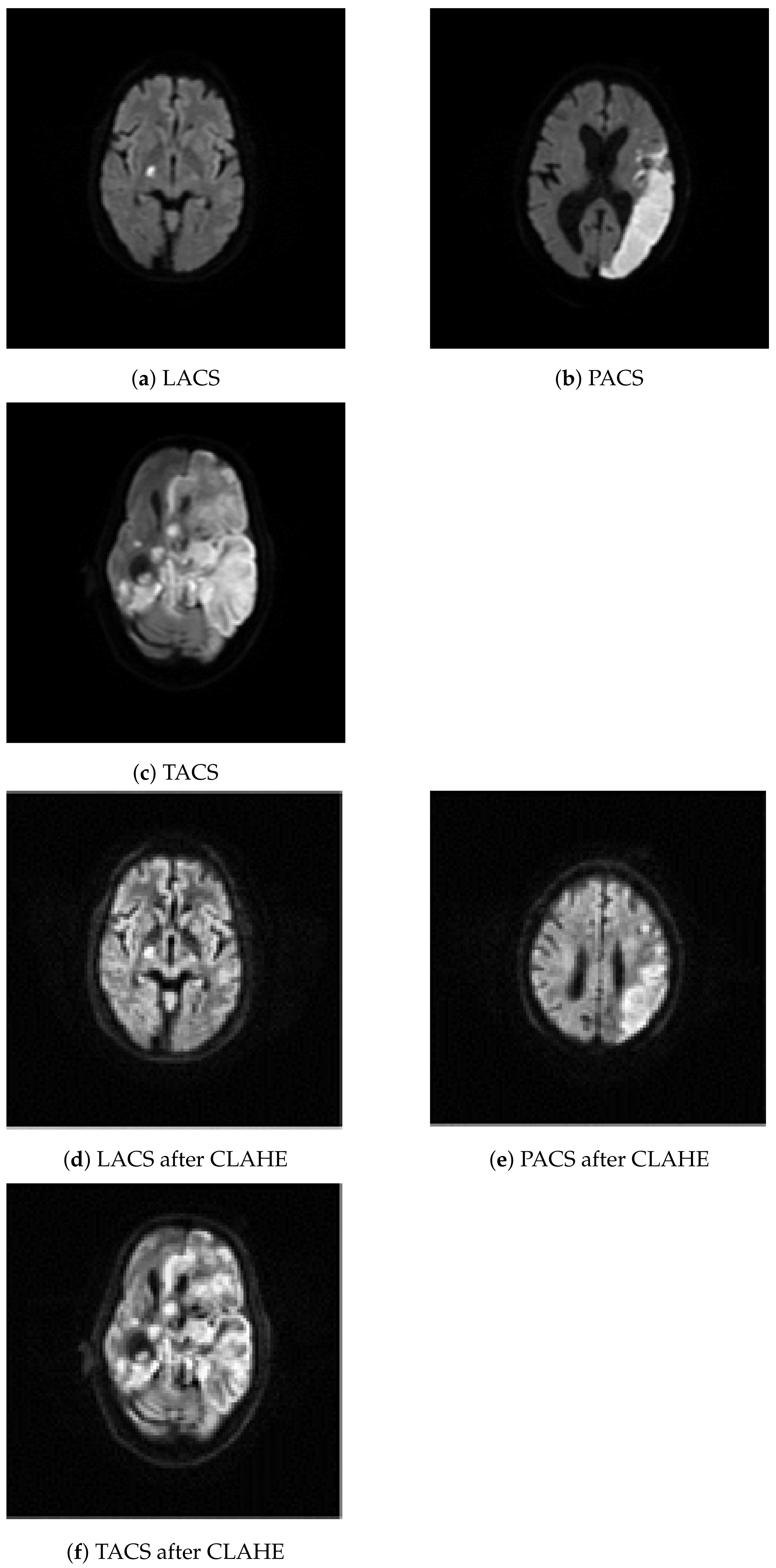
2.2. Image Resizing and Contrast Limited Adaptive Histogram Equalization
2.3. Adaptive Synthetic Sampling
2.4. Feature Extraction
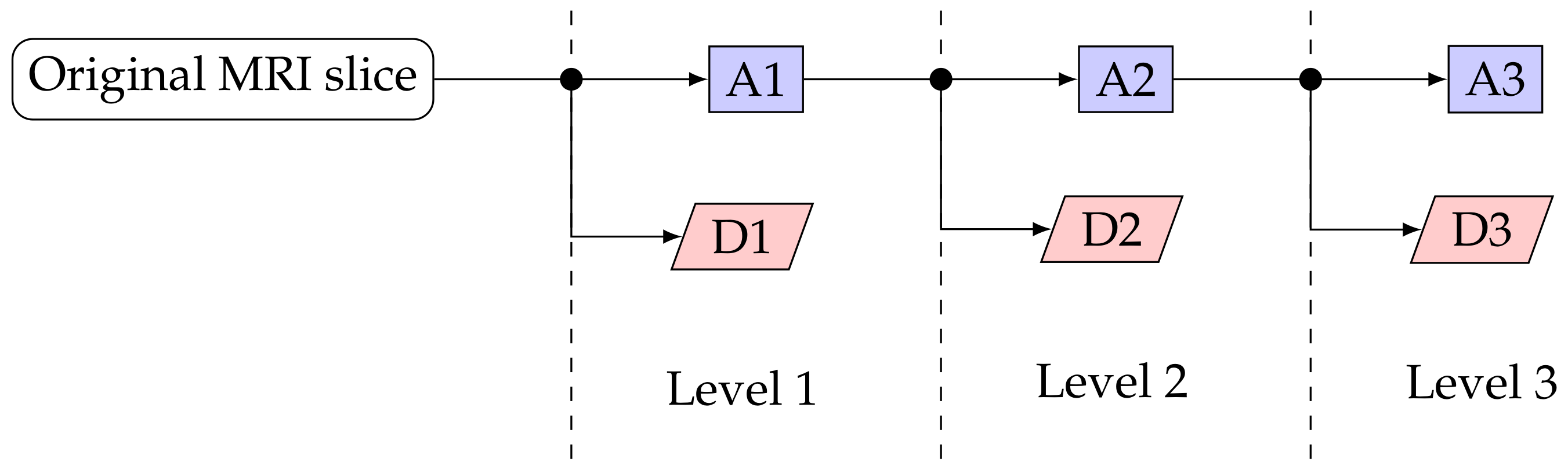
2.4.1. Discrete Wavelet Transform
2.4.2. Gray-Level Co-Occurrence Matrix
2.4.3. Gray-Level Run Length Matrix
2.4.4. Higher Order Spectra
2.5. Statistical Analysis
2.6. Classification
2.6.1. Support Vector Machine Classifier
2.6.2. Performance Measures
3. Results
4. Discussion
Limitations and Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACC | Accuracy |
| AdaSyn | Adaptive Synthetic |
| ANOVA | Analysis Of Variance |
| AI | Artificial Intelligence |
| CAD | Computer-Aided-Diagnosis |
| CLAHE | Contrast Limited Adaptive Histogram Equalization |
| CT | Computed Tomography |
| DL | Deep Learning |
| DWI | Diffusion-Weighted Imaging |
| DWT | Discrete Wavelet Transform |
| FN | False Negative |
| FP | False Positive |
| GLCM | Gray-Level Co-occurrence Matrix |
| GLRLM | Gray-Level Run Length Matrix |
| HOS | Higher Order Spectra |
| LACS | Lacunar Syndrome |
| MRI | Magnetic Resonance Imaging |
| PACS | Partial Anterior Circulation Syndrome |
| PPV | Positive Predictive Value |
| RBF | Radial Basis Function |
| SEN | Sensitivity |
| SPE | Specificity |
| SVM | Support Vector Machine |
| TACS | Total Anterior Circulation Stroke |
| TN | True Negative |
| TP | True Positive |
| VBM | Voxel-Based Morphometry |
References
- Bonita, R. Epidemiology of stroke. Lancet 1992, 339, 342–344. [Google Scholar] [CrossRef]
- Lopez, A.D.; Mathers, C.D.; Ezzati, M.; Jamison, D.T.; Murray, C.J. Global and regional burden of disease and risk factors, 2001: Systematic analysis of population health data. Lancet 2006, 367, 1747–1757. [Google Scholar] [CrossRef]
- Smith, S.C., Jr. Reducing the global burden of ischemic heart disease and stroke: A challenge for the cardiovascular community and the United Nations. Circulation 2011, 124, 278–279. [Google Scholar] [CrossRef]
- Feigin, V.L.; Lawes, C.M.; Bennett, D.A.; Anderson, C.S. Stroke epidemiology: A review of population-based studies of incidence, prevalence, and case-fatality in the late 20th century. Lancet Neurol. 2003, 2, 43–53. [Google Scholar] [CrossRef]
- Dirnagl, U.; Iadecola, C.; Moskowitz, M.A. Pathobiology of ischaemic stroke: An integrated view. Trends Neurosci. 1999, 22, 391–397. [Google Scholar] [CrossRef]
- van der Worp, H.B.; van Gijn, J. Acute ischemic stroke. N. Engl. J. Med. 2007, 357, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Latchaw, R.E.; Alberts, M.J.; Lev, M.H.; Connors, J.J.; Harbaugh, R.E.; Higashida, R.T.; Hobson, R.; Kidwell, C.S.; Koroshetz, W.J.; Mathews, V.; et al. Recommendations for imaging of acute ischemic stroke: A scientific statement from the American Heart Association. Stroke 2009, 40, 3646–3678. [Google Scholar] [CrossRef] [PubMed]
- Bates, E.; Wilson, S.M.; Saygin, A.P.; Dick, F.; Sereno, M.I.; Knight, R.T.; Dronkers, N.F. Voxel-based lesion–symptom mapping. Nat. Neurosci. 2003, 6, 448–450. [Google Scholar] [CrossRef] [PubMed]
- Dronkers, N.F.; Wilkins, D.P.; Van Valin, R.D., Jr.; Redfern, B.B.; Jaeger, J.J. Lesion analysis of the brain areas involved in language comprehension. Cognition 2004, 92, 145–177. [Google Scholar] [CrossRef] [PubMed]
- Fiez, J.A.; Damasio, H.; Grabowski, T.J. Lesion segmentation and manual warping to a reference brain: Intra-and interobserver reliability. Hum. Brain Mapp. 2000, 9, 192–211. [Google Scholar] [CrossRef]
- Lindgren, A.; Norrving, B.; Rudling, O.; Johansson, B.B. Comparison of clinical and neuroradiological findings in first-ever stroke. A population-based study. Stroke 1994, 25, 1371–1377. [Google Scholar] [CrossRef]
- Zaidi, S.F.; Aghaebrahim, A.; Urra, X.; Jumaa, M.A.; Jankowitz, B.; Hammer, M.; Nogueira, R.; Horowitz, M.; Reddy, V.; Jovin, T.G. Final infarct volume is a stronger predictor of outcome than recanalization in patients with proximal middle cerebral artery occlusion treated with endovascular therapy. Stroke 2012, 43, 3238–3244. [Google Scholar] [CrossRef]
- Vogt, G.; Laage, R.; Shuaib, A.; Schneider, A. Initial lesion volume is an independent predictor of clinical stroke outcome at day 90: An analysis of the Virtual International Stroke Trials Archive (VISTA) database. Stroke 2012, 43, 1266–1272. [Google Scholar] [CrossRef]
- Rangaraju, S.; Owada, K.; Noorian, A.R.; Nogueira, R.G.; Nahab, F.; Glenn, B.A.; Belagaje, S.R.; Anderson, A.M.; Frankel, M.R.; Gupta, R. Comparison of final infarct volumes in patients who received endovascular therapy or intravenous thrombolysis for acute intracranial large-vessel occlusions. JAMA Neurol. 2013, 70, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Seghier, M.L.; Ramlackhansingh, A.; Crinion, J.; Leff, A.P.; Price, C.J. Lesion identification using unified segmentation-normalisation models and fuzzy clustering. Neuroimage 2008, 41, 1253–1266. [Google Scholar] [CrossRef]
- Wilke, M.; de Haan, B.; Juenger, H.; Karnath, H.O. Manual, semi-automated, and automated delineation of chronic brain lesions: A comparison of methods. NeuroImage 2011, 56, 2038–2046. [Google Scholar] [CrossRef] [PubMed]
- Meyers, P.M.; Schumacher, H.C.; Connolly, E.S., Jr.; Heyer, E.J.; Gray, W.A.; Higashida, R.T. Current status of endovascular stroke treatment. Circulation 2011, 123, 2591–2601. [Google Scholar] [CrossRef]
- Ashton, E.A.; Takahashi, C.; Berg, M.J.; Goodman, A.; Totterman, S.; Ekholm, S. Accuracy and reproducibility of manual and semiautomated quantification of MS lesions by MRI. J. Magn. Reson. Imaging Off. J. Int. Soc. Magn. Reson. Med. 2003, 17, 300–308. [Google Scholar] [CrossRef]
- Filippi, M.; Horsfield, M.; Bressi, S.; Martinelli, V.; Baratti, C.; Reganati, P.; Campi, A.; Miller, D.; Comi, G. Intra-and inter-observer agreement of brain MRI lesion volume measurements in multiple sclerosis: A comparison of techniques. Brain 1995, 118, 1593–1600. [Google Scholar] [CrossRef]
- Lansberg, M.G.; Albers, G.W.; Beaulieu, C.; Marks, M.P. Comparison of diffusion-weighted MRI and CT in acute stroke. Neurology 2000, 54, 1557–1561. [Google Scholar] [CrossRef] [PubMed]
- Baliyan, V.; Das, C.J.; Sharma, R.; Gupta, A.K. Diffusion weighted imaging: Technique and applications. World J. Radiol. 2016, 8, 785. [Google Scholar] [CrossRef]
- Al-Khaled, M.; Matthis, C.; Münte, T.F.; Eggers, J.; QugSS2-Study. The incidence and clinical predictors of acute infarction in patients with transient ischemic attack using MRI including DWI. Neuroradiology 2013, 55, 157–163. [Google Scholar] [CrossRef]
- Lettau, M.; Laible, M. 3-T high-b-value diffusion-weighted MR imaging in hyperacute ischemic stroke. J. Neuroradiol. 2013, 40, 149–157. [Google Scholar] [CrossRef]
- Lutsep, H.L.; Albers, G.; DeCrespigny, A.; Kamat, G.; Marks, M.; Moseley, M. Clinical utility of diffusion-weighted magnetic resonance imaging in the assessment of ischemic stroke. Ann. Neurol. Off. J. Am. Neurol. Assoc. Child Neurol. Soc. 1997, 41, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Newcombe, V.; Das, T.; Cross, J. Diffusion imaging in neurological disease. J. Neurol. 2013, 260, 335–342. [Google Scholar] [CrossRef]
- De La Ossa, N.P.; Hernandez-Perez, M.; Domenech, S.; Cuadras, P.; Massuet, A.; Millan, M.; Gomis, M.; Lopez-Cancio, E.; Dorado, L.; Davalos, A. Hyperintensity of distal vessels on FLAIR is associated with slow progression of the infarction in acute ischemic stroke. Cerebrovasc. Dis. 2012, 34, 376–384. [Google Scholar] [CrossRef]
- Gitelman, D.R.; Ashburner, J.; Friston, K.J.; Tyler, L.K.; Price, C.J. Voxel-based morphometry of herpes simplex encephalitis. Neuroimage 2001, 13, 623–631. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mehta, S.; Grabowski, T.J.; Trivedi, Y.; Damasio, H. Evaluation of voxel-based morphometry for focal lesion detection in individuals. Neuroimage 2003, 20, 1438–1454. [Google Scholar] [CrossRef]
- Anbeek, P.; Vincken, K.L.; van Osch, M.J.; Bisschops, R.H.; van der Grond, J. Automatic segmentation of different-sized white matter lesions by voxel probability estimation. Med. Image Anal. 2004, 8, 205–215. [Google Scholar] [CrossRef]
- Capelle, A.S.; Colot, O.; Fernandez-Maloigne, C. Evidential segmentation scheme of multi-echo MR images for the detection of brain tumors using neighborhood information. Inf. Fusion 2004, 5, 203–216. [Google Scholar] [CrossRef]
- Colliot, O.; Bernasconi, N.; Khalili, N.; Antel, S.B.; Naessens, V.; Bernasconi, A. Individual voxel-based analysis of gray matter in focal cortical dysplasia. Neuroimage 2006, 29, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Colliot, O.; Mansi, T.; Bernasconi, N.; Naessens, V.; Klironomos, D.; Bernasconi, A. Segmentation of focal cortical dysplasia lesions on MRI using level set evolution. Neuroimage 2006, 32, 1621–1630. [Google Scholar] [CrossRef]
- Datta, S.; Sajja, B.R.; He, R.; Wolinsky, J.S.; Gupta, R.K.; Narayana, P.A. Segmentation and quantification of black holes in multiple sclerosis. Neuroimage 2006, 29, 467–474. [Google Scholar] [CrossRef]
- Fletcher-Heath, L.M.; Hall, L.O.; Goldgof, D.B.; Murtagh, F.R. Automatic segmentation of non-enhancing brain tumors in magnetic resonance images. Artif. Intell. Med. 2001, 21, 43–63. [Google Scholar] [CrossRef]
- Hojjatoleslami, S.; Kruggel, F. Segmentation of large brain lesions. IEEE Trans. Med. Imaging 2001, 20, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Prastawa, M.; Bullitt, E.; Ho, S.; Gerig, G. A brain tumor segmentation framework based on outlier detection. Med. Image Anal. 2004, 8, 275–283. [Google Scholar] [CrossRef]
- Sajja, B.R.; Datta, S.; He, R.; Mehta, M.; Gupta, R.K.; Wolinsky, J.S.; Narayana, P.A. Unified approach for multiple sclerosis lesion segmentation on brain MRI. Ann. Biomed. Eng. 2006, 34, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, E.A.; Tyler, L.K. Identifying lesions on structural brain images—Validation of the method and application to neuropsychological patients. Brain Lang. 2005, 94, 167–177. [Google Scholar] [CrossRef]
- Wu, Y.; Warfield, S.K.; Tan, I.L.; Wells, W.M., III; Meier, D.S.; van Schijndel, R.A.; Barkhof, F.; Guttmann, C.R. Automated segmentation of multiple sclerosis lesion subtypes with multichannel MRI. NeuroImage 2006, 32, 1205–1215. [Google Scholar] [CrossRef]
- Xie, K.; Yang, J.; Zhang, Z.; Zhu, Y. Semi-automated brain tumor and edema segmentation using MRI. Eur. J. Radiol. 2005, 56, 12–19. [Google Scholar] [CrossRef]
- Zhou, J.; Chan, K.; Chong, V.; Krishnan, S.M. Extraction of brain tumor from MR images using one-class support vector machine. In Proceedings of the 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference, Shanghai, China, 17–18 January 2006; pp. 6411–6414. [Google Scholar]
- Rekik, I.; Allassonnière, S.; Carpenter, T.K.; Wardlaw, J.M. Medical image analysis methods in MR/CT-imaged acute-subacute ischemic stroke lesion: Segmentation, prediction and insights into dynamic evolution simulation models. A critical appraisal. Neuroimage Clin. 2012, 1, 164–178. [Google Scholar] [CrossRef] [PubMed]
- Zuiderveld, K. Contrast Limited Adaptive Histogram Equalization. In Graphics Gems IV; Academic Press Professional, Inc.: Cambridge, MA, USA, 1994; pp. 474–485. [Google Scholar]
- He, H.; Garcia, E.A. Learning from imbalanced data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar]
- Pazzani, M.; Merz, C.; Murphy, P.; Ali, K.; Hume, T.; Brunk, C. Reducing misclassification costs. In Machine Learning Proceedings 1994; Elsevier: Amsterdam, The Netherlands, 1994; pp. 217–225. [Google Scholar]
- Domingos, P. MetaCost: A General Method for Making Classifiers Cost-Sensitive. In Proceedings of the Fifth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD ’99), New York, NY, USA, 1 August 1999; pp. 155–164. [Google Scholar] [CrossRef]
- Phankokkruad, M. Cost-Sensitive Extreme Gradient Boosting for Imbalanced Classification of Breast Cancer Diagnosis. In Proceedings of the 2020 10th IEEE International Conference on Control System, Computing and Engineering (ICCSCE), Penang, Malaysia, 21–22 August 2020; pp. 46–51. [Google Scholar]
- Kubat, M.; Matwin, S. Addressing the curse of imbalanced training sets: One-sided selection. ICML Citeseer 1997, 97, 179–186. [Google Scholar]
- Japkowicz, N. The class imbalance problem: Significance and strategies. In Proceedings of the 2000 International Conference on Artificial Intelligence (ICAI), Las Vegas, NV, USA, 1 July 2000; Volume 56. [Google Scholar]
- Susan, S.; Kumar, A. The balancing trick: Optimized sampling of imbalanced datasets—A brief survey of the recent State of the Art. Eng. Rep. 2020, 3, e12298. [Google Scholar] [CrossRef]
- He, H.; Bai, Y.; Garcia, E.A.; Li, S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–8 June 2008; pp. 1322–1328. [Google Scholar]
- Mishra, H.O.S.; Bhatnagar, S. MRI and CT image fusion based on wavelet transform. Int. J. Inf. Comput. Technol. 2014, 4, 47–52. [Google Scholar]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Ng, T.T.; Chang, S.F.; Sun, Q. Blind detection of photomontage using higher order statistics. In Proceedings of the 2004 IEEE International Symposium on Circuits and Systems (IEEE Cat. No. 04CH37512), Vancouver, BC, Canada, 23–26 May 2004; Volume 5. [Google Scholar] [CrossRef]
- Acharya, U.R.; Faust, O.; Sree, S.V.; Molinari, F.; Suri, J.S. ThyroScreen system: High resolution ultrasound thyroid image characterization into benign and malignant classes using novel combination of texture and discrete wavelet transform. Comput. Methods Programs Biomed. 2012, 107, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Faust, O.; Sree, S.V.; Molinari, F.; Garberoglio, R.; Suri, J. Cost-effective and non-invasive automated benign & malignant thyroid lesion classification in 3D contrast-enhanced ultrasound using combination of wavelets and textures: A class of ThyroScan™ algorithms. Technol. Cancer Res. Treat. 2011, 10, 371–380. [Google Scholar]
- Öztürk, Ş.; Akdemir, B. Application of feature extraction and classification methods for histopathological image using GLCM, LBP, LBGLCM, GLRLM and SFTA. Procedia Comput. Sci. 2018, 132, 40–46. [Google Scholar] [CrossRef]
- Rimavičius, T.; Gelžinis, A.; Verikas, A.; Vaičiukynas, E.; Bačauskienė, M.; Šaškov, A. Automatic benthic imagery recognition using a hierarchical two-stage approach. Signal Image Video Process. 2018, 12, 1107–1114. [Google Scholar] [CrossRef]
- Soh, L.K.; Tsatsoulis, C. Texture analysis of SAR sea ice imagery using gray level co-occurrence matrices. IEEE Trans. Geosci. Remote Sens. 1999, 37, 780–795. [Google Scholar] [CrossRef]
- Kannathal, N.; Choo, M.L.; Acharya, U.R.; Sadasivan, P. Entropies for detection of epilepsy in EEG. Comput. Methods Programs Biomed. 2005, 80, 187–194. [Google Scholar] [CrossRef]
- Mohanty, A.K.; Beberta, S.; Lenka, S.K. Classifying benign and malignant mass using GLCM and GLRLM based texture features from mammogram. Int. J. Eng. Res. Appl. 2011, 1, 687–693. [Google Scholar]
- Tang, X. Texture information in run-length matrices. IEEE Trans. Image Process. 1998, 7, 1602–1609. [Google Scholar] [CrossRef] [PubMed]
- Galloway, M.M. Texture analysis using grey level run lengths. NASA STI/Recon Tech. Rep. N 1974, 75, 18555. [Google Scholar]
- Xu, D.H.; Kurani, A.S.; Furst, J.D.; Raicu, D.S. Run-length encoding for volumetric texture. Heart 2004, 27, 452–458. [Google Scholar]
- Martis, R.J.; Acharya, U.R.; Mandana, K.; Ray, A.K.; Chakraborty, C. Cardiac decision making using higher order spectra. Biomed. Signal Process. Control 2013, 8, 193–203. [Google Scholar] [CrossRef]
- Noronha, K.P.; Acharya, U.R.; Nayak, K.P.; Martis, R.J.; Bhandary, S.V. Automated classification of glaucoma stages using higher order cumulant features. Biomed. Signal Process. Control 2014, 10, 174–183. [Google Scholar] [CrossRef]
- Swapna, G.; Rajendra Acharya, U.; VinithaSree, S.; Suri, J.S. Automated detection of diabetes using higher order spectral features extracted from heart rate signals. Intell. Data Anal. 2013, 17, 309–326. [Google Scholar] [CrossRef]
- Acharya, U.R.; Faust, O.; Molinari, F.; Sree, S.V.; Junnarkar, S.P.; Sudarshan, V. Ultrasound-based tissue characterization and classification of fatty liver disease: A screening and diagnostic paradigm. Knowl. Based Syst. 2015, 75, 66–77. [Google Scholar] [CrossRef]
- Acharya, U.R.; Raghavendra, U.; Fujita, H.; Hagiwara, Y.; Koh, J.E.; Hong, T.J.; Sudarshan, V.K.; Vijayananthan, A.; Yeong, C.H.; Gudigar, A.; et al. Automated characterization of fatty liver disease and cirrhosis using curvelet transform and entropy features extracted from ultrasound images. Comput. Biol. Med. 2016, 79, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, U.; Gudigar, A.; Maithri, M.; Gertych, A.; Meiburger, K.M.; Yeong, C.H.; Madla, C.; Kongmebhol, P.; Molinari, F.; Ng, K.H.; et al. Optimized multi-level elongated quinary patterns for the assessment of thyroid nodules in ultrasound images. Comput. Biol. Med. 2018, 95, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Assi, E.B.; Gagliano, L.; Rihana, S.; Nguyen, D.K.; Sawan, M. Bispectrum features and multilayer perceptron classifier to enhance seizure prediction. Sci. Rep. 2018, 8, 1–8. [Google Scholar]
- Hoaglin, D.C.; Welsch, R.E. The hat matrix in regression and ANOVA. Am. Stat. 1978, 32, 17–22. [Google Scholar]
- Last, M.; Kandel, A.; Maimon, O. Information-theoretic algorithm for feature selection. Pattern Recognit. Lett. 2001, 22, 799–811. [Google Scholar] [CrossRef]
- Berrar, D. Cross-validation. Encycl. Bioinform. Comput. Biol. 2019, 1, 542–545. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer Science & Business Media: Heidelberger/Berlin, Germany, 2009. [Google Scholar]
- Chang, Y.W.; Hsieh, C.J.; Chang, K.W.; Ringgaard, M.; Lin, C.J. Training and Testing Low-degree Polynomial Data Mappings via Linear SVM. J. Mach. Learn. Res. 2010, 11, 1471–1490. [Google Scholar]
- Ye, S.; Ye, J. Dice similarity measure between single valued neutrosophic multisets and its application in medical diagnosis. Neutrosophic Sets Syst. 2014, 6, 9. [Google Scholar]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Sarmento, R.M.; Vasconcelos, F.F.X.; Rebouças Filho, P.P.; Wu, W.; de Albuquerque, V.H.C. Automatic neuroimage processing and analysis in stroke—A systematic review. IEEE Rev. Biomed. Eng. 2019, 13, 130–155. [Google Scholar] [CrossRef] [PubMed]
- Kruggel, F.; Chalopin, C.; Descombes, X.; Kovalev, V.; Rajapakse, J. Segmentation of pathological features in MRI brain datasets. In Proceedings of the 9th International Conference on Neural Information Processing, 2002. ICONIP’02, Singapore, 18–22 November 2002; Volume 5, pp. 2673–2677. [Google Scholar]
- Agam, G.; Weiss, D.; Soman, M.; Arfanakis, K. Probabilistic brain lesion segmentation in DT-MRI. In Proceedings of the 2006 International Conference on Image Processing, Atlanta, GA, USA, 8–11 October 2006; pp. 89–92. [Google Scholar]
- Chen, L.; Bentley, P.; Rueckert, D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. NeuroImage: Clin. 2017, 15, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Acharya, U.R.; Tamura, T. Formal design methods for reliable computer-aided diagnosis: A review. IEEE Rev. Biomed. Eng. 2012, 5, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Debs, N.; Rasti, P.; Victor, L.; Cho, T.H.; Frindel, C.; Rousseau, D. Simulated perfusion MRI data to boost training of convolutional neural networks for lesion fate prediction in acute stroke. Comput. Biol. Med. 2020, 116, 103579. [Google Scholar] [CrossRef]
- Mitra, J.; Bourgeat, P.; Fripp, J.; Ghose, S.; Rose, S.; Salvado, O.; Connelly, A.; Campbell, B.; Palmer, S.; Sharma, G.; et al. Lesion segmentation from multimodal MRI using random forest following ischemic stroke. NeuroImage 2014, 98, 324–335. [Google Scholar] [CrossRef]
- Karthik, R.; Gupta, U.; Jha, A.; Rajalakshmi, R.; Menaka, R. A deep supervised approach for ischemic lesion segmentation from multimodal MRI using fully convolutional network. Appl. Soft Comput. 2019, 84, 105685. [Google Scholar] [CrossRef]
- Acharya, U.R.; Meiburger, K.M.; Faust, O.; Koh, J.E.W.; Oh, S.L.; Ciaccio, E.J.; Subudhi, A.; Jahmunah, V.; Sabut, S. Automatic detection of ischemic stroke using higher order spectra features in brain MRI images. Cogn. Syst. Res. 2019, 58, 134–142. [Google Scholar] [CrossRef]
- Vupputuri, A.; Ashwal, S.; Tsao, B.; Ghosh, N. Ischemic stroke segmentation in multi-sequence MRI by symmetry determined superpixel based hierarchical clustering. Comput. Biol. Med. 2020, 116, 103536. [Google Scholar] [CrossRef]
- Faust, O.; Hagiwara, Y.; Hong, T.J.; Lih, O.S.; Acharya, U.R. Deep learning for healthcare applications based on physiological signals: A review. Comput. Methods Programs Biomed. 2018, 161, 1–13. [Google Scholar] [CrossRef]
- Baygin, M.; Dogan, S.; Tuncer, T.; Barua, P.D.; Faust, O.; Arunkumar, N.; Abdulhay, E.W.; Palmer, E.E.; Acharya, U.R. Automated ASD detection using hybrid deep lightweight features extracted from EEG signals. Comput. Biol. Med. 2021, 134, 104548. [Google Scholar] [CrossRef]
| TP | TN | FP | FN | ACC % | PPV % | SEN % | SPE % | Dice |
|---|---|---|---|---|---|---|---|---|
| 37 | 21 | 1 | 6 | 89.23 | 97.37 | 86.05 | 95.45 | 0.91 |
| 41 | 19 | 4 | 2 | 90.91 | 91.11 | 95.35 | 82.61 | 0.93 |
| 38 | 17 | 5 | 6 | 83.33 | 88.37 | 86.36 | 77.27 | 0.87 |
| 40 | 21 | 1 | 4 | 92.42 | 97.56 | 90.91 | 95.45 | 0.94 |
| 39 | 17 | 5 | 5 | 84.85 | 88.64 | 88.64 | 77.27 | 0.89 |
| 39 | 18 | 4 | 5 | 86.36 | 90.70 | 88.64 | 81.82 | 0.90 |
| 40 | 19 | 3 | 4 | 89.39 | 93.02 | 90.91 | 86.36 | 0.92 |
| 36 | 20 | 2 | 8 | 84.85 | 94.74 | 81.82 | 90.91 | 0.88 |
| 38 | 20 | 2 | 6 | 87.88 | 95.00 | 86.36 | 90.91 | 0.90 |
| 36 | 21 | 1 | 7 | 87.69 | 97.30 | 83.72 | 95.45 | 0.90 |
| 384 | 193 | 28 | 53 | 87.69 | 93.38 | 87.88 | 87.35 | 0.90 |
| TP | TN | FP | FN | ACC % | PPV % | SEN % | SPE % | Dice |
|---|---|---|---|---|---|---|---|---|
| 37 | 22 | 0 | 6 | 90.77 | 100.00 | 86.05 | 100.00 | 0.93 |
| 39 | 23 | 0 | 4 | 93.94 | 100.00 | 90.70 | 100.00 | 0.95 |
| 40 | 20 | 2 | 4 | 90.91 | 95.24 | 90.91 | 90.91 | 0.93 |
| 39 | 22 | 0 | 5 | 92.42 | 100.00 | 88.64 | 100.00 | 0.94 |
| 43 | 21 | 1 | 1 | 96.97 | 97.73 | 97.73 | 95.45 | 0.98 |
| 40 | 22 | 0 | 4 | 93.94 | 100.00 | 90.91 | 100.00 | 0.95 |
| 41 | 21 | 1 | 3 | 93.94 | 97.62 | 93.18 | 95.45 | 0.95 |
| 36 | 22 | 0 | 8 | 87.88 | 100.00 | 81.82 | 100.00 | 0.90 |
| 40 | 22 | 0 | 4 | 93.94 | 100.00 | 90.91 | 100.00 | 0.95 |
| 40 | 22 | 0 | 3 | 95.38 | 100.00 | 93.02 | 100.00 | 0.96 |
| 395 | 217 | 4 | 42 | 93.01 | 99.06 | 90.39 | 98.18 | 0.94 |
| TP | TN | FP | FN | ACC % | PPV % | SEN % | SPE % | Dice |
|---|---|---|---|---|---|---|---|---|
| 38 | 22 | 0 | 5 | 92.31 | 100.00 | 88.37 | 100.00 | 0.94 |
| 38 | 23 | 0 | 5 | 92.42 | 100.00 | 88.37 | 100.00 | 0.94 |
| 40 | 21 | 1 | 4 | 92.42 | 97.56 | 90.91 | 95.45 | 0.94 |
| 39 | 22 | 0 | 5 | 92.42 | 100.00 | 88.64 | 100.00 | 0.94 |
| 42 | 21 | 1 | 2 | 95.45 | 97.67 | 95.45 | 95.45 | 0.97 |
| 41 | 22 | 0 | 3 | 95.45 | 100.00 | 93.18 | 100.00 | 0.96 |
| 38 | 20 | 2 | 6 | 87.88 | 95.00 | 86.36 | 90.91 | 0.90 |
| 37 | 22 | 0 | 7 | 89.39 | 100.00 | 84.09 | 100.00 | 0.91 |
| 37 | 22 | 0 | 7 | 89.39 | 100.00 | 84.09 | 100.00 | 0.91 |
| 41 | 22 | 0 | 2 | 96.92 | 100.00 | 95.35 | 100.00 | 0.98 |
| 391 | 217 | 4 | 46 | 92.41 | 99.02 | 89.48 | 98.18 | 0.94 |
| TP | TN | FP | FN | ACC % | PPV % | SEN % | SPE % | Dice |
|---|---|---|---|---|---|---|---|---|
| 38 | 21 | 1 | 5 | 90.77 | 97.44 | 88.37 | 95.45 | 0.93 |
| 40 | 23 | 0 | 3 | 95.45 | 100.00 | 93.02 | 100.00 | 0.96 |
| 41 | 18 | 4 | 3 | 89.39 | 91.11 | 93.18 | 81.82 | 0.92 |
| 40 | 22 | 0 | 4 | 93.94 | 100.00 | 90.91 | 100.00 | 0.95 |
| 43 | 20 | 2 | 1 | 95.45 | 95.56 | 97.73 | 90.91 | 0.97 |
| 40 | 21 | 1 | 4 | 92.42 | 97.56 | 90.91 | 95.45 | 0.94 |
| 42 | 21 | 1 | 2 | 95.45 | 97.67 | 95.45 | 95.45 | 0.97 |
| 39 | 22 | 0 | 5 | 92.42 | 100.00 | 88.64 | 100.00 | 0.94 |
| 41 | 22 | 0 | 3 | 95.45 | 100.00 | 93.18 | 100.00 | 0.96 |
| 40 | 22 | 0 | 3 | 95.38 | 100.00 | 93.02 | 100.00 | 0.96 |
| 404 | 212 | 9 | 33 | 93.62 | 97.93 | 92.44 | 95.91 | 0.95 |
| Classifier | Average SEN % | Average SPE % | Average PPV % | Average ACC % | Average Dice |
|---|---|---|---|---|---|
| SVM linear | 87.88 | 87.35 | 93.38 | 87.69 | 0.90 |
| SVM RBF | 92.44 | 95.91 | 97.93 | 93.62 | 0.95 |
| SVM polynomial 2 | 90.39 | 98.18 | 99.06 | 93.01 | 0.94 |
| SVM polynomial 3 | 89.48 | 98.18 | 99.02 | 92.41 | 0.94 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faust, O.; En Wei Koh, J.; Jahmunah, V.; Sabut, S.; Ciaccio, E.J.; Majid, A.; Ali, A.; Lip, G.Y.H.; Acharya, U.R. Fusion of Higher Order Spectra and Texture Extraction Methods for Automated Stroke Severity Classification with MRI Images. Int. J. Environ. Res. Public Health 2021, 18, 8059. https://doi.org/10.3390/ijerph18158059
Faust O, En Wei Koh J, Jahmunah V, Sabut S, Ciaccio EJ, Majid A, Ali A, Lip GYH, Acharya UR. Fusion of Higher Order Spectra and Texture Extraction Methods for Automated Stroke Severity Classification with MRI Images. International Journal of Environmental Research and Public Health. 2021; 18(15):8059. https://doi.org/10.3390/ijerph18158059
Chicago/Turabian StyleFaust, Oliver, Joel En Wei Koh, Vicnesh Jahmunah, Sukant Sabut, Edward J. Ciaccio, Arshad Majid, Ali Ali, Gregory Y. H. Lip, and U. Rajendra Acharya. 2021. "Fusion of Higher Order Spectra and Texture Extraction Methods for Automated Stroke Severity Classification with MRI Images" International Journal of Environmental Research and Public Health 18, no. 15: 8059. https://doi.org/10.3390/ijerph18158059
APA StyleFaust, O., En Wei Koh, J., Jahmunah, V., Sabut, S., Ciaccio, E. J., Majid, A., Ali, A., Lip, G. Y. H., & Acharya, U. R. (2021). Fusion of Higher Order Spectra and Texture Extraction Methods for Automated Stroke Severity Classification with MRI Images. International Journal of Environmental Research and Public Health, 18(15), 8059. https://doi.org/10.3390/ijerph18158059









