Activated Sludge Microbial Community and Treatment Performance of Wastewater Treatment Plants in Industrial and Municipal Zones
Abstract
1. Introduction
2. Materials and Methods
2.1. WWTP Operation Conditions and Sampling
2.2. DNA Extraction, PCR Amplification, and High-Throughput Sequencing
2.3. Taxonomy, Community Richness and Diversity, and Gene Function Prediction
2.4. Chemical Analysis
2.5. Statistical Analysis
3. Results
3.1. Pollutant Removal Efficiencies of the Three WWTPs
3.2. Microbial Community Structure Analysis
3.2.1. Microbial Community Richness and Diversity
3.2.2. Taxonomic Classification of the Bacterial Communities
3.3. KEGG Analysis of Dominant Taxonomic Groups and Related Metabolic Functions
3.4. Effects of Pollutant Concentration and Operation Condition on Microbial Communities
3.4.1. Pollutant Concentration
3.4.2. Operation Condition
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ye, C.; Yang, X.; Zhao, F.J.; Ren, L. The shift of the microbial community in activated sludge with calcium treatment and its implication to sludge settleability. Bioresour. Technol. 2016, 207, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Wen, X.; Zhang, B.; Yang, Y. Diversity and assembly patterns of activated sludge microbial communities: A review. Biotechnol. Adv. 2018, 36, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Zhang, H.; Han, L.; Mei, J.; Ge, Q.; Long, Z.; Yu, Y. Exploring bacterial communities and biodegradation genes in activated sludge from pesticide wastewater treatment plants via metagenomic analysis. Environ. Pollut. 2018, 243, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Peng, M.; Zhang, G.; Meng, F.; Zhang, Y.; Zou, Z. Effects of light-oxygen conditions on microbial community of photosynthetic bacteria during treating high-ammonia wastewater. Process Biochem. 2018, 72, 137–142. [Google Scholar] [CrossRef]
- Zeng, J.; Li, J.; Gou, M.; Xia, Z.-Y.; Sun, Z.-Y.; Tang, Y.-Q. Effective strategy for improving sludge treatment rate and microbial mechanisms during chromium bioleaching of tannery sludge. Process Biochem. 2019, 83, 159–167. [Google Scholar] [CrossRef]
- Feng, Q.; Sun, Y.; Wu, Y.; Xue, Z.; Luo, J.; Fang, F.; Li, C.; Cao, J. Physicochemical and Biological Effects on Activated Sludge Performance and Activity Recovery of Damaged Sludge by Exposure to CeO2 Nanoparticles in Sequencing Batch Reactors. Int. J. Environ. Res. Public Health 2019, 16, 4029. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Bu, X.; Chen, C.; Yang, X.; Lu, Y.; Liang, H.; Liu, M.; Lin, H.; Zhang, H.; Lin, Y.; et al. Impacts of sanitation improvement on reduction of nitrogen discharges entering the environment from human excreta in China. Sci. Total Environ. 2017, 593–594, 439–448. [Google Scholar] [CrossRef]
- Tong, Y.; Zhang, W.; Wang, X.; Couture, R.M.; Larssen, T.; Zhao, Y.; Li, J.; Liang, H.; Liu, X.; Bu, X.; et al. Decline in Chinese lake phosphorus concentration accompanied by shift in sources since 2006. Nat. Geosci. 2017, 10, 507–511. [Google Scholar] [CrossRef]
- Li, W.; Hua, T.; Zhou, Q.; Zhang, S.; Rong, W. Toxicity Identification and High-Efficiency Treatment of Aging Chemical Industrial Wastewater from the Hangu Reservoir, China. J. Environ. Qual. 2011, 40, 1714–1721. [Google Scholar] [CrossRef]
- Wu, P.; Jiang, L.Y.; He, Z.; Song, Y. Treatment of metallurgical industry wastewater for organic contaminant removal in China: Status, challenges, and perspectives. Environ. Sci. Water Res. Technol. 2017, 3, 1015–1031. [Google Scholar] [CrossRef]
- Elawwad, A.; Sandner, H.; Kappelmeyer, U.; Koeser, H. Long-term starvation and subsequent recovery of nitrifiers in aerated submerged fixed-bed biofilm reactors. Environ. Technol. 2013, 34, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Shahi, A.; Ozbayram, E.G.; Ince, B.; Ince, O. Use of PCR-DGGE based molecular methods to assessment of microbial diversity during anaerobic treatment of antibiotic combinations. Bioresour. Technol. 2015, 192, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Torà, J.A.; Lafuente, J.; Baeza, J.A.; Carrera, J. Combined effect of inorganic carbon limitation and inhibition by free ammonia and free nitrous acid on ammonia oxidizing bacteria. Bioresour. Technol. 2010, 101, 6051–6058. [Google Scholar] [CrossRef]
- Miao, Y.; Liao, R.; Zhang, X.X.; Liu, B.; Li, Y.; Wu, B.; Li, A. Metagenomic insights into salinity effect on diversity and abundance of denitrifying bacteria and genes in an expanded granular sludge bed reactor treating high-nitrate wastewater. Chem. Eng. J. 2015, 277, 116–123. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, S.; Wang, J.; Song, Q.; Zhang, W.; He, Q.; Song, J.; Ma, F. Comparison of performance and microbial communities in a bioelectrochemical system for simultaneous denitrification and chromium removal: Effects of pH. Process Biochem. 2018, 73, 154–161. [Google Scholar] [CrossRef]
- Zhao, L.; Ji, Y.; Sun, P.; Li, R.; Xiang, F.; Wang, H.; Ruiz-Martinez, J.; Yang, Y. Effects of individual and complex ciprofloxacin, fullerene C60, and ZnO nanoparticles on sludge digestion: Methane production, metabolism, and microbial community. Bioresour. Technol. 2018, 267, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Waste Water, 20th ed.; American Public Health Association: Washington, DC, USA, 1999. [Google Scholar]
- Long, S.; Zhao, L.; Liu, H.; Li, J.; Zhou, X.; Liu, Y.; Qiao, Z.; Zhao, Y.; Yang, Y. A Monte Carlo-based integrated model to optimize the cost and pollution reduction in wastewater treatment processes in a typical comprehensive industrial park in China. Sci. Total Environ. 2019, 647, 1–10. [Google Scholar] [CrossRef]
- Ibarbalz, F.M.; Orellana, E.; Figuerola, E.L.M.; Erijman, L. Shotgun metagenomic profiles have a high capacity to discriminate samples of activated sludge according to wastewater type. Appl. Environ. Microbiol. 2016, 82, 5186–5196. [Google Scholar] [CrossRef]
- Zhang, T.; Shao, M.F.; Ye, L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012, 6, 1137–1147. [Google Scholar] [CrossRef]
- Zhang, B.; Yu, Q.; Yan, G.; Zhu, H.; Xu, X.Y.; Zhu, L. Seasonal bacterial community succession in four typical wastewater treatment plants: Correlations between core microbes and process performance. Sci. Rep. 2018, 8, 4566. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.S.; Wells, G.F. Regional synchrony in full-scale activated sludge bioreactors due to deterministic microbial community assembly. ISME J. 2017, 11, 500–511. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, L.; Tian, X.; Yin, Y. Seasonal variation of bacterial community in biological aerated filter for ammonia removal in drinking water treatment. Water Res. 2017, 123, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Zhao, C.; Ma, B.; Li, S.; She, Z.; Guo, L.; Zhang, Q.; Zhao, Y.; Jin, C.; Gao, M. Impact of ampicillin on the nitrogen removal, microbial community and enzymatic activity of activated sludge. Bioresour. Technol. 2018, 272, 337–345. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Y.; Chen, X.; Li, Y. Effects of carbon sources on sludge performance and microbial community for 4-chlorophenol wastewater treatment in sequencing batch reactors. Bioresour. Technol. 2018, 255, 22–28. [Google Scholar] [CrossRef]
- Ou, D.; Li, H.; Li, W.; Wu, X.; Wang, Y.Q.; Liu, Y.D. Salt-tolerance aerobic granular sludge: Formation and microbial community characteristics. Bioresour. Technol. 2018, 249, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Vasiliadou, I.A.; Molina, R.; Martinez, F.; Melero, J.A.; Stathopoulou, P.M.; Tsiamis, G. Toxicity assessment of pharmaceutical compounds on mixed culture from activated sludge using respirometric technique: The role of microbial community structure. Sci. Total Environ. 2018, 630, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Ji, B.; Zhang, S.; Kong, Z. Study on the bacterial and archaeal community structure and diversity of activated sludge from three wastewater treatment plants. Mar. Pollut. Bull. 2018, 135, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Yang, L.; Wang, J. Denitrification performance and microbial diversity in a packed-bed bioreactor using PCL as carbon source and biofilm carrier. Appl. Microbiol. Biotechnol. 2013, 97, 2725–2733. [Google Scholar] [CrossRef]
- Tong, J.; Tang, A.; Wang, H.; Liu, X.; Huang, Z.; Wang, Z.; Zhang, J.; Wei, Y.; Su, Y.; Zhang, Y. Microbial community evolution and fate of antibiotic resistance genes along six different full-scale municipal wastewater treatment processes. Bioresour. Technol. 2019, 272, 489–500. [Google Scholar] [CrossRef]
- Xu, J.; He, J.; Wang, M.; Li, L. Cultivation and stable operation of aerobic granular sludge at low temperature by sieving out the batt-like sludge. Chemosphere 2018, 211, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Li, S.; Dong, Q.; Huang, X.; Liu, Y. Effect of blending landfill leachate with activated sludge on the domestic wastewater treatment process. Environ. Sci. Water Res. Technol. 2019, 5, 268–276. [Google Scholar] [CrossRef]
- Liu, X.; Yin, H.; Tang, S.; Feng, M.; Peng, H.; Lu, G.; Liu, Z.; Dang, Z. Effects of single and combined copper/perfluorooctane sulfonate on sequencing batch reactor process and microbial community in activated sludge. Bioresour. Technol. 2017, 238, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Ou, D.; Li, W.; Li, H.; Wu, X.; Li, C.; Zhuge, Y.; Liu, Y.D. Enhancement of the removal and settling performance for aerobic granular sludge under hypersaline stress. Chemosphere 2018, 212, 400–407. [Google Scholar] [CrossRef]
- Delforno, T.P.; Lacerda, G.V.; Sierra-Garcia, I.N.; Okada, D.Y.; Macedo, T.Z.; Varesche, M.B.A.; Oliveira, V.M. Metagenomic analysis of the microbiome in three different bioreactor configurations applied to commercial laundry wastewater treatment. Sci. Total Environ. 2017, 587–588, 389–398. [Google Scholar] [CrossRef]
- Chen, S.; Li, N.; Dong, B.; Zhao, W.; Dai, L.; Dai, X. New insights into the enhanced performance of high solid anaerobic digestion with dewatered sludge by thermal hydrolysis: Organic matter degradation and methanogenic pathways. J. Hazard. Mater. 2018, 342, 1–9. [Google Scholar] [CrossRef]
- More, R.P.; Mitra, S.; Raju, S.C.; Kapley, A.; Purohit, H.J. Mining and assessment of catabolic pathways in the metagenome of a common effluent treatment plant to induce the degradative capacity of biomass. Bioresour. Technol. 2014, 153, 137–146. [Google Scholar] [CrossRef]
- Liu, X.; Yang, X.; Hu, X.; He, Q.; Zhai, J.; Chen, Y.; Xiong, Q.; Vymazal, J. Comprehensive metagenomic analysis reveals the effects of silver nanoparticles on nitrogen transformation in constructed wetlands. Chem. Eng. J. 2019, 358, 1552–1560. [Google Scholar] [CrossRef]
- Li, X.; Meng, D.; Li, J.; Yin, H.; Liu, H.; Liu, X.; Cheng, C.; Xiao, Y.; Liu, Z.; Yan, M. Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environ. Pollut. 2017, 231, 908–917. [Google Scholar] [CrossRef]
- Lu, M.; Luo, X.; Jiao, J.J.; Li, H.; Wang, X.; Gao, J.; Zhang, X.; Xiao, K. Nutrients and heavy metals mediate the distribution of microbial community in the marine sediments of the Bohai Sea, China. Environ. Pollut. 2019, 255, 113069. [Google Scholar] [CrossRef]
- Guo, H.; Nasir, M.; Lv, J.; Dai, Y.; Gao, J. Understanding the variation of microbial community in heavy metals contaminated soil using high throughput sequencing. Ecotoxicol. Environ. Saf. 2017, 144, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Majeed, S.; Xu, R.; Zhang, K.; Kakade, A.; Khan, A.; Hafeez, F.Y.; Mao, C.; Liu, P.; Li, X. Heavy metals interact with the microbial community and affect biogas production in anaerobic digestion: A review. J. Environ. Manag. 2019, 240, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, Y.; Xu, T.; Zheng, K.; Zhang, R.; Peng, Z.; Zhang, H. Toxic effects of CuO, ZnO and TiO2 nanoparticles in environmental concentration on the nitrogen removal, microbial activity and community of Anammox process. Chem. Eng. J. 2018, 332, 42–48. [Google Scholar] [CrossRef]
- Miura, Y.; Watanabe, Y.; Okabe, S. Membrane biofouling in pilot-scale membrane bioreactors (MBRs) treating municipal wastewater: Impact of biofilm formation. Environ. Sci. Technol. 2007, 41, 632–638. [Google Scholar] [CrossRef]
- Jo, S.J.; Kwon, H.; Jeong, S.Y.; Lee, C.H.; Kim, T.G. Comparison of microbial communities of activated sludge and membrane biofilm in 10 full-scale membrane bioreactors. Water Res. 2016, 101, 214–225. [Google Scholar] [CrossRef]
- Han, H.; Zhang, Y.; Cui, C.; Zheng, S. Effect of COD level and HRT on microbial community in a yeast-predominant activated sludge system. Bioresour. Technol. 2010, 101, 3463–3465. [Google Scholar] [CrossRef]
- Gao, P.; Xu, W.; Sontag, P.; Li, X.; Xue, G.; Liu, T.; Sun, W. Correlating microbial community compositions with environmental factors in activated sludge from four full-scale municipal wastewater treatment plants in Shanghai, China. Appl. Microbiol. Biotechnol. 2016, 100, 4663–4673. [Google Scholar] [CrossRef]
- Zhang, Q.; Ji, F.Y.; Fu, X.F.; Chen, Q.K. Effects of Gas/Water Ratio on the Characteristics of Nitrogen Removal and the Microbial Community in Post Solid-Phase Denitrification Biofilter Process. Huanjing Kexue Environ. Sci. 2018, 39, 3297–3305. [Google Scholar]
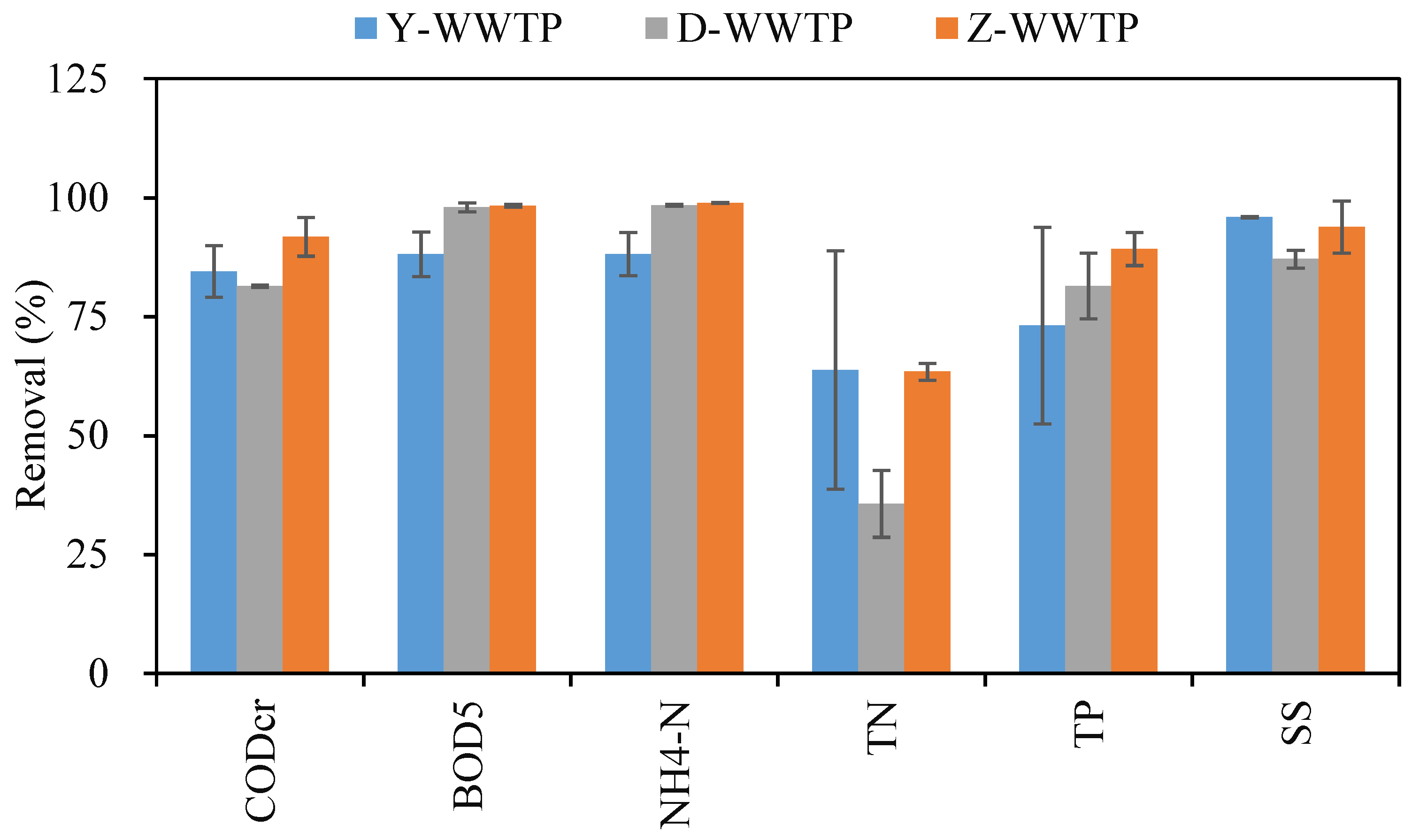



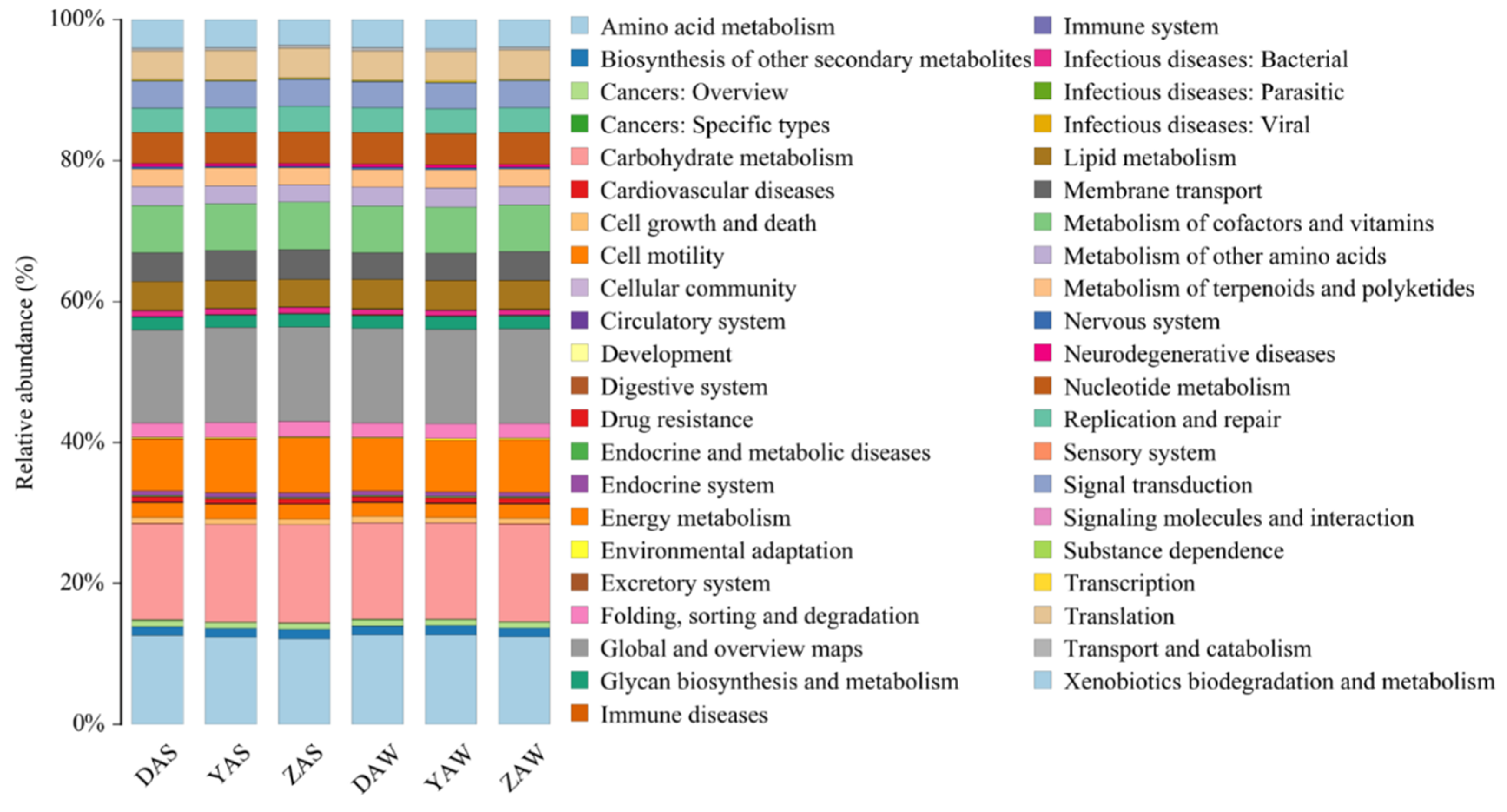

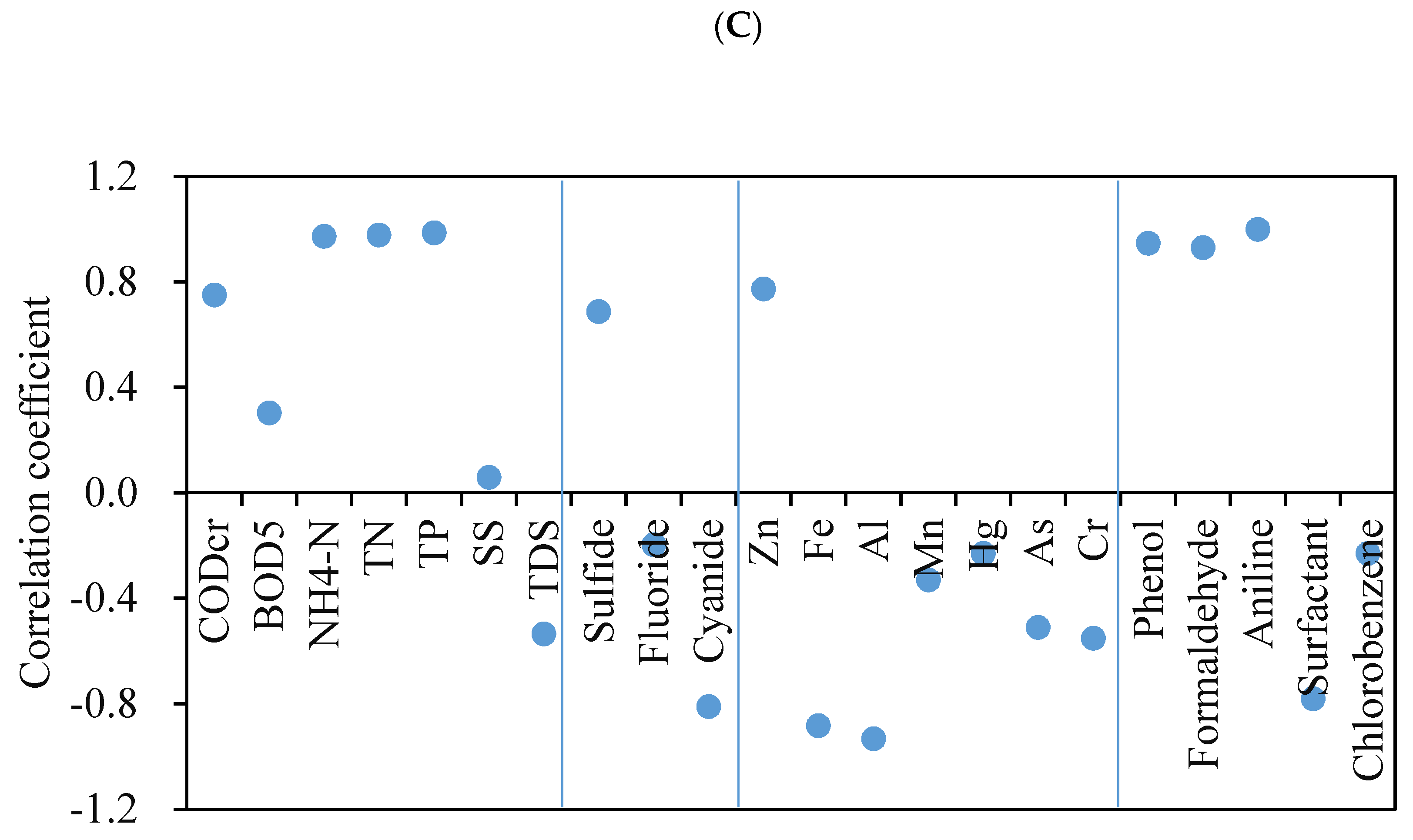
| Pollutants | D-WWTP | Y-WWTP | Z-WWTP |
|---|---|---|---|
| CODcr (mg/L) | 185.17 | 221.00 | 234.00 |
| BOD5 (mg/L) | 47.87 | 49.64 | 41.80 |
| NH4-N (mg/L) | 13.87 | 26.28 | 36.60 |
| TN (mg/L) | 19.20 | 39.81 | 45.60 |
| TP (mg/L) | 2.40 | 3.95 | 4.36 |
| Suspended solids (mg/L) | 45.67 | 88.17 | 63.00 |
| Total dissolved solids (mg/L) | 2578.33 | 3748.33 | 770.00 |
| Sulfide (mg/L) | 3.322 | 0.922 | 8.860 |
| Fluoride (mg/L) | 1.170 | 0.882 | 1.080 |
| Cyanide (μg/L) | 4.3 | 61.7 | 4.0 |
| Zn (mg/L) | 0.192 | 0.126 | 0.428 |
| Fe (mg/L) | 3.128 | 3.329 | 0.622 |
| Al (mg/L) | 1.413 | 1.519 | 0.172 |
| Mn (mg/L) | 0.293 | 0.323 | 0.180 |
| Hg (μg/L) | 0.038 | 0.248 | 0.040 |
| As (μg/L) | 0.600 | 0.783 | 0.500 |
| Cr (mg/L) | 0.030 | 0.060 | 0.004 |
| Phenol (mg/L) | 0.014 | 0.015 | 0.028 |
| Formaldehyde (mg/L) | 0.057 | 0.050 | 0.140 |
| Aniline (mg/L) | 0.082 | 0.062 | 0.220 |
| Surfactant (mg/L) | 0.834 | 0.473 | 0.170 |
| Chlorobenzene (μg/L) | 1.000 | 159.867 | 1.000 |
| Sampling Date | Sludge Sampling Site | OTU | ACE | Chao1 | Shannon | Simpson |
|---|---|---|---|---|---|---|
| D-WWTP | 873 a | 956 a | 985 a | 5.694 b | 0.007 c | |
| November. | Y-WWTP | 759 b | 864 b | 871 b | 5.344 c | 0.010 a |
| Z-WWTP | 888 a | 945 a | 957 a | 5.763 a | 0.008 b | |
| D-WWTP | 962 a | 1010 a | 1020 a | 5.271 c | 0.024 a | |
| September. | Y-WWTP | 921 b | 989 a | 1022 a | 5.372 b | 0.011 b |
| Z-WWTP | 968 a | 1007 a | 1017 a | 5.612 a | 0.010 b |
| Bacteria | Sludge Sample (Summer) | Sludge Sample (Winter) | ||||
|---|---|---|---|---|---|---|
| DAS | YAS | ZAS | DAW | YAW | ZAW | |
| Acidobacteria | A | B | C | AB | A | B |
| Actinobacteria | B | A | B | B | A | A |
| Aminicenantes | B | A | B | A | A | A |
| Armatimonadetes | A | C | B | A | B | B |
| BRC1 | B | A | A | B | A | B |
| Bacteroidetes | A | B | A | A | B | C |
| Chlamydiae | B | C | A | A | C | B |
| Chlorobi | B | C | A | B | C | A |
| Chloroflexi | C | B | A | B | C | A |
| Cyanobacteria | A | C | B | A | C | B |
| Deferribacteres | A | B | B | A | A | A |
| Elusimicrobia | B | B | A | B | B | A |
| Fibrobacteres | A | B | A | A | C | B |
| Firmicutes | C | A | B | B | B | A |
| Fusobacteria | A | A | A | A | A | A |
| Gemmatimonadetes | B | A | A | C | A | B |
| Gracilibacteria | A | A | B | A | C | B |
| Hydrogenedentes | A | B | B | A | B | B |
| Ignavibacteriae | C | B | A | B | A | A |
| Latescibacteria | B | B | A | A | A | B |
| Microgenomates | A | B | B | A | B | B |
| Nitrospirae | C | A | B | B | C | A |
| Parcubacteria | C | B | A | B | C | A |
| Peregrinibacteria | A | A | A | B | A | B |
| Planctomycetes | AB | B | A | A | A | B |
| Proteobacteria | A | B | C | B | C | A |
| RBG-1[Zixibacteria] | B | B | A | B | B | A |
| SR1[Absconditabacteria] | B | B | A | A | A | A |
| Saccharibacteria | B | A | B | B | A | C |
| Spirochaetae | B | B | A | B | C | A |
| Synergistetes | A | A | A | A | A | A |
| TM6[Dependentiae] | A | C | B | A | AB | B |
| Verrucomicrobia | A | B | A | A | A | A |
| Class 1 | Class 2 | DAS | YAS | ZAS | DAW | YAW | ZAW |
|---|---|---|---|---|---|---|---|
| Metabolism | Carbohydrate metabolism | A | A | A | A | A | A |
| Metabolism | Lipid metabolism | A | A | A | A | A | A |
| Metabolism | Metabolism of cofactors and vitamins | A | A | A | A | A | A |
| Metabolism | Energy metabolism | A | A | A | A | A | A |
| Metabolism | Nucleotide metabolism | A | A | A | A | A | A |
| Metabolism | Biosynthesis of other secondary metabolites | A | A | A | A | A | A |
| Metabolism | Metabolism of terpenoids and polyketides | A | A | A | A | A | A |
| Metabolism | Glycan biosynthesis and metabolism | AB | B | A | A | A | A |
| Metabolism | Global and overview maps | A | A | A | A | A | A |
| Metabolism | Amino acid metabolism | A | A | A | A | AB | B |
| Metabolism | Xenobiotics biodegradation and metabolism | A | A | B | A | A | B |
| Metabolism | Metabolism of other amino acids | A | AB | B | A | AB | B |
| Environmental Information Processing | Membrane transport | A | A | A | A | A | A |
| Environmental Information Processing | Signal transduction | A | A | A | A | A | A |
| Cellular Processes | Cell motility | A | A | A | A | A | A |
| Cellular Processes | Transport and catabolism | A | A | A | A | A | A |
| Cellular Processes | Cell growth and death | A | B | B | A | A | B |
| Cellular Processes | Cellular community | A | B | C | AB | B | A |
| Genetic Information Processing | Folding, sorting and degradation | B | AB | A | A | A | A |
| Genetic Information Processing | Transcription | A | A | A | A | A | A |
| Genetic Information Processing | Translation | A | A | A | A | A | A |
| Genetic Information Processing | Replication and repair | A | A | A | A | A | A |
| Organismal Systems | Endocrine system | A | A | A | AB | A | B |
| Organismal Systems | Circulatory system | A | A | A | A | A | A |
| Organismal Systems | Immune system | B | B | A | A | A | A |
| Organismal Systems | Environmental adaptation | A | B | B | A | AB | B |
| Organismal Systems | Nervous system | A | B | B | A | A | B |
| Organismal Systems | Sensory system | A | B | B | A | B | A |
| Organismal Systems | Excretory system | B | B | A | AB | A | B |
| Organismal Systems | Digestive system | A | B | AB | A | A | B |
| Human Diseases | Drug resistance | A | A | A | A | AB | B |
| Human Diseases | Endocrine and metabolic diseases | A | A | A | A | A | A |
| Human Diseases | Cancers: Overview | A | A | A | A | A | A |
| Human Diseases | Infectious diseases: Bacterial | A | A | A | A | A | A |
| Human Diseases | Neurodegenerative diseases | A | B | B | A | AB | B |
| Human Diseases | Substance dependence | A | B | B | A | B | C |
| Human Diseases | Infectious diseases: Parasitic | A | C | B | A | B | B |
| Human Diseases | Infectious diseases: Viral | A | B | B | A | B | B |
| Human Diseases | Cancers: Specific types | A | B | B | A | A | A |
| Human Diseases | Immune diseases | A | B | B | A | A | B |
| Human Diseases | Cardiovascular diseases | A | B | C | A | B | B |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Wang, L.; Xiang, F.; Zhao, L.; Qiao, Z. Activated Sludge Microbial Community and Treatment Performance of Wastewater Treatment Plants in Industrial and Municipal Zones. Int. J. Environ. Res. Public Health 2020, 17, 436. https://doi.org/10.3390/ijerph17020436
Yang Y, Wang L, Xiang F, Zhao L, Qiao Z. Activated Sludge Microbial Community and Treatment Performance of Wastewater Treatment Plants in Industrial and Municipal Zones. International Journal of Environmental Research and Public Health. 2020; 17(2):436. https://doi.org/10.3390/ijerph17020436
Chicago/Turabian StyleYang, Yongkui, Longfei Wang, Feng Xiang, Lin Zhao, and Zhi Qiao. 2020. "Activated Sludge Microbial Community and Treatment Performance of Wastewater Treatment Plants in Industrial and Municipal Zones" International Journal of Environmental Research and Public Health 17, no. 2: 436. https://doi.org/10.3390/ijerph17020436
APA StyleYang, Y., Wang, L., Xiang, F., Zhao, L., & Qiao, Z. (2020). Activated Sludge Microbial Community and Treatment Performance of Wastewater Treatment Plants in Industrial and Municipal Zones. International Journal of Environmental Research and Public Health, 17(2), 436. https://doi.org/10.3390/ijerph17020436






