Evaluation of Methicillin-Resistant Staphylococcus aureus Carriage and High Livestock Production Areas in North Carolina through Active Case Finding at a Tertiary Care Hospital
Abstract
1. Introduction
2. Materials and Methods
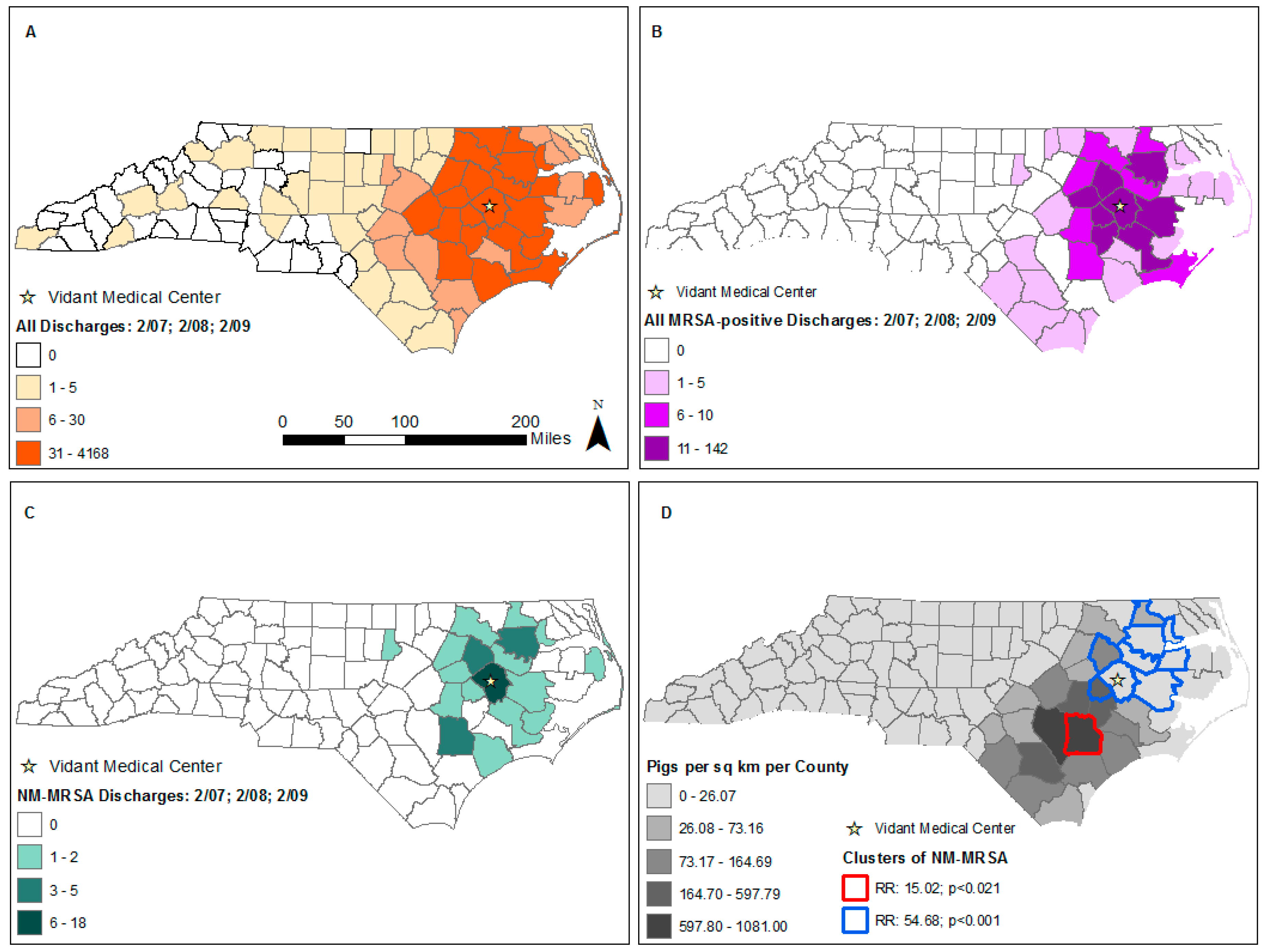
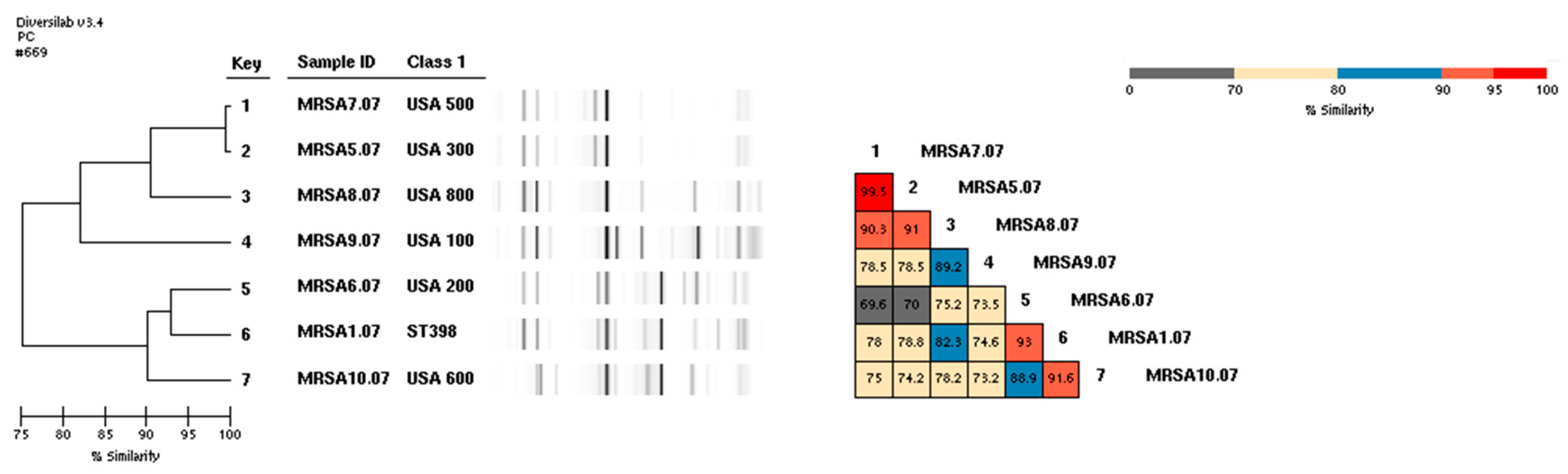
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention (CDC). Methicillin-Resistant Staphylococcus Aureus (MRSA) Infections. Available online: http://www.cdc.gov/mrsa/healthcare/index.html (accessed on 28 December 2014).
- Kinross, P.; Petersen, A.; Skov, R.; Van Hauwermeiren, E.; Pantosti, A.; Laurent, F.; Voss, A.; Kluytmans, J.; Struelens, M.J.; Heuer, O.; et al. Livestock-associated meticillin-resistant Staphylococcus aureus (MRSA) among human MRSA isolates, European Union/European Economic Area countries, 2013. Eurosurveillance 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Cuny, C.; Wieler, L.H.; Witte, W. Livestock-Associated MRSA: The Impact on Humans. Antibiotics 2015, 4, 521–543. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.C. Livestock-associated Staphylococcus aureus: The United States experience. PLoS Pathog. 2015, 11, e1004564. [Google Scholar] [CrossRef] [PubMed]
- Van Loo, I.; Huijsdens, X.; Tiemersma, E.; de Neeling, A.; van de Sande-Bruinsma, N.; Beaujean, D.; Voss, A.; Kluytmans, J. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg. Infect. Dis. 2007, 13, 1834–1839. [Google Scholar] [CrossRef] [PubMed]
- Neyra, R.C.; Frisancho, J.A.; Rinsky, J.L.; Resnick, C.; Carroll, K.C.; Rule, A.M.; Ross, T.; You, Y.; Price, L.B.; Silbergeld, E.K. Multidrug-resistant and methicillin-resistant Staphylococcus aureus (MRSA) in hog slaughter and processing plant workers and their community in North Carolina (USA). Environ. Health Perspect. 2014, 122, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Nadimpalli, M.; Rinsky, J.L.; Wing, S.; Hall, D.; Stewart, J.; Larsen, J.; Nachman, K.E.; Love, D.C.; Pierce, E.; Pisanic, N.; et al. Persistence of livestock-associated antibiotic-resistant Staphylococcus aureus among industrial hog operation workers in North Carolina over 14 days. Occup. Environ. Med. 2015, 72, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Pofahl, W.E.; Goettler, C.E.; Ramsey, K.M.; Cochran, M.K.; Nobles, D.L.; Rotondo, M.F. Active surveillance screening of MRSA and eradication of the carrier state decreases surgical-site infections caused by MRSA. J. Am. Coll. Surg. 2009, 208, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C.; Gay, E.A.; Frye, S.; Eells, S.J.; Healy, M.; McGowan, J.E., Jr. Comparison of typing results obtained for methicillin-resistant Staphylococcus aureus isolates with the DiversiLab system and pulsed-field gel electrophoresis. J. Clin. Microbiol. 2009, 47, 2452–2457. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feingold, B.J.; Kluytmans, J.A.; van Cleef, B.A.; Curriero, F.C.; Silbergeld, E.K. Livestock Density as Risk Factor for Livestock-associated Methicillin-Resistant Staphylococcus aureus, the Netherlands. Emerg. Infect. Dis. 2012, 19, 1552. [Google Scholar] [CrossRef] [PubMed]
- Schinasi, L.; Wing, S.; Augustino, K.L.; Ramsey, K.M.; Nobles, D.L.; Richardson, D.B.; Price, L.B.; Aziz, M.; MacDonald, P.D.; Stewart, J.R. A case control study of environmental and occupational exposures associated with methicillin resistant Staphylococcus aureus nasal carriage in patients admitted to a rural tertiary care hospital in a high density swine region. Environ. Health 2014, 13, 54. [Google Scholar] [CrossRef] [PubMed]
- Kulldorff, M. SaTScan—Software for the Spatial, Temporal and Space-Time Scan Statistics, version 9.0; Information Management Services, Inc.: Calverton, MD, USA, 2010; Available online: http://www.satscan.org/ (accessed on 11 September 2019).
- Grisold, A.J.; Zarfel, G.; Hoenigl, M.; Krziwanek, K.; Feierl, G.; Masoud, L.; Leitner, E.; Wagner-Eibel, U.; Badura, A.; Marth, E. Occurrence and genotyping using automated repetitive-sequence-based PCR of methicillin-resistant Staphylococcus aureus ST398 in Southeast Austria. Diagn. Microbiol. Infect. Dis. 2010, 66, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Freitas, E.A.; Harris, R.M.; Blake, R.K.; Salgado, C.D. Prevalence of USA300 strain type of methicillin-resistant Staphylococcus aureus among patients with nasal colonization identified with active surveillance. Infect. Control Hosp. Epidemiol. 2010, 31, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Bal, A.M.; Coombs, G.W.; Holden, M.T.G.; Lindsay, J.A.; Nimmo, G.R.; Tattevin, P.; Skov, R.L. Genomic insights into the emergence and spread of international clones of healthcare-, community- and livestock-associated meticillin-resistant Staphylococcus aureus: Blurring of the traditional definitions. J. Glob. Antimicrob. Resist. 2016, 6, 95–101. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feingold, B.J.; Augustino, K.L.; Curriero, F.C.; Udani, P.C.; Ramsey, K.M. Evaluation of Methicillin-Resistant Staphylococcus aureus Carriage and High Livestock Production Areas in North Carolina through Active Case Finding at a Tertiary Care Hospital. Int. J. Environ. Res. Public Health 2019, 16, 3418. https://doi.org/10.3390/ijerph16183418
Feingold BJ, Augustino KL, Curriero FC, Udani PC, Ramsey KM. Evaluation of Methicillin-Resistant Staphylococcus aureus Carriage and High Livestock Production Areas in North Carolina through Active Case Finding at a Tertiary Care Hospital. International Journal of Environmental Research and Public Health. 2019; 16(18):3418. https://doi.org/10.3390/ijerph16183418
Chicago/Turabian StyleFeingold, Beth J., Kerri L. Augustino, Frank C. Curriero, Paras C. Udani, and Keith M. Ramsey. 2019. "Evaluation of Methicillin-Resistant Staphylococcus aureus Carriage and High Livestock Production Areas in North Carolina through Active Case Finding at a Tertiary Care Hospital" International Journal of Environmental Research and Public Health 16, no. 18: 3418. https://doi.org/10.3390/ijerph16183418
APA StyleFeingold, B. J., Augustino, K. L., Curriero, F. C., Udani, P. C., & Ramsey, K. M. (2019). Evaluation of Methicillin-Resistant Staphylococcus aureus Carriage and High Livestock Production Areas in North Carolina through Active Case Finding at a Tertiary Care Hospital. International Journal of Environmental Research and Public Health, 16(18), 3418. https://doi.org/10.3390/ijerph16183418




