Spatial Distribution of Toxic Metal(loid)s and Microbial Community Analysis in Soil Vertical Profile at an Abandoned Nonferrous Metal Smelting Site
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Soil Sampling
2.2. Soil Properties and Metal(loid)s Determination
2.3. DNA Extraction, PCR Amplification, and High Throughput Sequencing
2.4. Data Process and Statistical Analyses
3. Results and Discussion
3.1. Soils Characteristics and Vertical Distribution of Metal(loid)s
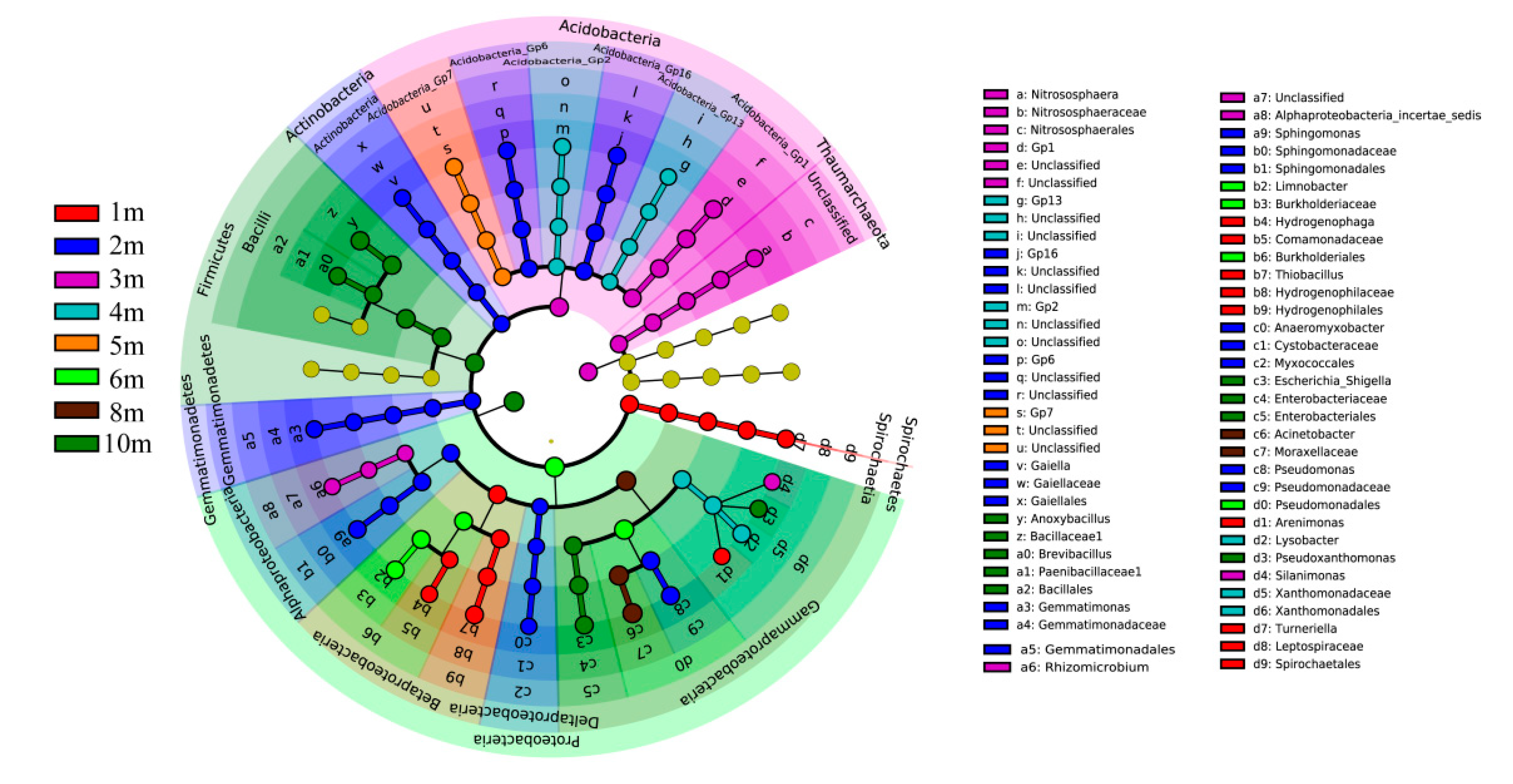
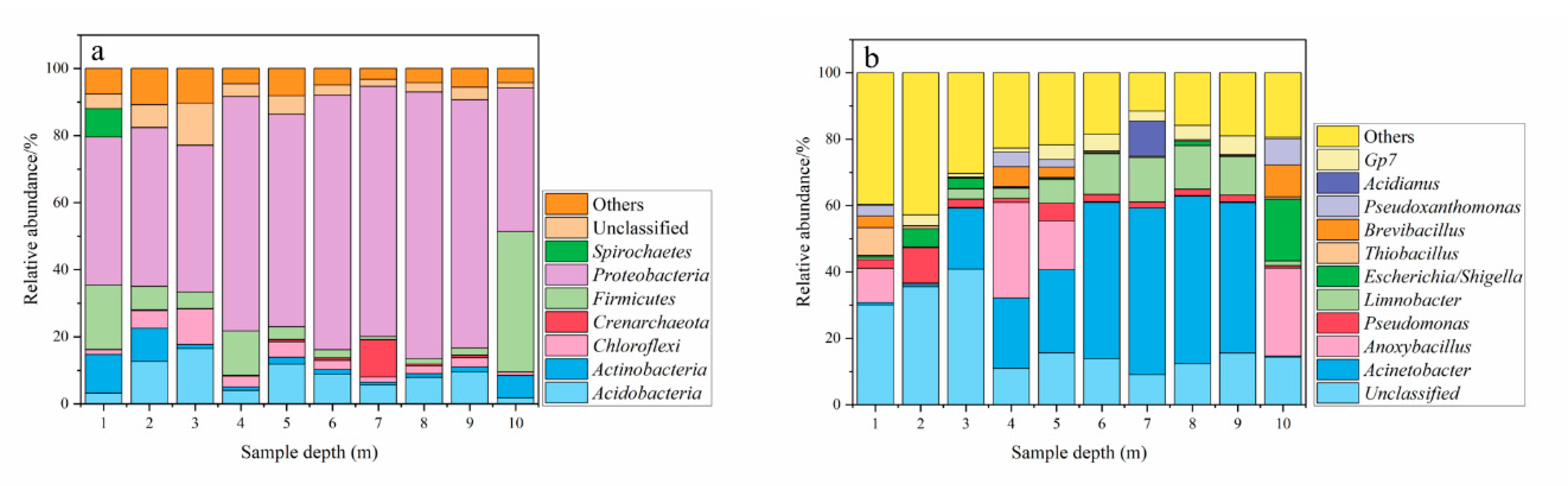
3.2. Alpha-Diversity and Composition of Archaea and Bacterial Communities
3.3. Relationship between the Environmental Variables and Microbial Communities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Singh, B.K. Chapter 1—Soil Carbon: Introduction, Importance, Status, Threat, and Mitigation. In Soil Carbon Storage; Academic Press: Cambridge, MA, USA, 2018; pp. 1–28. [Google Scholar]
- Race, M.; Marotta, R.; Fabbricino, M.; Pirozzi, F.; Andreozzi, R.; Guida, M.; Siciliano, A. Assessment of optimal conditions for the restoration and recovery of agricultural soil. J. Hazard. Mater. 2019, 373, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Kasemodel, M.; Sakamoto, I.; Varesche, M.; Rodrigues, V. Potentially toxic metal contamination and microbial community analysis in an abandoned Pb and Zn mining waste deposit. Sci. Total Environ. 2019, 675, 367–379. [Google Scholar] [CrossRef] [PubMed]
- MEP. China Soil Pollution Survey Communique in Chinese [EB/OL]. 110998. 2014. Available online: http://www.mee.gov.cn/gkml/sthjbgw/qt/201404/t20140417_270670.htm (accessed on 17 April 2014).
- Luo, Y.; Wu, Y.; Shu, J.; Wu, Z. Effect of particulate organic matter fractions on the distribution of heavy metals with aided phytostabilization at a zinc smelting waste slag site. Environ. Pollut. 2019, 253, 330–341. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Wang, X.; Lei, J.; Ran, Q.; Ren, Y.; Zhou, J. Spatial distribution and source identification of heavy metals in a typical Pb/Zn smelter in an arid area of northwest China. Hum. Ecol. Risk Assess. Int. J. 2019, 25, 1661–1687. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, X.; Yin, H.; Liang, Y.; Liu, H.; Miao, B.; Peng, Q.; Meng, D.; Wang, S.; Yang, J.; et al. The utilization of biomineralization technique based on microbial induced phosphate precipitation in remediation of potentially toxic ions contaminated soil: A mini review. Ecotoxicol. Environ. Saf. 2020, 191, 110009. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, L.; Xu, L.; Hao, X.; Zhang, S.; Xu, M.; Zhu, P.; Fu, S.; Liang, Y.; Yin, H.; et al. Spatial distribution and risk assessment of heavy metals in contaminated paddy fields—A case study in Xiangtan City, southern China. Ecotoxicol. Environ. Saf. 2019, 171, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Adedeji, O.H.; Olayinka, O.O.; Tope-Ajayi, O.O. Spatial distribution and health risk assessment of soil pollution by heavy metals in Ijebu-Ode, Nigeria. J. Health Pollut. 2019, 9, 190601. [Google Scholar] [CrossRef]
- Naderi, A.; Delavar, M.A.; Kaboudin, B.; Askari, M.S. Erratum to: Assessment of spatial distribution of soil heavy metals using ANN-GA, MSLR and satellite imagery. Environ. Monit. Assess. 2017, 189, 291. [Google Scholar] [CrossRef]
- Ding, Q.; Cheng, G.; Wang, Y.; Zhuang, D. Effects of natural factors on the spatial distribution of heavy metals in soils surrounding mining regions. Sci. Total Environ. 2017, 578, 577–585. [Google Scholar] [CrossRef]
- Qu, M.; Li, W.; Zhang, C. Spatial distribution and uncertainty assessment of potential ecological risks of heavy metals in soil using sequential gaussian simulation. Hum. Ecol. Risk Assess. Int. J. 2014, 20, 764–778. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, S.; Xiao, L.; Zhong, Q.; Li, L.; Xu, G.; Deng, O.; Pu, Y. Heavy metals in soils from a typical industrial area in Sichuan, China: Spatial distribution, source identification, and ecological risk assessment. Environ. Sci. Pollut. Res. 2017, 24, 16618–16630. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, N.; Hooke, A.M. Toxicity of heavy metals for microorganisms isolated from slow sand filter schmutzdecke. Environ. Technol. 2003, 24, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.K.; Singh, N.R.; Kumar, U.; Mishra, S.R.; Perumal, R.C.; Benny, J.; Thatoi, H. Illumina MiSeq based assessment of bacterial community structure and diversity along the heavy metal concentration gradient in Sukinda chromite mine area soils, India. Ecol. Genet. Genom. 2020, 15, 100054. [Google Scholar] [CrossRef]
- Li, X.; Meng, D.; Li, J.; Yin, H.; Liu, H.; Liu, X.; Cheng, C.; Xiao, Y.; Liu, Z.; Yan, M. Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environ. Pollut. 2017, 231, 908–917. [Google Scholar] [CrossRef]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.M.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef]
- Van Wesemael, B.; Poesen, J.; de Figueiredo, T. Effects of rock fragments on physical degradation of cultivated soils by rainfall. Soil Tillage Res. 1995, 33, 229–250. [Google Scholar] [CrossRef]
- Hao, X.; Zhu, P.; Zhang, H.; Liang, Y.; Yin, H.; Liu, X.; Bai, L.; Liu, H.; Jiang, H. Mixotrophic acidophiles increase cadmium soluble fraction and phytoextraction efficiency from cadmium contaminated soils. Sci. Total Environ. 2019, 655, 347–355. [Google Scholar] [CrossRef]
- Wei, W.; Yang, M.; Liu, Y.; Huang, H.; Ye, C.; Zheng, J.; Guo, C.; Hao, M.; He, X.; Zhu, S. Fertilizer N application rate impacts plant-soil feedback in a sanqi production system. Sci. Total Environ. 2018, 633, 796–807. [Google Scholar] [CrossRef]
- Pueyo, M.; Mateu, J.; Rigol, A.; Vidal, M.; Lopezsanchez, J.F.; Rauret, G. Use of the modified BCR three-step sequential extraction procedure for the study of trace element dynamics in contaminated soils. Environ. Pollut. 2008, 152, 330–341. [Google Scholar] [CrossRef]
- Li, Z.W.; Zhang, Q.; Fang, Y.; Yang, X.C.; Yuan, Q.S. Examining social-economic factors in spatial and temporal change of water quality in red soil hilly region of South China: A case study in Hunan Province. Int. J. Environ. Pollut. 2010, 42, 184–198. [Google Scholar] [CrossRef]
- Hossain, S.; Ishiyama, T.; Hachinohe, S.; Oguchi, C.T. Leaching Behavior of As, Pb, Ni, Fe, and Mn from subsurface marine and nonmarine depositional environment in Central Kanto Plain, Japan. Geoences 2019, 9, 435. [Google Scholar] [CrossRef]
- Jetten, M.S.M. New pathways for ammonia conversion in soil and aquatic systems. Plant Soil 2001, 230, 9–19. [Google Scholar] [CrossRef]
- Li, T.; Liu, Y.; Lin, S.; Liu, Y.; Xie, Y. Soil pollution management in China: A brief introduction. Sustainability 2019, 11, 556. [Google Scholar] [CrossRef]
- Liu, B.; Ai, S.; Zhang, W.; Huang, D.; Zhang, Y. Assessment of the bioavailability, bioaccessibility and transfer of heavy metals in the soil-grain-human systems near a mining and smelting area in NW China. Sci. Total Environ. 2017, 609, 822–829. [Google Scholar] [CrossRef]
- Yuan, C.; Zhang, L.; Wang, J.; Teng, W.; Hu, H.; Shen, J.; He, J. Limited effects of depth (0–80 cm) on communities of archaea, bacteria and fungi in paddy soil profiles. Eur. J. Soil Sci. 2020. [Google Scholar] [CrossRef]
- Kim, H.M.; Lee, M.J.; Jung, J.Y.; Hwang, C.Y.; Kim, M.; Ro, H.-M.; Chun, J.; Lee, Y.K. Vertical distribution of bacterial community is associated with the degree of soil organic matter decomposition in the active layer of moist acidic tundra. J. Microbiol. 2016, 54, 713–723. [Google Scholar] [CrossRef]
- Beattie, R.E.; Henke, W.; Campa, M.F.; Hazen, T.C.; Mcaliley, L.R.; Campbell, J.H. Variation in microbial community structure correlates with heavy-metal contamination in soils decades after mining ceased. Soil Biol. Biochem. 2018, 126, 57–63. [Google Scholar] [CrossRef]
- Jin, H.U.; Meng, D.; Liu, X.; Liang, Y.; Yin, H.; Liu, H. Response of soil fungal community to long-term chromium contamination. Trans. Nonferrous Met. Soc. China 2018, 28, 1838–1846. [Google Scholar]
- Spain, A.M.; Krumholz, L.R.; Elshahed, M.S. Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 2009, 3, 992–1000. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.; Zhang, W.; Wang, H.; Fu, X.; Le, Y. The variability of soil microbial community composition of different types of tidal wetland in Chongming Dongtan and its effect on soil microbial respiration. Ecol. Eng. 2011, 37, 1276–1282. [Google Scholar] [CrossRef]
- Navarro, A.; Faria, M.; Barata, C.; Pina, B. Transcriptional response of stress genes to metal exposure in zebra mussel larvae and adults. Environ. Pollut. 2011, 159, 100–107. [Google Scholar] [CrossRef]
- Spang, A.; Poehlein, A.; Offre, P.; Zumbragel, S.; Haider, S.; Rychlik, N.; Nowka, B.; Schmeisser, C.; Lebedeva, E.V.; Rattei, T. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: Insights into metabolic versatility and environmental adaptations. Environ. Microbiol. 2012, 14, 3122–3145. [Google Scholar] [CrossRef]
- Mangold, S.; Potrykus, J.; Bjorn, E.; Lovgren, L.; Dopson, M. Extreme zinc tolerance in acidophilic microorganisms from the bacterial and archaeal domains. Extremophiles 2013, 17, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Albalawi, T.H.A.; Wood, A.L.; Solis, A.; Cooper, T.; Barabote, R.D. Anoxybacillus sp. strain UARK-01, a new thermophilic soil bacterium with hyperthermostable alkaline laccase activity. Curr. Microbiol. 2017, 74, 762–771. [Google Scholar]
- Luo, L.Y.; Xie, L.L.; Jin, D.C.; Mi, B.B.; Wang, D.H.; Li, X.F.; Dai, X.Z.; Zou, X.X.; Zhang, Z.; Ma, Y.Q.; et al. Bacterial community response to cadmium contamination of agricultural paddy soil. Appl. Soil Ecol. 2019, 139, 100–106. [Google Scholar] [CrossRef]
- Hemmatjou, M.H.; Safarisinegani, A.A.; Mirzaieasl, A.; Tahmourespour, A. Analysis of microbial communities in heavy metals-contaminated soils using the metagenomic approach. Ecotoxicology 2018, 27, 1281–1291. [Google Scholar] [CrossRef]
- Zhao, X.; Huang, J.; Lu, J.; Sun, Y. Study on the influence of soil microbial community on the long-term heavy metal pollution of different land use types and depth layers in mine. Ecotoxicol. Environ. Saf. 2019, 170, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Niu, J.; Ren, Y.; Cong, J.; Zhang, X.; Fan, F.; Xiao, Y.; Zhang, X.; Deng, J.; Xie, M. An integrated insight into the response of sedimentary microbial communities to heavy metal contamination. Sci. Rep. 2015, 5, 14266. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Su, G.; Yang, Y.; Yao, Y.; Huang, Y.; Hu, L.; Zhong, H.; He, Z. Vertical distribution of microbial communities in chromium-contaminated soil and isolation of Cr(VI)—Reducing strains. Ecotoxicol. Environ. Saf. 2019, 180, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Yu, Z.; Ji, J.; Khan, A.; Li, X. Microbial community structure and function indicate the severity of chromium contamination of the Yellow river. Front. Microbiol. 2018, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Rossel, R.A.V.; Li, S.; Bissett, A.; Lee, J.; Shi, Z.; Behrens, T.; Court, L.N. Soil bacterial abundance and diversity better explained and predicted with spectro-transfer functions. Soil Biol. Biochem. 2019, 129, 29–38. [Google Scholar] [CrossRef]
- Wolinska, A.; Kuźniar, A.; Zielenkiewicz, U.; Izak, D.; Szafraneknakonieczna, A.; Banach, A.; Blaszczyk, M. Bacteroidetes as a sensitive biological indicator of agricultural soil usage revealed by a culture-independent approach. Appl. Soil Ecol. 2017, 119, 128–137. [Google Scholar] [CrossRef]
- Solyanikova, I.P.; Suzina, N.E.; Golovleva, L.A. The role of non-spore-forming actinobacteria in cleaning up sites contaminated by persistent pollutants and the ability of these microorganisms to survive under unfavourable conditions. Microbiol. Aust. 2018, 39, 141–144. [Google Scholar] [CrossRef]
- Jeong, H.; Rha, E.; Kim, H.; Lee, S.-G. Complete genome sequence of the soil bacterium Pseudomonas kribbensis strain 46-2(T). Microbiol. Resour. Announc. 2018, 7, e01161-18. [Google Scholar] [CrossRef]
- Zhang, C.; Tayyab, M.; Abubakar, A.Y.; Yang, Z.; Pang, Z.; Islam, W.; Lin, Z.; Li, S.; Luo, J.; Fan, X. Bacteria with different assemblages in the soil profile drive the diverse nutrient cycles in the sugarcane straw retention ecosystem. Diversity 2019, 11, 194. [Google Scholar] [CrossRef]
- Kemmitt, S.J.; Wright, D.L.; Goulding, K.W.T.; Jones, D.L. pH regulation of carbon and nitrogen dynamics in two agricultural soils. Soil Biol. Biochem. 2006, 38, 898–911. [Google Scholar] [CrossRef]
- Peralta, A.L.; Ludmer, S.; Matthews, J.W.; Kent, A.D. Bacterial community response to changes in soil redox potential along a moisture gradient in restored wetlands. Ecol. Eng. 2014, 73, 246–253. [Google Scholar] [CrossRef]
- Cui, B.; Zhao, H.; Li, X.; Zhang, K.; Ren, H.; Bai, J. Temporal and spatial distributions of soil nutrients in Hani terraced paddy fields, Southwestern China. Procedia Environ. Sci. 2010, 2, 1032–1042. [Google Scholar] [CrossRef][Green Version]
- Baz, S.E.; Baz, M.; Barakate, M.; Hassani, L.; Gharmali, A.E.; Imziln, B. Resistance to and accumulation of heavy metals by actinobacteria isolated from abandoned mining areas. Sci. World J. 2015, 2015, 761834. [Google Scholar]
- Huaidong, H.E.; Waichin, L.I.; Riqing, Y.U.; Zhihong, Y.E. Illumina-based analysis of bulk and rhizosphere soil bacterial communities in paddy fields under mixed heavy metal contamination. Pedosphere 2017, 27, 569–578. [Google Scholar]
- Liu, J.; Li, C.; Jing, J.; Zhao, P.; Luo, Z.; Cao, M.; Ma, Z.; Jia, T.; Chai, B. Ecological patterns and adaptability of bacterial communities in alkaline copper mine drainage. Water Res. 2018, 133, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhang, Y.; Zhang, X.; Qian, L.; Sun, M.; Yang, Y.; Zhang, Y.; Wang, J.; Kim, H.; Qiu, G. The dissolution and passivation mechanism of chalcopyrite in bioleaching: An overview. Miner. Eng. 2019, 136, 140–154. [Google Scholar] [CrossRef]
- Wu, Z.; Gao, G.; Wang, Y. Effects of soil properties, heavy metals, and PBDEs on microbial community of e-waste contaminated soil. Ecotoxicol. Environ. Saf. 2019, 180, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Kamitani, T.; Oba, H.; Kaneko, N. Microbial biomass and tolerance of microbial community on an aged heavy metal polluted floodplain in Japan. Water Air Soil Pollut. 2006, 172, 185–200. [Google Scholar] [CrossRef]
- Pan, X.; Zhang, S.; Zhong, Q.; Gong, G.; Wang, G.; Guo, X.; Xu, X. Effects of soil chemical properties and fractions of Pb, Cd, and Zn on bacterial and fungal communities. Sci. Total Environ. 2020, 715, 136904. [Google Scholar] [CrossRef]
- García-Carmona, M.; Romero-Freire, A.; Sierra Aragón, M.; Martín Peinado, F.J. Effectiveness of ecotoxicological tests in relation to physicochemical properties of Zn and Cu polluted Mediterranean soils. Geoderma 2019, 338, 259–268. [Google Scholar] [CrossRef]
- Pérez-Esteban, J.; Escolástico, C.; Masaguer, A.; Moliner, A. Effects of sheep and horse manure and pine bark amendments on metal distribution and chemical properties of contaminated mine soils. Eur. J. Soil Sci. 2012, 63, 733–742. [Google Scholar] [CrossRef]
- Mei, J.; He, H.; Hong, F.F.; Leng, Y.; Zhao, Y.B.; Zheng, L.; Ma, C.; Tao, X. Microbial community structures and sulfur speciation characteristics in soil sample around the Xiang-tan Liejiaqiao coal gangue dump, Hunan Province in South of China. Energy Sources Part A Recovery Util. Environ. Eff. 2019, 1–11. [Google Scholar] [CrossRef]
- Kumar, R.N.; Nagendran, R. Influence of initial pH on bioleaching of heavy metals from contaminated soil employing indigenous Acidithiobacillus thiooxidans. Chemosphere 2007, 66, 1775–1781. [Google Scholar] [CrossRef]
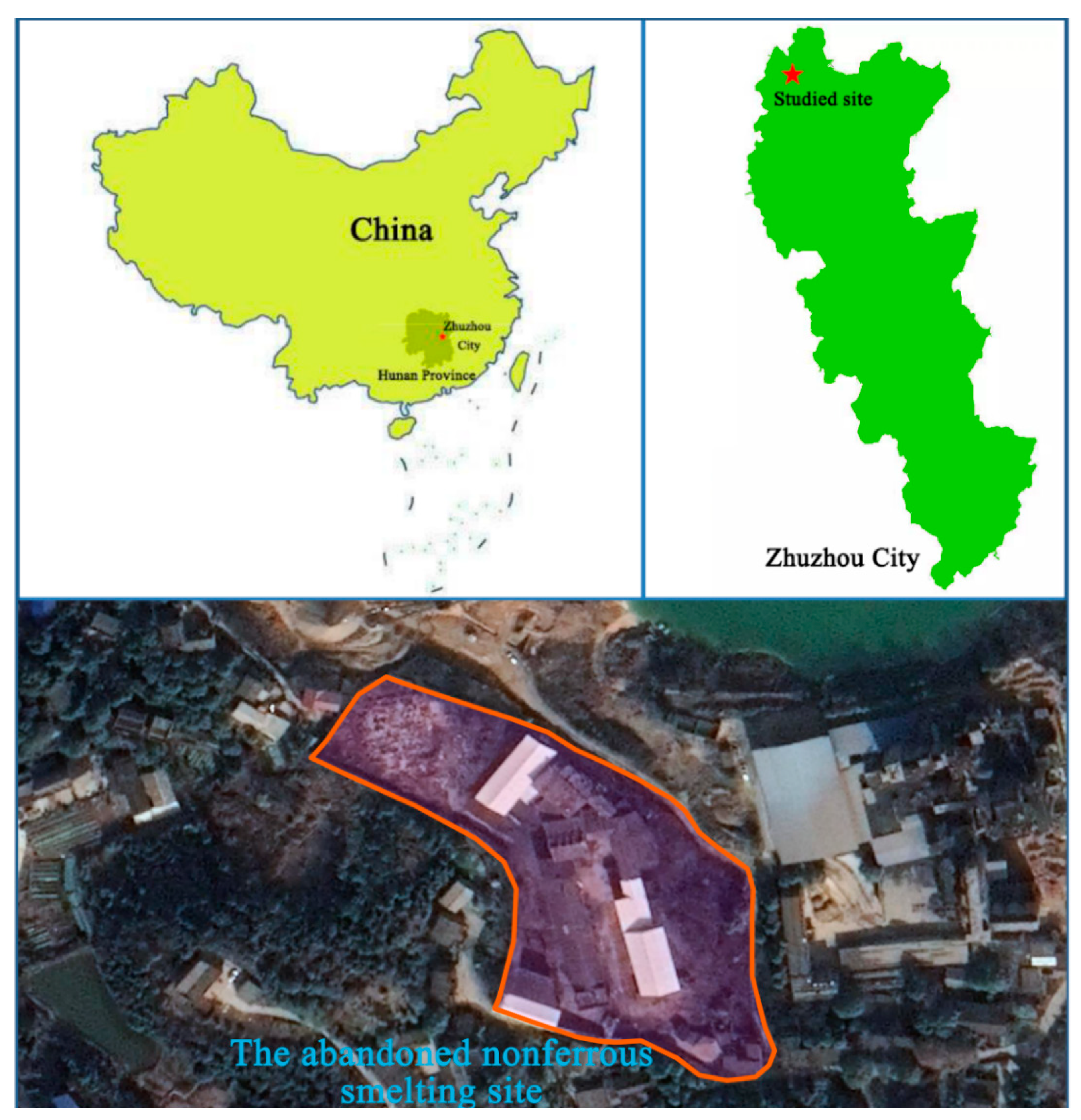

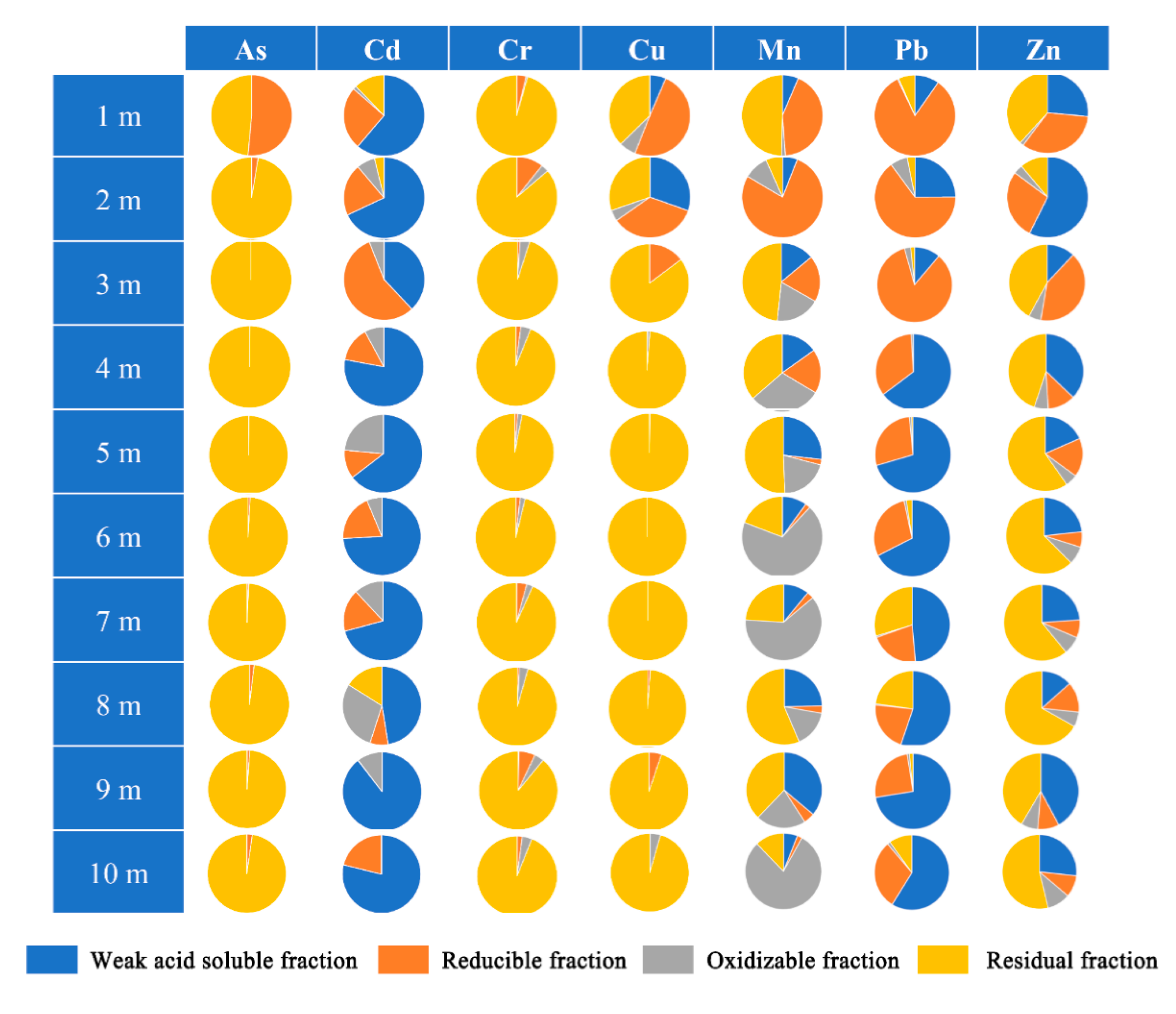
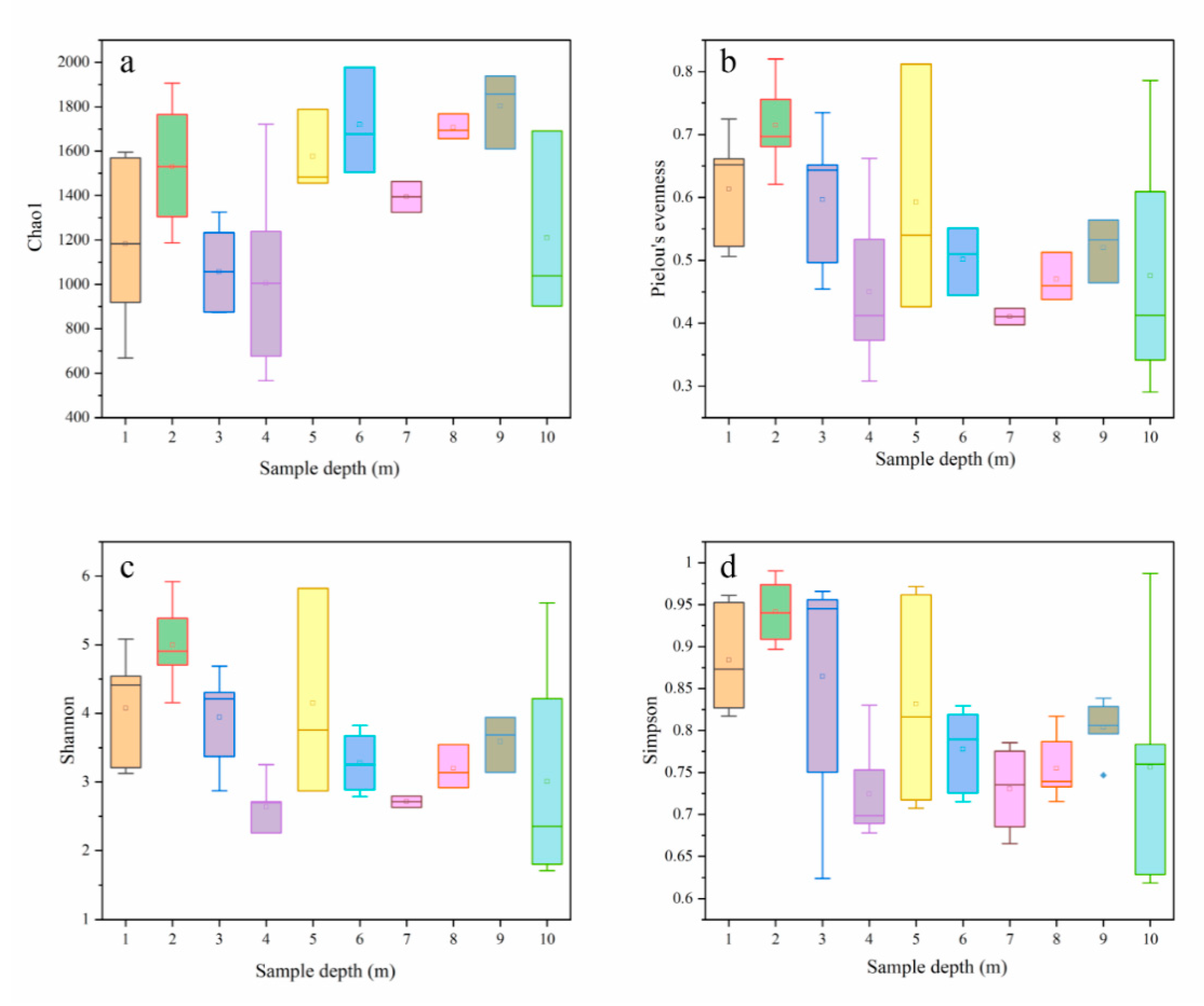
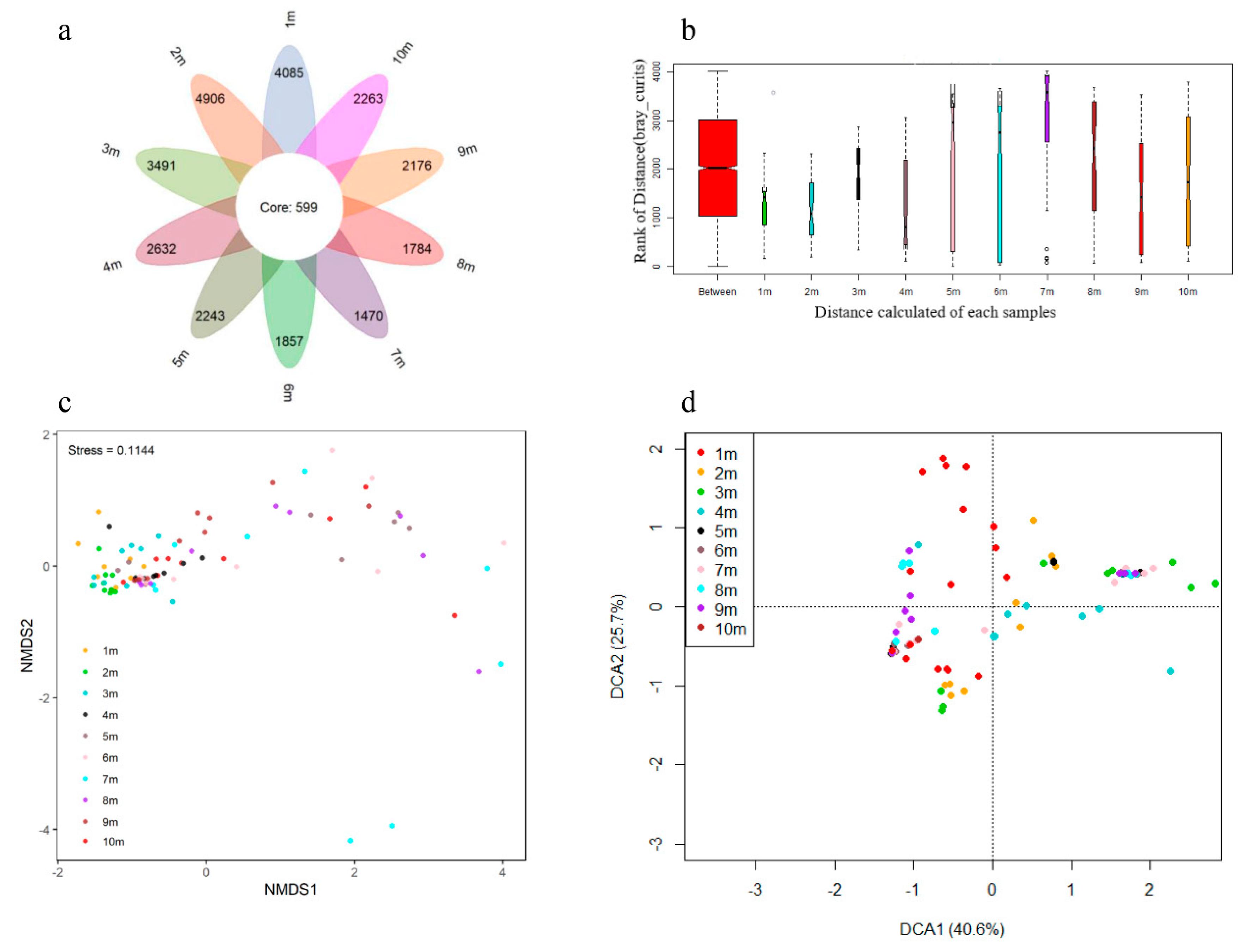

| Depth (m) | pH | ORP (mV) | Sand (%) | Silt (%) | Clay (%) | MC (%) | SOM (%) | AN (mg/kg) | NN (mg/kg) | AP (mg/kg) | AK (mg/kg) | TK (mg/kg) | CEC (cmol(+)/kg) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7.5 ± 0.3 a | 205 ± 38 b | 54.3 ± 5.9 a | 26.6 ± 1.6 a | 19.1 ± 6.7 b | 19.4 ± 2.9 a | 6.5 ± 1.4 a | 45.2 ± 18.1 a | 51.6 ± 2.3 a | 20.6 ± 9 a | 410 ± 1.8 a | 9157 ± 2588 d | 6 ± 1.3 a,b,c |
| 2 | 7.5 ± 0 a | 229 ± 13 a,b | 19.8 ± 7.3 b | 35.4 ± 10.4 a | 44.9 ± 11.5 a,b | 21.9 ± 2.4 a | 2.2 ± 0.6 b | 55.7 ± 23.5 a | 45.7 ± 2.2 a | 9.1 ± 5.7 a,b | 246.7 ± 132.7 a,b | 15709 ± 427 c | 8.2 ± 0.5 a |
| 3 | 7.3 ± 0.1 a | 243 ± 6 a,b | 13.4 ± 2.6 b | 35 ± 17.4 a | 51.6 ± 17.1 a | 23 ± 0.9 a | 0.8 ± 0.2 b | 56.6 ± 15.2 a | 46.3 ± 2.5 a | 4.2 ± 2 a,b | 238.4 ± 137 a,b | 18444 ± 589 c | 7.9 ± 1.4 a,b |
| 4 | 6.6 ± 0.1 a | 280 ± 33 a,b | 14.8 ± 4.2 b | 49.3 ± 2.5 a | 35.9 ± 6.6 a,b | 22.9 ± 1.8 a | 0.9 ± 0.8 b | 65.7 ± 17.2 a | 48.6 ± 4.1 a | 5.4 ± 5.6 a,b | 162.2 ± 95.1 a,b | 26331 ± 2218 b | 5.1 ± 1.1 b,c,d |
| 5 | 5 ± 0.4 b | 301 ± 44 a,b | 21 ± 6.7 b | 54.2 ± 9.2 a | 24.8 ± 4 b | 21.9 ± 0.8 a | 0.3 ± 0.1 b | 52.8 ± 9.5 a | 48.9 ± 4.5 a | 6.7 ± 7.7 a,b | 125.6 ± 59.3 b | 28155 ± 1581 a,b | 3.4 ± 0.6 c,d |
| 6 | 4.5 ± 0.1 b | 323 ± 36 a | 26.1 ± 4.6 b | 47 ± 7.1 a | 26.9 ± 2.5 a,b | 20.6 ± 0.8 a | 0.4 ± 0.1 b | 59.2 ± 15.3 a | 55.2 ± 9.9 a | 3.6 ± 3.7 a,b | 80.8 ± 16.3 b | 28456 ± 1572 a,b | 3.6 ± 0.4 c,d |
| 7 | 4.5 ± 0.2 b | 306 ± 15 a,b | 27.1 ± 6.9 b | 46.5 ± 5.2 a | 26.4 ± 4.2 a,b | 20.7 ± 1.4 a | 0.3 ± 0 b | 58.3 ± 10.4 a | 50.2 ± 3.2 a | 2.4 ± 2.5 b | 77.2 ± 14.2 b | 29470 ± 1267 a,b | 3 ± 0.5 d |
| 8 | 4.9 ± 0.4 b | 290 ± 8 a,b | 23.9 ± 2.7 b | 51.7 ± 2.8 a | 24.4 ± 0.9 b | 20.4 ± 1 a | 0.5 ± 0.3 b | 73.6 ± 6.5 a | 45.9 ± 1.6 a | 2.7 ± 1.5 b | 66.8 ± 21 b | 31158 ± 547 a,b | 3.3 ± 0.5 c,d |
| 9 | 4.8 ± 0.4 b | 278 ± 16 a,b | 26.3 ± 8.3 b | 45.1 ± 4.3 a | 28.6 ± 4 a,b | 18.7 ± 1.8 a | 0.5 ± 0.1 b | 70 ± 11.5 a | 47.8 ± 9.2 a | 4.7 ± 3.7 a,b | 77 ± 8.5 b | 32678 ± 497 a | 3.1 ± 0.3 d |
| 10 | 4.6 ± 0.6 b | 272 ± 42 a,b | 26.6 ± 2.4 b | 49.4 ± 2.7 a | 24 ± 0.6 b | 17.9 ± 0.9 a | 0.8 ± 0.6 b | 48.9 ± 1.5 a | 47.2 ± 5.9 a | 3 ± 1.8 b | 90.1 ± 10.8 b | 31309 ± 1184 a,b | 3.2 ± 0.5 c,d |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Wang, S.; Guo, Z.; Deng, Y.; Xu, M.; Zhang, S.; Yin, H.; Liang, Y.; Liu, H.; Miao, B.; et al. Spatial Distribution of Toxic Metal(loid)s and Microbial Community Analysis in Soil Vertical Profile at an Abandoned Nonferrous Metal Smelting Site. Int. J. Environ. Res. Public Health 2020, 17, 7101. https://doi.org/10.3390/ijerph17197101
Yang J, Wang S, Guo Z, Deng Y, Xu M, Zhang S, Yin H, Liang Y, Liu H, Miao B, et al. Spatial Distribution of Toxic Metal(loid)s and Microbial Community Analysis in Soil Vertical Profile at an Abandoned Nonferrous Metal Smelting Site. International Journal of Environmental Research and Public Health. 2020; 17(19):7101. https://doi.org/10.3390/ijerph17197101
Chicago/Turabian StyleYang, Jiejie, Siqi Wang, Ziwen Guo, Yan Deng, Menglong Xu, Siyuan Zhang, Huaqun Yin, Yili Liang, Hongwei Liu, Bo Miao, and et al. 2020. "Spatial Distribution of Toxic Metal(loid)s and Microbial Community Analysis in Soil Vertical Profile at an Abandoned Nonferrous Metal Smelting Site" International Journal of Environmental Research and Public Health 17, no. 19: 7101. https://doi.org/10.3390/ijerph17197101
APA StyleYang, J., Wang, S., Guo, Z., Deng, Y., Xu, M., Zhang, S., Yin, H., Liang, Y., Liu, H., Miao, B., Meng, D., Liu, X., & Jiang, L. (2020). Spatial Distribution of Toxic Metal(loid)s and Microbial Community Analysis in Soil Vertical Profile at an Abandoned Nonferrous Metal Smelting Site. International Journal of Environmental Research and Public Health, 17(19), 7101. https://doi.org/10.3390/ijerph17197101





