A Network-Based Bioinformatics Approach to Identify Molecular Biomarkers for Type 2 Diabetes that Are Linked to the Progression of Neurological Diseases
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets Employed in This Study
2.2. Preprocessing and Identification of Differentially Expressed Genes
2.3. Identification of Molecular Pathway and Gene Ontology
2.4. Protein-Protein Interactions Analysis
2.5. Transcription Factors-microRNA Interactions Analysis
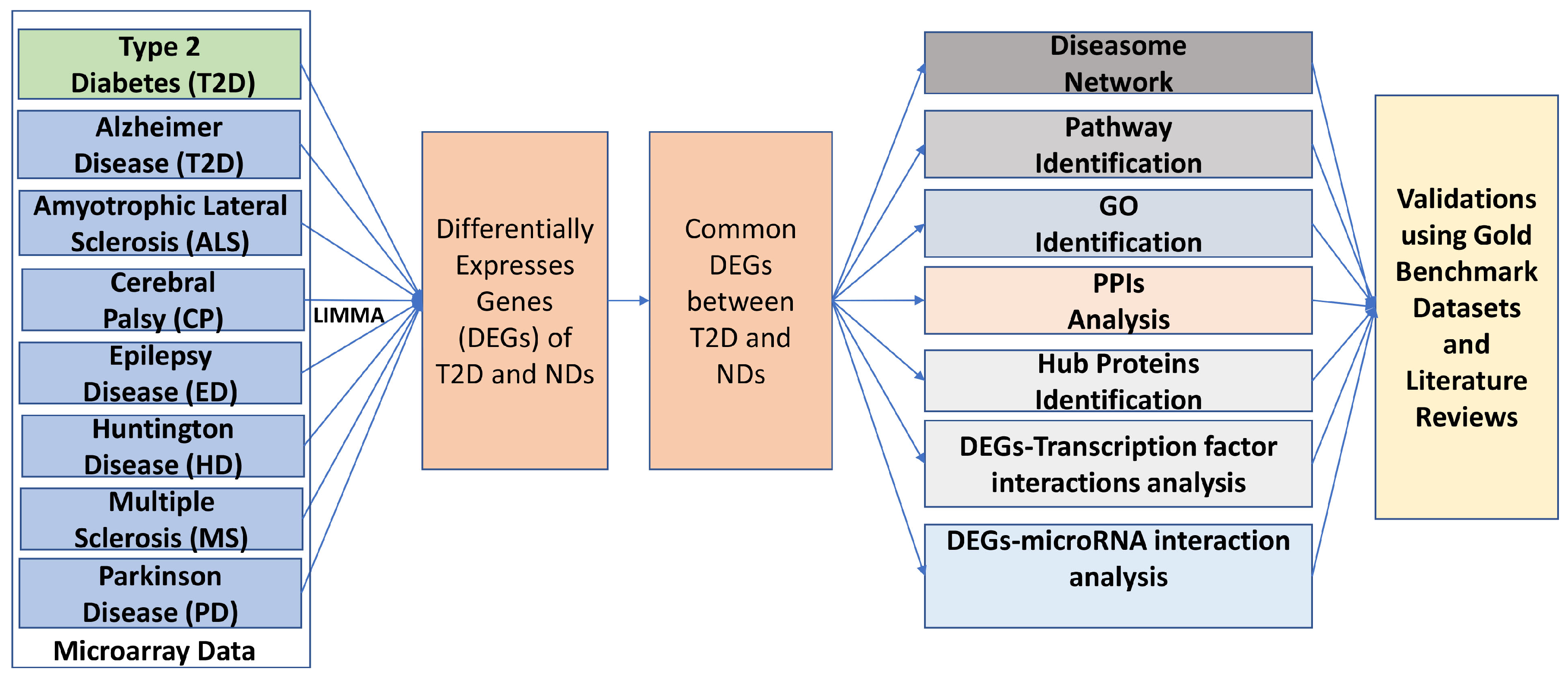
2.6. An Overview of the Analytical Approach
3. Results
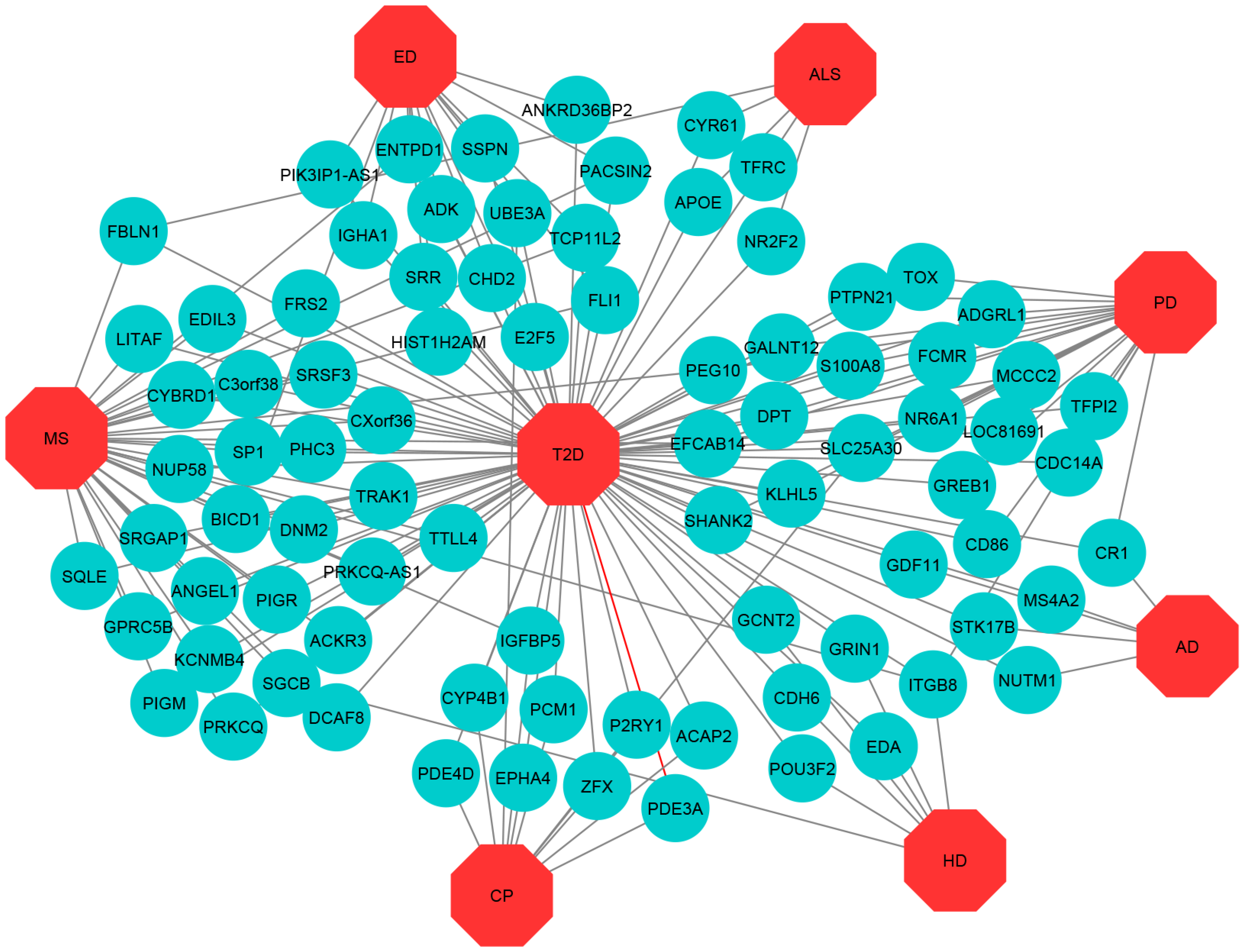
3.1. Gene Expression Analysis
3.2. Pathway and Functional Association Analysis
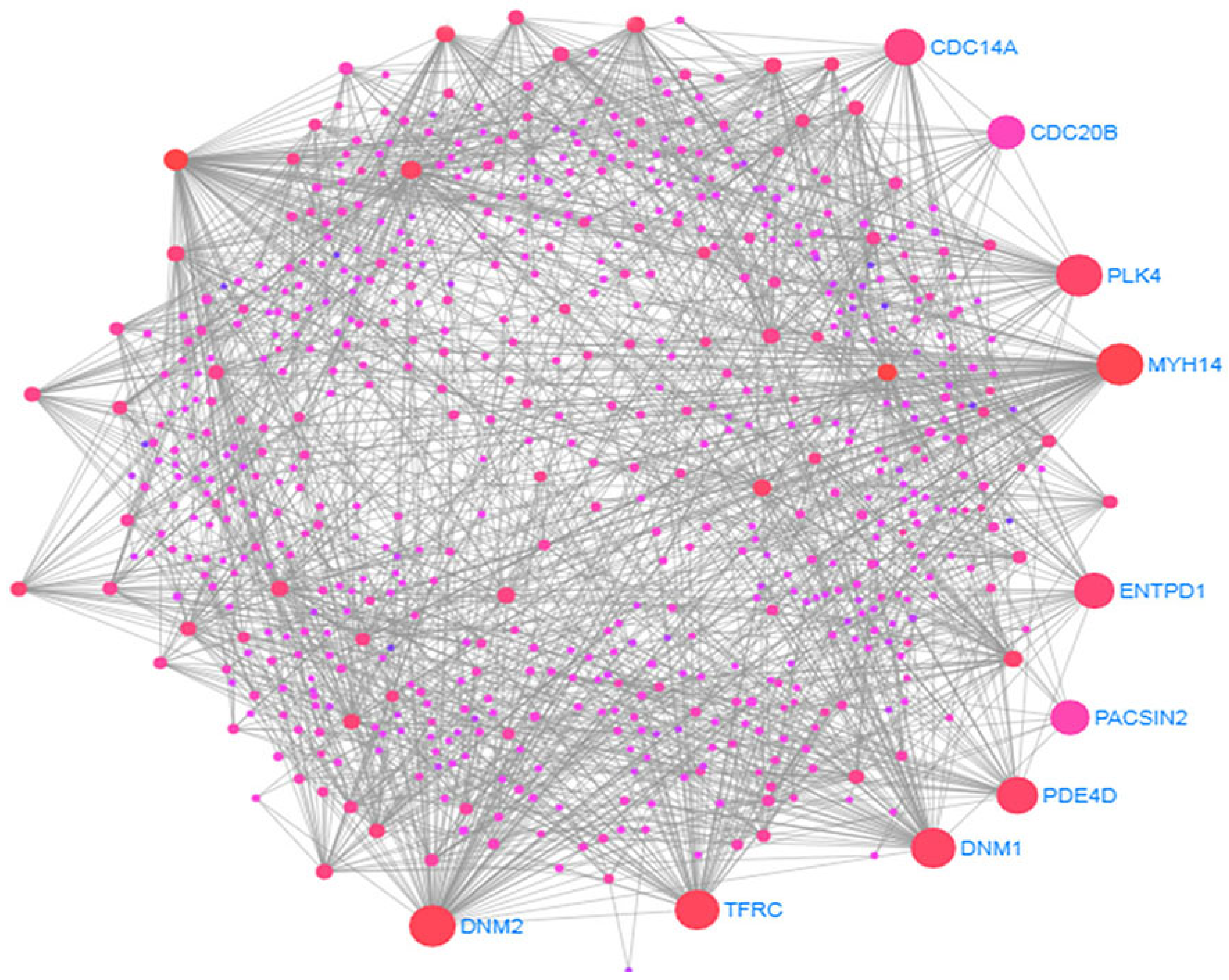
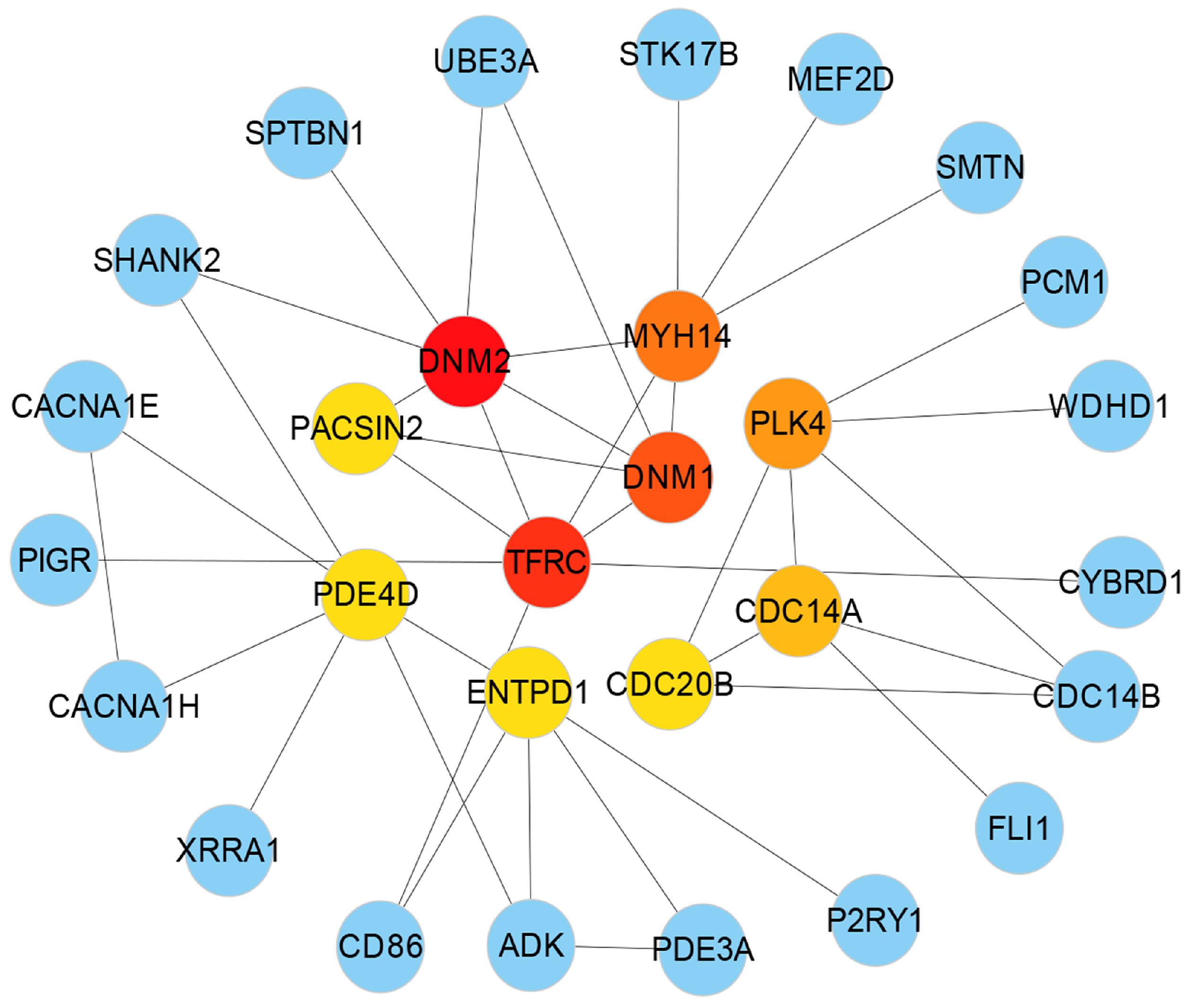
3.3. Protein-Protein Interactions (PPIs) Analysis
3.4. Identification of Transcriptional and Post-Transcriptional Regulators of the Differentially Expressed Genes
3.5. Validating Potential Targets Using Gold Benchmark Databases and Literatures
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- DeFronzo, R.A.; Ferrannini, E.; Groop, L.; Henry, R.R.; Herman, W.H.; Holst, J.J.; Hu, F.B.; Kahn, C.R.; Raz, I.; Shulman, G.I.; et al. Type 2 diabetes mellitus. Nat. Rev. Dis. Primers 2008, 1, 15019. [Google Scholar] [CrossRef]
- Mallorquí-Bagué, N.; Lozano-Madrid, M.; Toledo, E.; Corella, D.; Salas-Salvadó, J.; Cuenca-Royo, A.; Vioque, J.; Romaguera, D.; Martínez, J.A.; Wärnberg, J.; et al. Type 2 diabetes and cognitive impairment in an older population with overweight or obesity and metabolic syndrome: Baseline cross-sectional analysis of the predimed-plus study. Sci. Rep. 2018, 8, 16128. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Huang, X.; Ma, J.; Huang, J.; Fan, Y.; Li, H.; Qiu, J.; Zhang, H.; Huang, W. Value of three-dimensional strain parameters for predicting left ventricular remodeling after ST-elevation myocardial infarction. Int. J. Cardiovasc. Imaging 2017, 33, 663–673. [Google Scholar] [CrossRef]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2004, 27, S5eS10. [Google Scholar]
- Xu, L.; Zhao, H.; Qiu, J.; Zhu, W.; Lei, H.; Cai, Z.; Lin, W.H.; Huang, W.; Zhang, H.; Zhang, Y.T. The different effects of BMI and WC on organ damage in patients from a cardiac rehabilitation program after acute coronary syndrome. BioMed Res. Int. 2015, 2015, 942695. [Google Scholar] [CrossRef]
- Mota, M.; Banini, B.A.; Cazanave, S.C.; Sanyal, A.J. Molecular mechanisms of lipotoxicity and glucotoxicity in nonalcoholic fatty liver disease. Metabolism 2016, 65, 1049–1061. [Google Scholar] [CrossRef] [PubMed]
- Dolan, C.; Glynn, R.; Griffin, S.; Conroy, C.; Loftus, C.; Wiehe, P.C.; Healy, M.L.; Lawlor, B. Brain complications of diabetes mellitus: A cross-sectional study of awareness among individuals with diabetes and the general population in Ireland. Diabet. Med. 2018, 35, 871–879. [Google Scholar] [CrossRef]
- Mushtaq, G.; Khan, J.A.; Kumosani, T.A.; Kamal, M.A. Alzheimer’s disease and type 2 diabetes via chronic inflammatory mechanisms. Saudi J. Biol. Sci. 2015, 22, 4–13. [Google Scholar] [CrossRef]
- Verdile, G.; Fuller, S.J.; Martins, R.N. The role of type 2 diabetes in neurodegeneration. Neurobiol. Dis. 2015, 84, 22–38. [Google Scholar] [CrossRef]
- Bharadwaj, P.; Wijesekara, N.; Liyanapathirana, M.; Newsholme, P.; Ittner, L.; Fraser, P.; Verdile, G. The link between type 2 diabetes and neurodegeneration: Roles for amyloid-β, amylin, and tau proteins. J. Alzheimer’s Dis. 2017, 59, 421–432. [Google Scholar] [CrossRef]
- Porte, D., Jr.; Baskin, D.G.; Schwartz, M.W. Insulin signaling in the central nervous system: A critical role in metabolic homeostasis and disease from C. elegans to humans. Diabetes 2005, 54, 1264–1276. [Google Scholar] [CrossRef] [PubMed]
- Morsi, M.; Kobeissy, F.; Magdeldin, S.; Maher, A.; Aboelmagd, O.; Johar, D.; Bernstein, L. A shared comparison of diabetes mellitus and neurodegenerative disorders. J. Cell. Biochem. 2019, 120, 4318–14325. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Mudher, A. Alzheimer’s disease and type 2 diabetes: A critical assessment of the shared pathological traits. Front. Neurosci. 2018, 12, 383. [Google Scholar] [CrossRef]
- Martinez-Valbuena, I.; Valenti-Azcarate, R.; Amat-Villegas, I.; Riverol, M.; Marcilla, I.; de Andrea, C.E.; Sánchez-Arias, J.A.; del Mar Carmona-Abellan, M.; Marti, G.; Erro, M.E.; et al. Amylin as a potential link between type 2 diabetes and alzheimer disease. Ann. Neurol. 2019, 86, 539–551. [Google Scholar] [CrossRef] [PubMed]
- D’Ovidio, F.; d’Errico, A.; Carnà, P.; Calvo, A.; Costa, G.; Chiò, A. The role of pre-morbid diabetes on developing amyotrophic lateral sclerosis. Eur. J. Neurol. 2018, 25, 164–170. [Google Scholar]
- Crisham Janik, M.D.; Newman, T.B.; Cheng, Y.W.; Xing, G.; Gilbert, W.M.; Wu, Y.W. Maternal diagnosis of obesity and risk of cerebral palsy in the child. J. Pediatr. 2013, 163, 1307–1312. [Google Scholar] [CrossRef]
- Lu, C.L.; Chang, Y.H.; Sun, Y.; Li, C.Y. A population-based study of epilepsy incidence in association with type 2 diabetes and severe hypoglycaemia. Diabetes Res. Clin. Pract. 2018, 140, 97–106. [Google Scholar] [CrossRef]
- Montojo, M.T.; Aganzo, M.; González, N. Huntington’s disease and diabetes: Chronological sequence of its association. J. Huntington’s Dis. 2017, 6, 179–188. [Google Scholar] [CrossRef]
- Ruiz-Argüelles, A.; Méndez-Huerta, M.A.; Lozano, C.D.; Ruiz-Argüelles, G.J. Metabolomic profile of insulin resistance in patients with multiple sclerosis is associated to the severity of the disease. Mult. Scler. Relat. Disord. 2018, 25, 316–321. [Google Scholar] [CrossRef]
- Shaw, K. Type 2 diabetes and Parkinson’s disease. Pract. Diabetes 2019, 36, 115–116. [Google Scholar] [CrossRef]
- Schmitz, T.W.; Nathan Spreng, R.; Alzheimer’s Disease Neuroimaging Initiative. Basal forebrain degeneration precedes and predicts the cortical spread of Alzheimer’s pathology. Nat. Commun. 2016, 7, 13249. [Google Scholar] [CrossRef] [PubMed]
- Livingston, G.; Sommerlad, A.; Orgeta, V.; Costafreda, S.G.; Huntley, J.; Ames, D.; Ballard, C.; Banerjee, S.; Burns, A.; Cohen-Mansfield, J.; et al. Dementia prevention, intervention, and care. Lancet 2017, 390, 2673–2734. [Google Scholar] [CrossRef]
- Al-Chalabi, A.; Hardiman, O. The epidemiology of ALS: A conspiracy of genes, environment and time. Nat. Rev. Neurol. 2013, 9, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, L.; Pradat, P.F.; Ludolph, A.C.; Loeffler, J.P. Energy metabolism in amyotrophic lateral sclerosis. Lancet Neurol. 2011, 10, 75–82. [Google Scholar] [CrossRef]
- Desport, J.C.; Torny, F.; Lacoste, M.; Preux, P.M.; Couratier, P. Hypermetabolism in ALS: Correlations with clinical and paraclinical parameters. Neurodegener. Dis. 2005, 2, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Kioumourtzoglou, M.A.; Rotem, R.S.; Seals, R.M.; Gredal, O.; Hansen, J.; Weisskopf, M.G. Diabetes mellitus, obesity, and diagnosis of amyotrophic lateral sclerosis: A population-based study. JAMA Neurol. 2015, 72, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Cerebral Palsy Guidance. Cerebral Palsy and Diabetes. 2019. Available online: https://www.cerebralpalsyguidance.com/cerebral-palsy/associated-disorders/diabetes/ (accessed on 11 April 2019).
- Schendel, D.E.; Schuchat, A.; Thorsen, P. Public health issues related to infection in pregnancy and cerebral palsy. Ment. Retard. Dev. Disabil. Res. Rev. 2002, 8, 39–45. [Google Scholar] [CrossRef]
- Marcovecchio, M.L.; Petrosino, M.I.; Chiarelli, F. Diabetes and epilepsy in children and adolescents. Curr. Diabetes Rep. 2015, 15, 21. [Google Scholar] [CrossRef]
- Soltesz, G.; Acsadi, G. Association between diabetes, severe hypoglycemia, and electroencephalographic abnormalities. Arch. Dis. Child. 1989, 64, 992–996. [Google Scholar] [CrossRef]
- Ferlazzo, E.; Gasparini, S.; Beghi, E.; Sueri, C.; Russo, E.; Leo, A.; Labate, A.; Gambardella, A.; Belcastro, V.; Striano, P.; et al. Epilepsy in cerebrovascular diseases: Review of experimental and clinical data with meta-analysis of risk factors. Epilepsia 2016, 57, 1205–1214. [Google Scholar] [CrossRef]
- Schönberger, S.J.; Jezdic, D.; Faull, R.L.; Cooper, G.J. Proteomic analysis of the human brain in Huntington’s Disease indicates pathogenesis by molecular processes linked to other neurodegenerative diseases and to type-2 diabetes. J. Huntington’s Dis. 2013, 2, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Lalić, N.M.; Marić, J.; Svetel, M.; Jotić, A.; Stefanova, E.; Lalić, K.; Dragašević, N.; Miličić, T.; Lukić, L.; Kostić, V.S. Glucose homeostasis in Huntington disease: Abnormalities in insulin sensitivity and early-phase insulin secretion. Arch. Neurol. 2008, 65, 476–480. [Google Scholar] [CrossRef]
- Hou, W.H.; Li, C.Y.; Chang, H.H.; Sun, Y.; Tsai, C.C. A population-based cohort study suggests an increased risk of multiple sclerosis incidence in patients with type 2 diabetes mellitus. J. Epidemiol. 2017, 27, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Biosa, A.; Outeiro, T.F.; Bubacco, L.; Bisaglia, M. Diabetes Mellitus as a Risk Factor for Parkinson’s Disease: A Molecular Point of View. Mol. Neurobiol. 2018, 55, 8754–8763. [Google Scholar] [CrossRef]
- Green, H.; Tsitsi, P.; Markaki, I.; Aarsland, D.; Svenningsson, P. Novel Treatment Opportunities Against Cognitive Impairment in Parkinson’s Disease with an Emphasis on Diabetes-Related Pathways. CNS Drugs 2019, 33, 143–160. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI geo: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Misu, H.; Takamura, T.; Takayama, H.; Hayashi, H.; Matsuzawa-Nagata, N.; Kurita, S.; Ishikura, K.; Ando, H.; Takeshita, Y.; Ota, T.; et al. A liver-derived secretory protein, selenoprotein p, causes insulin resistance. Cell Metab. 2010, 12, 483–495. [Google Scholar] [CrossRef]
- Blalock, E.M.; Buechel, H.M.; Popovic, J.; Geddes, J.W.; Landfield, P.W. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient alzheimer’s disease. J. Chem. Neuroanat. 2011, 42, 118–126. [Google Scholar] [CrossRef]
- Dangond, F.; Hwang, D.; Camelo, S.; Pasinelli, P.; Frosch, M.P.; Stephanopoulos, G.; Stephanopoulos, G.; Brown, R.H., Jr.; Gullans, S.R. Molecular signature of late-stage human als revealed by expression profiling of postmortem spinal cord gray matter. Physiol. Genom. 2004, 16, 229–239. [Google Scholar] [CrossRef]
- Smith, L.R.; Chambers, H.G.; Subramaniam, S.; Lieber, R.L. Transcriptional abnormalities of hamstring muscle contractures in children with cerebral palsy. PLoS ONE 2012, 7, e40686. [Google Scholar] [CrossRef]
- Carlet, M.; Janjetovic, K.; Rainer, J.; Schmidt, S.; Panzer-Grümayer, R.; Mann, G.; Prelog, M.; Meister, B.; Ploner, C.; Kofler, R. Expression, regulation and function of phosphofructo-kinase/fructose-biphosphatases (pfkfbs) in glucocorticoid-induced apoptosis of acute lymphoblastic leukemia cells. BMC Cancer 2010, 10, 638. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Borovecki, F.; Lovrecic, L.; Zhou, J.; Jeong, H.; Then, F.; Rosas, H.D.; Hersch, S.M.; Hogarth, P.; Bouzou, B.; Jensen, R.V.; et al. Genome-wide expression profiling of human blood reveals biomarkers for huntington’s disease. Proc. Natl. Acad. Sci. USA 2005, 102, 11023–11028. [Google Scholar] [CrossRef] [PubMed]
- Han, M.H.; Lundgren, D.H.; Jaiswal, S.; Chao, M.; Graham, K.L.; Garris, C.S.; Axtell, R.C.; Ho, P.P.; Lock, C.B.; Woodard, J.I.; et al. Janus-like opposing roles of cd47 in autoimmune brain inflammation in humans and mice. J. Exp. Med. 2012, 209, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Lewandowski, N.M.; Ju, S.; Verbitsky, M.; Ross, B.; Geddie, M.L.; Rockenstein, E.; Adame, A.; Muhammad, A.; Vonsattel, J.P.; Ringe, D.; et al. Polyamine pathway contributes to the pathogenesis of parkinson disease. Proc. Natl. Acad. Sci. USA 2010, 107, 16970–16975. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Moni, M.A.; Liò, P. Genetic profiling and comorbidities of zika infection. J. Infect. Dis. 2017, 216, 703–712. [Google Scholar] [CrossRef]
- Moni, M.A.; Liò, P. comor: A software for disease comorbidity risk assessment. J. Clin. Bioinform. 2014, 4, 1. [Google Scholar] [CrossRef]
- Moni, M.A.; Liò, P. How to build personalized multi-omics comorbidity profiles. Front. Cell Dev. Biol. 2015, 3, 28. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Jin, L.; Zuo, X.-Y.; Su, W.-Y.; Zhao, X.-L.; Yuan, M.-Q.; Han, L.-Z.; Zhao, X.; Chen, Y.-D.; Rao, S.-Q. Pathway-based analysis tools for complex diseases: A review. Genom. Proteomics Bioinform. 2014, 12, 210–220. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. Gene ontology consortium: Going forward. Nucleic Acids Res. 2014, 43, D1049–D1056. [Google Scholar]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. Kegg for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011, 40, D109–D114. [Google Scholar] [CrossRef]
- Croft, D.; O’kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B.; et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2010, 39, D691–D697. [Google Scholar] [CrossRef]
- Krupa, S.; Anthony, K.; Buchoff, J.R.; Day, M.; Hannay, T.; Schaefer, C.F. The nci-nature pathway interaction database: A cell signaling resource. Nat. Preced. 2007, 2, 1. [Google Scholar] [CrossRef]
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Mélius, J.; Cirillo, E.; Coort, S.L.; Digles, D.; et al. Wikipathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2017, 46, D661–D667. [Google Scholar] [CrossRef]
- BioCarta, N.D. Biotech Software & Internet Report. RG J. 2001, 2, 117–120. [Google Scholar] [CrossRef]
- Mi, H.; Thomas, P. PANTHER pathway: An ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. 2009, 563, 123–140. [Google Scholar]
- Trupp, M.; Altman, T.; Fulcher, C.A.; Caspi, R.; Krummenacker, M.; Paley, S.; Karp, P.D. Beyond the genome (BTG) is a (PGDB) pathway genome database: HumanCyc. Genome Biol. 2010, 11, O12. [Google Scholar] [CrossRef]
- De Las Rivas, J.; Fontanillo, C. Protein–protein interactions essentials: Key concepts to building and analyzing interactome networks. PLoS Comput. Biol. 2010, 6, e1000807. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. String v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; van der Lee, R.; Bessy, A.; Cheneby, J.; Kulkarni, S.R.; Tan, G.; et al. Jaspar 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2017, 46, D260–D266. [Google Scholar] [CrossRef] [PubMed]
- Sethupathy, P.; Corda, B.; Hatzigeorgiou, A.G. Tarbase: A comprehensive database of experimentally supported animal microrna targets. RNA 2006, 12, 192–197. [Google Scholar] [CrossRef]
- Hsu, S.-D.; Lin, F.-M.; Wu, W.-Y.; Liang, C.; Huang, W.-C.; Chan, W.-L.; Tsai, W.-T.; Chen, G.-Z.; Lee, C.-J.; Chiu, C.M.; et al. mirtarbase: A database curates experimentally validated microrna–target interactions. Nucleic Acids Res. 2010, 39 (Suppl. 1), D163–D169. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Gill, E.E.; Hancock, R.E.W. Networkanalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015, 10, 823. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Gentleman, R.; Carey, V.; Huber, W.; Hahne, F. Genefilter: Methods for Filtering Genes from High-Throughput Experiments. 2015. Bioconductor. Available online: https://bioconductor.riken.jp/packages/3.0/bioc/html/genefilter.html (accessed on 5 February 2020).
- Chen, S.-H.; Chin, C.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cyto-hubba: A cytoscape plug-in for hub object analysis in network biology. In Proceedings of the 20th International Conference on Genome Informatics, Yokohama, Japan, 14–16 December 2009. [Google Scholar]
- Cheng, C.; Alexander, R.; Min, R.; Leng, J.; Yip, K.Y.; Rozowsky, J.; Yan, K.K.; Dong, X.; Djebali, S.; Ruan, Y.; et al. Understanding transcriptional regulation by integrative analysis of transcription factor binding data. Genome Res. 2012, 22, 1658–1667. [Google Scholar] [CrossRef]
- Moradifard, S.; Hoseinbeyki, M.; Ganji, S.M.; Minuchehr, Z. Analysis of microRNA and gene expression profiles in Alzheimer’s disease: A meta-analysis approach. Sci. Rep. 2018, 8, 4767. [Google Scholar] [CrossRef]
- Van Cauwenberghe, C.; Van Broeckhoven, C.; Sleegers, K. The genetic landscape of Alzheimer disease: Clinical implications and perspectives. Genet. Med. 2016, 18, 421–430. [Google Scholar] [CrossRef]
- Anjana, M.; Ahuja, Y.R. Genes associated with Alzheimer Disease. Neurol. Asia 2010, 15, 109–118. [Google Scholar]
- Eykens, C.; Robberecht, W. The Genetic basis of amyotrophic lateral sclerosis: Recent breakthroughs. Adv. Genom. Genet. 2015, 5, 327. [Google Scholar]
- Fahey, M.C.; Maclennan, A.H.; Kretzschmar, D.; Gecz, J.; Kruer, M.C. The genetic basis of cerebral palsy. Dev. Med. Child Neurol. 2017, 59, 462–469. [Google Scholar] [CrossRef] [PubMed]
- GeneDx. Genetic Testing for Epilepsy: A Guide for Patients. Available online: https://www.genedx.com/wpcontent/uploads/crm_docs/91040_Epilepsy-Patient-Guide.pdf (accessed on 26 October 2019).
- Myers, C.T.; Mefford, H.C. Advancing epilepsy genetics in the genomic era. Genome Med. 2015, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Arning, L.; Epplen, J.T. Genetic modifiers of Huntington’s disease: Beyond CAG. Future Neurol. 2012, 7, 93–109. [Google Scholar] [CrossRef]
- Baranzini, S.E. Revealing the genetic basis of multiple sclerosis: Are we there yet? Curr. Opin. Genet. Dev. 2011, 21, 317–324. [Google Scholar] [CrossRef]
- Redenšek, S.; Trošt, M.; Dolžan, V. Genetic determinants of Parkinson’s disease: Can they help to stratify the patients based on the underlying molecular defect? Front. Aging Neurosci. 2017, 9, 20. [Google Scholar] [CrossRef]
- Antoni Romeu and Lluís Arola. Classical dynamin dnm1 and dnm3 genes attain maximum expression in the normal human central nervous system. BMC Res. Notes 2014, 7, 188. [Google Scholar]
- Sidiropoulos, P.N.M.; Miehe, M.; Bock, T.; Tinelli, E.; Oertli, C.I.; Kuner, R.; Meijer, D.; Wollscheid, B.; Niemann, A.; Suter, U. Dynamin 2 mutations in charcot–marie–tooth neuropathy highlight the importance of clathrin-mediated endocytosis in myelination. Brain 2012, 135, 1395–1411. [Google Scholar] [CrossRef]
- Dumont, V.; Tolvanen, T.A.; Kuusela, S.; Wang, H.; Nyman, T.A.; Lindfors, S.; Tienari, J.; Nisen, H.; Suetsugu, S.; Plomann, M.; et al. Pacsin2 accelerates nephrin trafficking and is up-regulated in diabetic kidney disease. FASEB J. 2017, 31, 3978–3990. [Google Scholar] [CrossRef]
- Borie, C.; Gasparini, F.; Verpillat, P.; Bonnet, A.-M.; Agid, Y.; Hetet, G.; Brice, A.; Dürr, A.; Grandchamp, B.; French Parkinson’s Disease Genetic Study Group; et al. Association study between iron-related genes polymorphisms and parkinson’s disease. J. Neurol. 2002, 249, 801–804. [Google Scholar] [CrossRef]
- Rahman, M.; Islam, T.; Shahjaman, M.; Zaman, T.; Faruquee, H.M.; Jamal, M.A.H.M.; Huq, F.; Quinn, J.M.W.; Moni, M.A. Discovering biomarkers and pathways shared by alzheimer’s disease and ischemic stroke to identify novel therapeutic targets. Medicina 2019, 55, 191. [Google Scholar] [CrossRef] [PubMed]
- Mamelona, J.; Crapoulet, N.; Marrero, A. A new case of spastic paraplegia type 64 due to a missense mutation in the entpd1 gene. Hum. Genome Var. 2019, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.R.; Islam, T.; Turanli, B.; Zaman, T.; Faruquee, H.M.; Rahman, M.M.; Mollah, M.N.H.; Nanda, R.K.; Arga, K.Y.; Gov, E.; et al. Network-based approach to identify molecular signatures and therapeutic agents in alzheimer’s disease. Comput. Biol. Chem. 2019, 78, 431–439. [Google Scholar] [CrossRef]
- Godlewski, J.; Lenart, J.; Salinska, E. Microrna in brain pathology: Neurodegeneration the other side of the brain cancer. Non-Coding RNA 2019, 5, 20. [Google Scholar] [CrossRef]
- Persengiev, S.P.; Kondova, I.I.; Bontrop, R.E. The impact of micrornas on brain aging and neurodegeneration. Curr. Gerontol. Geriatr. Res. 2012, 2012, 359369. [Google Scholar] [CrossRef]
- Liu, W.; Liu, C.; Zhu, J.; Shu, P.; Yin, B.; Gong, Y.; Qiang, B.; Yuan, J.; Peng, X. Microrna-16 targets amyloid precursor protein to potentially modulate alzheimer’s-associated pathogenesis in samp8 mice. Neurobiol. Aging 2012, 33, 522–534. [Google Scholar] [CrossRef]
- Cloonan, N.; Brown, M.K.; Steptoe, A.L.; Wani, S.; Chan, W.L.; Forrest, A.R.R.; Kolle, G.; Gabrielli, B.; Grimmond, S.M. The mir-17-5p microrna is a key regulator of the g1/s phase cell cycle transition. Genome Biol. 2008, 9, R127. [Google Scholar] [CrossRef]



| Disease Name | GEO Platform | Tissues/Cells | GEO Accession | Raw Genes | Case Samples | Control Samples | UP Reg. Genes | Down Reg. Genes |
|---|---|---|---|---|---|---|---|---|
| Type 2 Diabetes (T2D) | Affymetrix Human Genome U133 Plus 2.0 Array | Liver | GSE23343 | 54613 | 10 | 7 | 622 | 698 |
| Alzheimer’s disease | Affymetrix Human Genome U133 Plus 2.0 Array | CA1 tissue | GSE28146 | 54675 | 22 | 8 | 847 | 759 |
| Amyotrophic lateral sclerosis | Affymetrix Human Full Length HuGeneFL Array | Spinal cord | GSE833 | 22277 | 7 | 5 | 735 | 2166 |
| Cerebral palsy | Affymetrix Human Genome U133 Plus 2.0 Array | Muscle | GSE31243 | 22277 | 20 | 20 | 243 | 345 |
| Epilepsy disease | Affymetrix Human Genome U133 Plus 2.0 Array | Peripheral Blood | GSE22779 | 54675 | 12 | 4 | 882 | 1007 |
| Huntington’s disease | Affymetrix Human Genome U133 Plus 2.0 Array | Whole Blood | GSE1751 | 22283 | 17 | 14 | 365 | 973 |
| Multiple sclerosis | Affymetrix Human Genome U133 Plus 2.0 Array | Brain | GSE38010 | 33398 | 5 | 2 | 3987 | 3476 |
| Parkinson’s disease | Affymetrix Human Genome U133A 2.0 Array | Brain | GSE19587 | 22277 | 12 | 10 | 1167 | 422 |
| (a) Common significant pathway common between T2D and AD | ||
| Pathway Name | p-Value | Source |
| Neuroregulin receptor degredation protein-1 Controls ErbB3 receptor recycling | 3.78 | Biocarta |
| Cytokine-cytokine receptor interaction | 1.17 | KEGG |
| Toll-like receptor signaling pathway | 2.92 | KEGG |
| Glycosphingolipid biosynthesis | 3.36 | KEGG |
| IL1-mediated signaling events | 1.89 | NCI-Nature |
| TCR signaling in naive CD8+ T cells | 4.53 | NCI-Nature |
| Ubiquitin proteasome pathway | 3.09 | Panther |
| Ionotropic glutamate receptor pathway | 3.36 | Panther |
| Glutamate Neurotransmitter Release Cycle | 1.02 | Reactome |
| Signaling by Interleukins | 1.32 | Reactome |
| Cytokine Signaling in the Immune system | 1.83 | Reactome |
| Oxidative Stress-Induced Senescence | 2.07 | Reactome |
| Neurotransmitter Release Cycle | 4.23 | Reactome |
| Transmission across Chemical Synapses | 4.68 | Reactome |
| Apoptosis Modulation and Signaling | 2.97 | Wiki |
| (b) Common significant pathway common between T2D and ALS | ||
| Pathway Name | p-Value | Source |
| Neuroregulin receptor degredation protein-1 Controls ErbB3 receptor recycling | 1.48 | Biocarta |
| Cytokine-cytokine receptor interaction | 3.10 | KEGG |
| Glycosphingolipid biosynthesis | 1.05 | KEGG |
| Ubiquitin mediated proteolysis | 1.28 | KEGG |
| Glutamatergic synapse | 2.85 | KEGG |
| Immune System | 3.74 | Reactome |
| Innate Immune System | 7.98 | Reactome |
| Insulin receptor signaling cascade | 8.94 | Reactome |
| Cytokine Signaling in the Immune system | 1.05 | Reactome |
| Neuronal System | 1.17 | Reactome |
| Neurotransmitter Receptor Binding And Downstream Transmission in The Postsynaptic Cell | 1.47 | Reactome |
| Transmission across Chemical Synapses | 2.05 | Reactome |
| Adaptive Immune System | 2.30 | Reactome |
| NO/cGMP/PKG mediated Neuroprotection | 1.19 | Wiki |
| Toll-like Receptor Signaling | 3.90 | Wiki |
| (c) Common significant pathway common between T2D and CP | ||
| Pathway Name | p-Value | Source |
| Focal adhesion | 4.47 | KEGG |
| Dopaminergic synapse | 9.70 | KEGG |
| Cell adhesion molecules (CAMs) | 1.28 | KEGG |
| Rapid glucocorticoid signaling | 2.65 | NCI-Nature |
| Inflammation mediated by chemokine and cytokine signaling pathway | 4.19 | Panther |
| Adaptive Immune System | 7.50 | Reactome |
| Immune System | 1.47 | Reactome |
| Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell | 1.21 | Reactome |
| Electric Transmission Across Gap Junctions | 1.66 | Reactome |
| Transmission across Electrical Synapses | 1.66 | Reactome |
| Neuronal System | 1.84 | Reactome |
| Neurofascin interactions | 2.32 | Reactome |
| Transmission across Chemical Synapses | 3.39 | Reactome |
| Inflammatory Response Pathway | 4.53 | Wiki |
| Insulin signaling in human adipocytes | 2.65 | Wiki |
| Toll-like Receptor Signaling Pathway | 4.68 | Wiki |
| (d) Common significant pathway common between T2D and ED | ||
| Pathway Name | p-Value | Source |
| Cell adhesion molecules (CAMs) | 1.14 | KEGG |
| Ubiquitin mediated proteolysis | 4.95 | KEGG |
| TCR signaling in naive CD8+ T cells | 4.03 | NCI-Nature |
| Rapid glucocorticoid signaling | 4.70 | NCI-Nature |
| Apoptosis signaling pathway | 2.35 | Panther |
| Ubiquitin proteasome pathway | 2.75 | Panther |
| Adaptive Immune System | 6.15 | Reactome |
| Oxidative Stress-Induced Senescence | 1.75 | Reactome |
| Immune System | 2.26 | Reactome |
| Deposition of new CENPA-containing nucleosomes at the centromere | 3.90 | Reactome |
| Spinal Cord Injury | 3.42 | Wiki |
| (e) Common significant pathway common between T2D and HD | ||
| Pathway Name | p-Value | Source |
| Neuroregulin receptor degredation protein-1 Controls ErbB3 receptor recycling | 2.59 | Biocarta |
| Glycosphingolipid biosynthesis | 1.54 | KEGG |
| Cell adhesion molecules (CAMs) | 2.32 | KEGG |
| Cytokine-cytokine receptor interaction | 3.53 | KEGG |
| Neurotrophic factor-mediated Trk receptor signaling | 2.72 | NCI-Nature |
| Ubiquitin proteasome pathway | 1.41 | Panther |
| Cholesterol biosynthesis | 4.53 | Panther |
| Immune System | 1.82 | Reactome |
| Cytokine Signaling in the Immune system | 1.54 | Reactome |
| Innate Immune System | 2.00 | Reactome |
| Adaptive Immune System | 4.10 | Reactome |
| Toll-Like Receptors Cascades | 2.12 | Reactome |
| NO/cGMP/PKG mediated Neuroprotection | 1.67 | Wiki |
| Apoptosis | 4.87 | Wiki |
| Tryptophan metabolism | 1.60 | Wiki |
| (f) Common significant pathway common between T2D and MS | ||
| Pathway Name | p-Value | Source |
| Neurotrophin signaling pathway | 1.17 | KEGG |
| Glycosphingolipid biosynthesis | 1.36 | KEGG |
| Adipocytokine signaling pathway | 1.39 | KEGG |
| Autophagy | 1.69 | KEGG |
| Glutamatergic synapse | 3.07 | KEGG |
| GABAergic synapse | 3.68 | KEGG |
| Neurotrophic factor-mediated Trk receptor signaling | 8.37 | NCI-Nature |
| Insulin/IGF pathway-protein kinase B signaling cascade | 5.05 | Panther |
| Apoptosis signaling pathway | 5.17 | Panther |
| Ubiquitin proteasome pathway | 1.16 | Panther |
| Ionotropic glutamate receptor pathway | 1.36 | Panther |
| Transmission across Chemical Synapses | 8.34 | Reactome |
| Neuronal System | 1.71 | Reactome |
| Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell | 1.77 | Reactome |
| Insulin receptor signaling cascade | 2.65 | Reactome |
| Oxidative Stress-Induced Senescence | 1.13 | Reactome |
| Brain-Derived Neurotrophic Factor (BDNF) signaling pathway | 2.98 | Wiki |
| (g) Common significant pathway common between T2D and PD | ||
| Pathway Name | p-Value | Source |
| Neuroregulin receptor degredation protein-1 Controls ErbB3 receptor recycling | 4.04 | Biocarta |
| Toll-like receptor signaling pathway | 2.21 | KEGG |
| Cell adhesion molecules (CAMs) | 7.20 | KEGG |
| Allograft rejection | 1.70 | KEGG |
| Graft-versus-host disease | 1.96 | KEGG |
| Intestinal immune network for IgA production | 2.63 | KEGG |
| Innate Immune System | 1.07 | Reactome |
| Insulin receptor signaling cascade | 4.07 | Reactome |
| Immune System | 1.50 | Reactome |
| Chemokine receptors bind chemokines | 3.50 | Reactome |
| Adaptive Immune System | 4.73 | Reactome |
| Transmission across Electrical Synapses | 2.60 | Reactome |
| (a) Common significant GOs of T2D and AD | ||
| GO ID | Pathway | p-Value |
| GO:0032000 | positive regulation of fatty acid beta-oxidation | 5.99 |
| GO:0016064 | immunoglobulin mediated immune response | 6.73 |
| GO:2000269 | regulation of fibroblast apoptotic process | 6.73 |
| GO:0031998 | regulation of fatty acid beta-oxidation | 8.22 |
| GO:0051588 | regulation of neurotransmitter transport | 1.05 |
| GO:0007498 | mesoderm development | 1.64 |
| GO:0051961 | negative regulation of nervous system development | 1.71 |
| GO:0046928 | regulation of neurotransmitter secretion | 2.08 |
| (b) Common significant GOs of T2D and ALS | ||
| GO ID | Pathway | p-Value |
| GO:0006352 | DNA-templated transcription, initiation | 2.46 |
| GO:0050870 | positive regulation of T cell activation | 4.48 |
| GO:0048935 | peripheral nervous system neuron development | 1.01 |
| GO:0021522 | spinal cord motor neuron differentiation | 1.01 |
| GO:0021559 | trigeminal nerve development | 1.01 |
| GO:2000145 | regulation of cell motility | 1.10 |
| GO:0048665 | neuron fate specification | 1.15 |
| GO:1902692 | regulation of neuroblast proliferation | 1.58 |
| GO:0048663 | neuron fate commitment | 2.01 |
| GO:2000177 | regulation of neural precursor cell proliferation | 2.15 |
| GO:0014033 | neural crest cell differentiation | 2.30 |
| GO:0045597 | positive regulation of cell differentiation | 3.23 |
| GO:0051961 | negative regulation of nervous system development | 3.28 |
| GO:0019228 | neuronal action potential | 3.42 |
| GO:0021953 | central nervous system neuron differentiation | 4.68 |
| GO:0050768 | negative regulation of neurogenesis | 4.82 |
| (c) Common significant GOs of T2D and CP | ||
| GO ID | Pathway | p-Value |
| GO:0071363 | cellular response to growth factor stimulus | 5.47 |
| GO:0048681 | negative regulation of axon regeneration | 6.38 |
| GO:0070571 | negative regulation of neuron projection regeneration | 7.18 |
| GO:0099590 | neurotransmitter receptor internalization | 8.77 |
| GO:0048679 | regulation of axon regeneration | 8.77 |
| GO:0021952 | central nervous system projection neuron axonogenesis | 8.77 |
| GO:0008045 | motor neuron axon guidance | 8.77 |
| GO:0106030 | neuron projection fasciculation | 8.77 |
| GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 1.67 |
| GO:0071875 | adrenergic receptor signaling pathway | 1.67 |
| GO:1990090 | cellular response to nerve growth factor stimulus | 1.82 |
| GO:0010977 | negative regulation of neuron projection development | 4.08 |
| GO:0016192 | vesicle-mediated transport | 4.18 |
| GO:0001934 | positive regulation of protein phosphorylation | 4.22 |
| (d) Common significant GOs of T2D and ED | ||
| GO ID | Pathway | p-Value |
| GO:0006897 | endocytosis | 6.45 |
| GO:0007599 | hemostasis | 1.15 |
| GO:0090286 | cytoskeletal anchoring at the nuclear membrane | 1.30 |
| GO:0034214 | protein hexamerization | 1.44 |
| GO:0034063 | stress granule assembly | 1.73 |
| GO:0097205 | renal filtration | 1.73 |
| GO:0035278 | miRNA mediated inhibition of translation | 1.73 |
| GO:0071470 | cellular response to osmotic stress | 2.15 |
| GO:0034656 | nucleobase-containing small molecule catabolic process | 2.30 |
| GO:0021953 | central nervous system neuron differentiation | 4.68 |
| (e) Common significant GOs of T2D and HD | ||
| GO ID | Pathway | p-Value |
| GO:0042127 | regulation of cell proliferation | 6.92 |
| GO:0022409 | positive regulation of cell-cell adhesion | 2.16 |
| GO:0048665 | neuron fate specification | 5.19 |
| GO:0048663 | neuron fate commitment | 9.06 |
| GO:0014033 | neural crest cell differentiation | 1.04 |
| GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 1.49 |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 2.65 |
| GO:0007399 | nervous system development | 3.43 |
| (f) Common significant GOs of T2D and MS | ||
| GO ID | Pathway | p-Value |
| GO:0070229 | negative regulation of lymphocyte apoptotic process | 4.32 |
| GO:0099590 | neurotransmitter receptor internalization | 5.27 |
| GO:0051588 | regulation of neurotransmitter transport | 8.67 |
| GO:0050804 | modulation of chemical synaptic transmission | 2.29 |
| GO:0046928 | regulation of neurotransmitter secretion | 3.50 |
| GO:2000146 | negative regulation of cell motility | 3.66 |
| GO:0090181 | regulation of cholesterol metabolic process | 7.75 |
| GO:0072657 | protein localization to membrane | 1.43 |
| GO:0007005 | mitochondrion organization | 1.60 |
| GO:0010595 | positive regulation of endothelial cell migration | 2.11 |
| GO:0010646 | regulation of cell communication | 2.17 |
| GO:0050658 | RNA transport | 2.70 |
| GO:0010628 | positive regulation of gene expression | 3.43 |
| GO:2000145 | regulation of cell motility | 4.71 |
| GO:0014033 | neural crest cell differentiation | 4.92 |
| (g) Common significant GOs of T2D and PD | ||
| GO ID | Pathway | p-Value |
| GO:0070486 | leukocyte aggregation | 9.16 |
| GO:0070584 | mitochondrion morphogenesis | 1.14 |
| GO:0090128 | regulation of synapse maturation | 1.26 |
| GO:0045624 | positive regulation of T-helper cell differentiation | 1.37 |
| GO:0048854 | brain morphogenesis | 1.37 |
| GO:0019228 | neuronal action potential | 2.73 |
| GO:0097061 | dendritic spine organization | 2.84 |
| GO:0030199 | collagen fibril organization | 3.40 |
| GO:0061448 | connective tissue development | 3.51 |
| GO:0048813 | dendrite morphogenesis | 3.73 |
| GO:0099601 | regulation of neurotransmitter receptor activity | 3.84 |
| Protein Symbol | Degree | Description | Feature |
|---|---|---|---|
| DNM1 | 65 | Dynamin 1 | GTP binding |
| DNM2 | 84 | Dynamin 2 | GTP binding and GTPase activity |
| MYH14 | 82 | Myosin Heavy Chain 14 | Calmodulin binding and motor activity |
| PACSIN2 | 60 | Protein Kinase C And Casein Kinase Substrate In Neurons | Identical protein binding and lipid binding |
| TFRC | 10 | Transferrin Receptor | Double-stranded RNA binding |
| PDE4D | 39 | Phosphodiesterase 4D | Enzyme binding and protein domain specific binding |
| ENTPD1 | 32 | Ectonucleoside Triphosphate Diphosphohydrolase 1 | Hydrolase activity and nucleoside-diphosphatase activity |
| PLK4 | 45 | Polo Like Kinase 4 | Identical protein binding and protein kinase activity |
| CDC20B | 20 | Cell Division Cycle 20B | Pathways related to DNA damage response |
| CDC14A | 36 | Cell Division Cycle 14A | Phosphatase activity and phosphoprotein phosphatase activity |
| (a) Regulatory Transcription Factors | ||
| Symbol | Description | Feature |
| FOXC1 | Forkhead Box C1 | NA-binding transcription factor activity and transcription factor binding |
| GATA2 | GATA binding protein 2 | DNA-binding transcription factor activity and chromatin binding |
| FOXL1 | Forkhead Box L1 | NA-binding transcription factor activity and DNA-binding transcription factor activity, RNA polymerase II-specific |
| YY1 | YY1 Transcription Factor | NA-binding transcription factor activity and transcription coactivator activity |
| E2F1 | E2F transcription factor 1 | DNA-binding transcription factor activity and transcription factor binding. |
| NFIC | Nuclear Factor I C | DNA-binding transcription factor activity and proximal promoter DNA-binding transcription activator activity, RNA polymerase II-specific |
| NFYA | Nuclear Transcription Factor Y Subunit Alpha | DNA-binding transcription factor activity and transcription regulatory region DNA binding |
| USF2 | Upstream Transcription Factor 2, C-Fos Interacting | DNA-binding transcription factor activity and sequence-specific DNA binding |
| HINFP | Histone H4 Transcription Factor | DNA-binding transcription factor activity and enzyme binding |
| MEF2A | Myocyte Enhancer Factor 2A | DNA-binding transcription factor activity and protein heterodimerization activity |
| SRF | Serum Response Factor | DNA-binding transcription factor activity and sequence-specific DNA binding |
| NFKB1 | Nuclear Factor Kappa B Subunit 1 | DNA-binding transcription factor activity and sequence-specific DNA binding |
| PDE4D | Phosphodiesterase 4D | enzyme binding and protein domain specific binding |
| CREB1 | CAMP Responsive Element Binding Protein 1 | DNA-binding transcription factor activity and enzyme binding |
| SP1 | Sp1 Transcription Factor | DNA-binding transcription factor activity and sequence-specific DNA binding |
| HOXA5 | Homeobox A5 | DNA-binding transcription factor activity and RNA polymerase II proximal promoter sequence-specific DNA binding |
| SREBF1 | Sterol Regulatory Element Binding Transcription Factor 1 | DNA-binding transcription factor activity and chromatin binding |
| TFAP2A | Transcription Factor AP-2 Alpha | DNA-binding transcription factor activity and sequence-specific DNA binding |
| STAT3 | Signal Transducer And Activator Of Transcription 3 | DNA-binding transcription factor activity and sequence-specific DNA binding |
| POU2F2 | POU Class 2 Homeobox 2 | DNA-binding transcription factor activity and protein domain specific binding |
| TP53 | Tumor Protein P53 | NA-binding transcription factor activity and protein heterodimerization activity |
| PPARG | Peroxisome Proliferator Activated Receptor Gamma | DNA-binding transcription factor activity and chromatin binding |
| JUN | Jun Proto-Oncogene, AP-1 Transcription Factor Subunit | sequence-specific DNA binding |
| (b) Regulatory microRNAs | ||
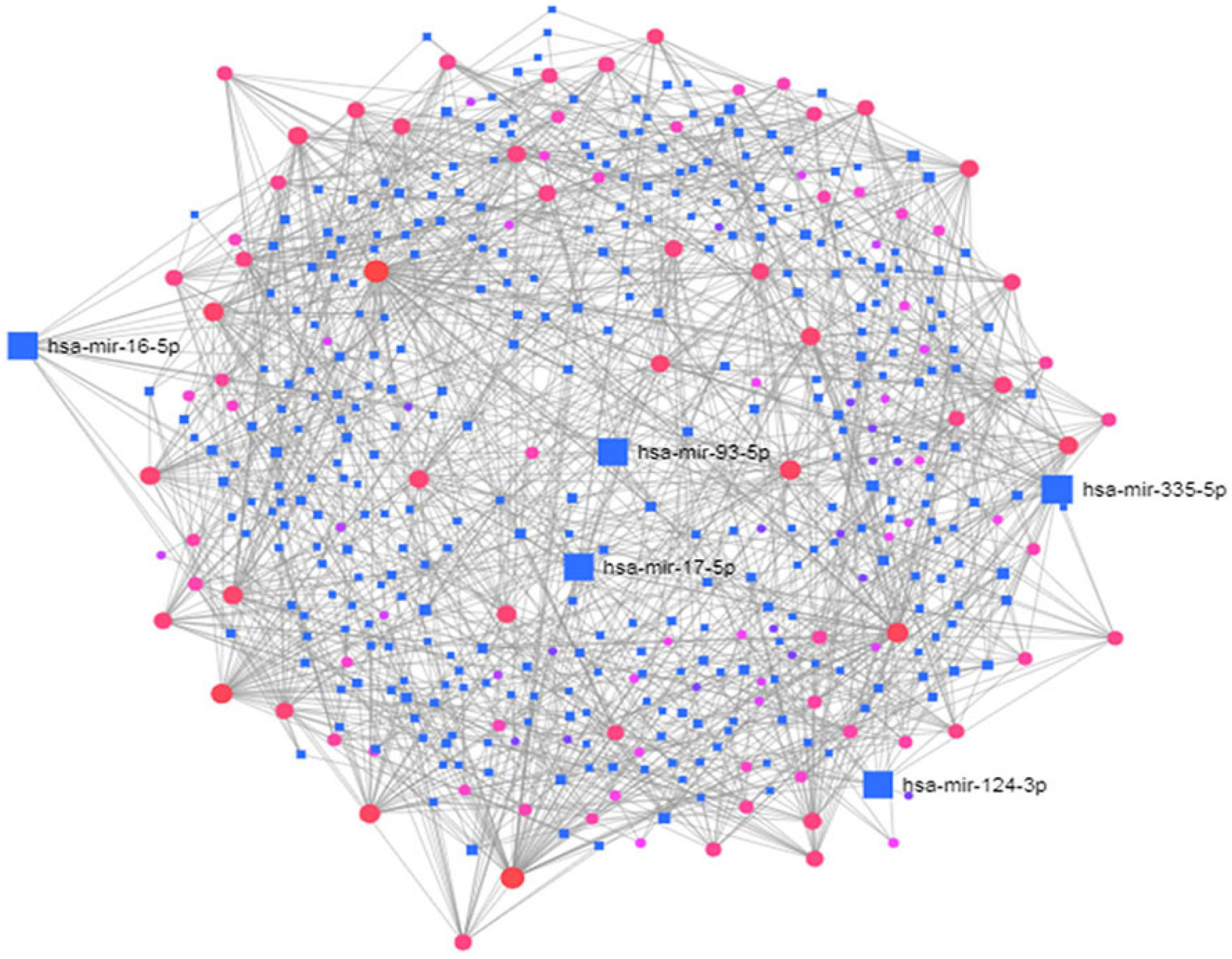
| Symbol | Description | Feature |
| mir-335-5p | MicroRNA 335 | Afflicted with Alzheimer’s disease |
| mir-16-5p | MicroRNA 16 | Afflicted with apoptosis of neural cells |
| mir-93-5p | MicroRNA 93 | Involved in DNA damage pathways |
| mir-17-5p | MicroRNA 17 | Act as oncogene or tumour suppressor gene depending on the cellular context |
| mir-124-3p | MicroRNA 124 | Abundant in the brain and involved in neurodegenerative disease |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, M.H.; Peng, S.; Hu, X.; Chen, C.; Rahman, M.R.; Uddin, S.; Quinn, J.M.W.; Moni, M.A. A Network-Based Bioinformatics Approach to Identify Molecular Biomarkers for Type 2 Diabetes that Are Linked to the Progression of Neurological Diseases. Int. J. Environ. Res. Public Health 2020, 17, 1035. https://doi.org/10.3390/ijerph17031035
Rahman MH, Peng S, Hu X, Chen C, Rahman MR, Uddin S, Quinn JMW, Moni MA. A Network-Based Bioinformatics Approach to Identify Molecular Biomarkers for Type 2 Diabetes that Are Linked to the Progression of Neurological Diseases. International Journal of Environmental Research and Public Health. 2020; 17(3):1035. https://doi.org/10.3390/ijerph17031035
Chicago/Turabian StyleRahman, Md Habibur, Silong Peng, Xiyuan Hu, Chen Chen, Md Rezanur Rahman, Shahadat Uddin, Julian M.W. Quinn, and Mohammad Ali Moni. 2020. "A Network-Based Bioinformatics Approach to Identify Molecular Biomarkers for Type 2 Diabetes that Are Linked to the Progression of Neurological Diseases" International Journal of Environmental Research and Public Health 17, no. 3: 1035. https://doi.org/10.3390/ijerph17031035
APA StyleRahman, M. H., Peng, S., Hu, X., Chen, C., Rahman, M. R., Uddin, S., Quinn, J. M. W., & Moni, M. A. (2020). A Network-Based Bioinformatics Approach to Identify Molecular Biomarkers for Type 2 Diabetes that Are Linked to the Progression of Neurological Diseases. International Journal of Environmental Research and Public Health, 17(3), 1035. https://doi.org/10.3390/ijerph17031035






