Molecular Characterization and Antimicrobial Resistance Pattern of Escherichia coli Recovered from Wastewater Treatment Plants in Eastern Cape South Africa
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Collection of Water Samples
2.2. Bacterial Isolation
2.3. Extraction of Bacterial DNA
2.4. Molecular Confirmation of E. coli and E. coli Pathotypes
2.5. Antimicrobial Susceptibility Testing
3. Results
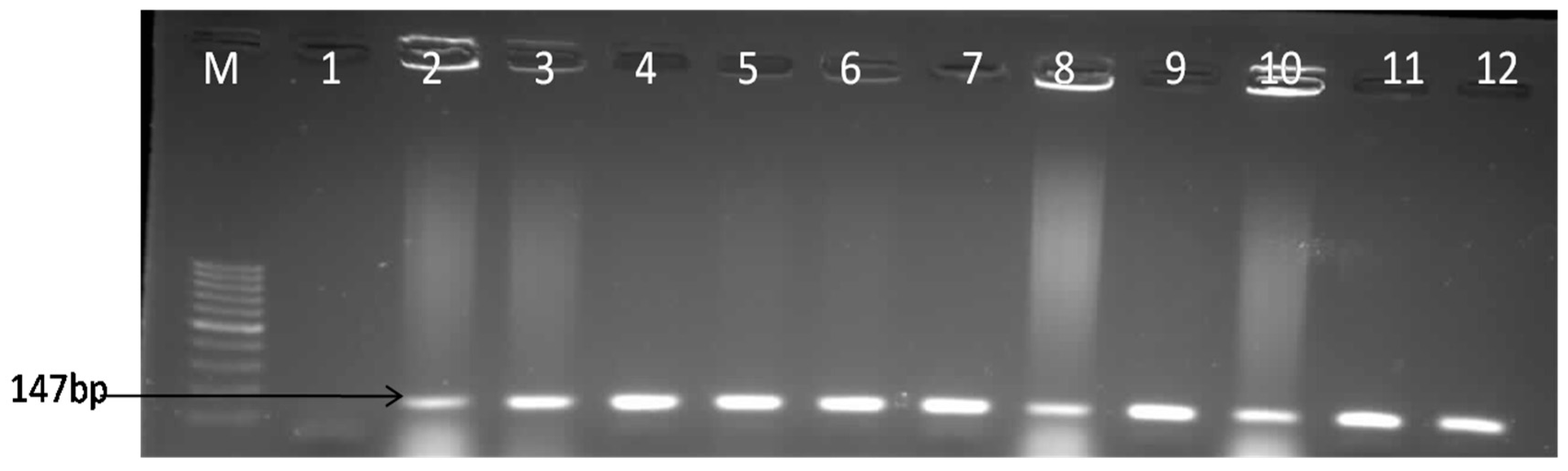
3.1. Molecular Confirmation of E. coli Isolates
3.2. Molecular Confirmation of E. coli Pathotypes
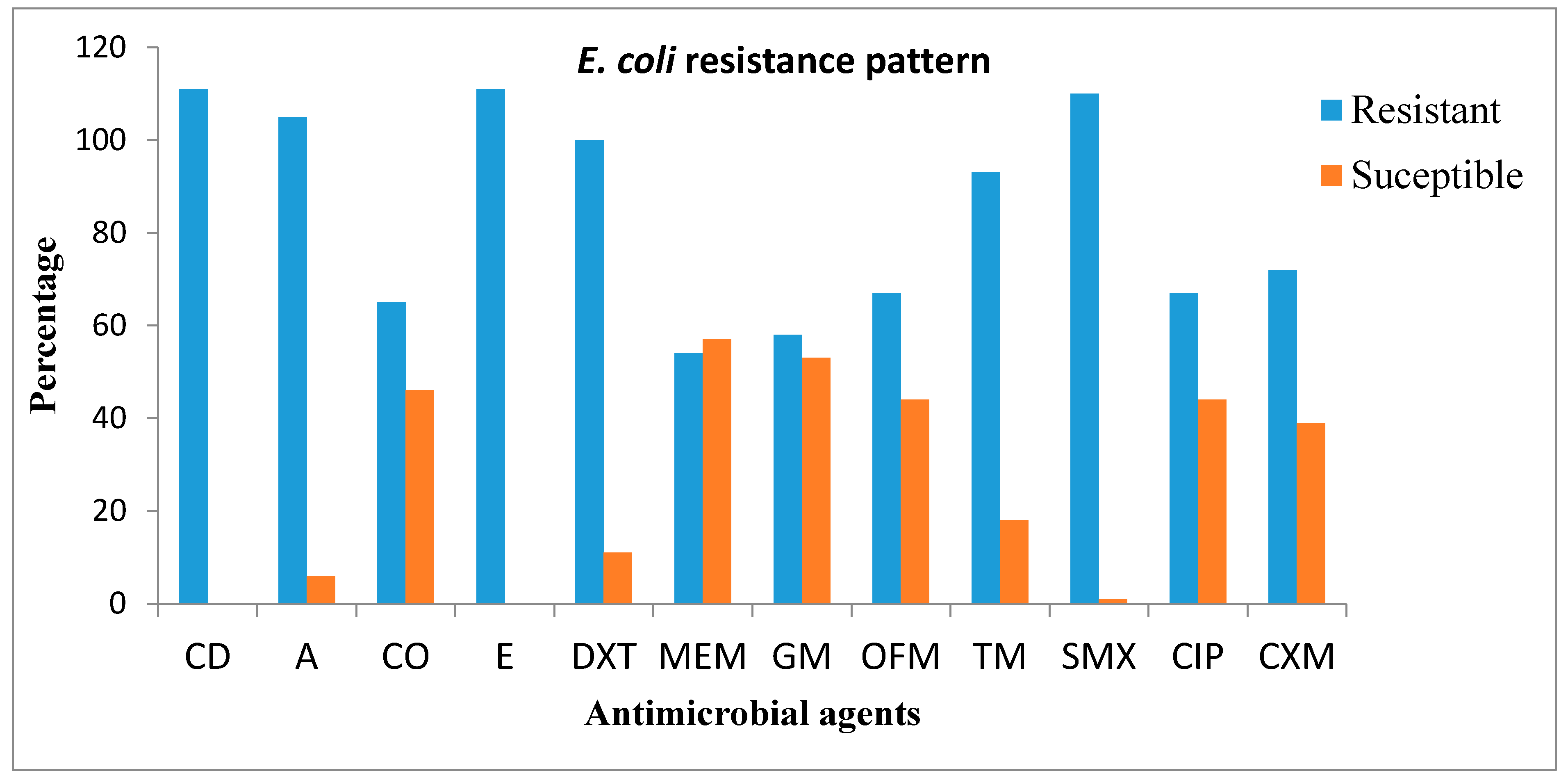
3.3. Antimicrobial Susceptibility Pattern of the Confirmed E. coli Isolates
3.4. Resistance Determinants among the Isolates
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martínez, J.L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, J.; Seo, K. Presence of Stenotrophomonas maltophilia exhibiting high genetic similarity to clinical isolates in final effluents of pig farm wastewater treatment plants. Int. J. Hyg. Environ. Health 2018, 221, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, V.; Stabnikov, V.; Zhuang, W.Q.; Tay, J.H.; Tay, S.T.L. Phosphate removal from the returned liquor of municipal wastewater treatment plant using iron-reducing bacteria. J. Appl. Microbiol. 2005, 98, 1152–1161. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P.J. Antibiotic-resistance genes in waste water. Trends Microbol. 2018, 26, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Sanders, E.C.; Yuan, Y.; Pitchford, A. Fecal coliform and E. coli concentrations in effluent-dominated streams of the upper Santa Cruz Watershed. Water 2013, 5, 243–261. [Google Scholar] [CrossRef]
- Diallo, A.A.; Bibbal, D.; Lô, F.T.; Mbengue, M.; Sarr, M.M.; Diouf, M.; Sambe, Y.; Kérourédan, M.; Alambédji, R.; Thiongane, Y.; Oswald, E.; Brugère, H. Prevalence of pathogenic and antibiotics resistant Escherichia coli from effluents of a slaughterhouse and a municipal wastewater treatment plant in Dakar. Afr. J. Microbiol. Res. 2017, 11, 1035–1042. [Google Scholar]
- Amos, G.C.A.; Ploumakis, S.; Zhang, L.; Hawkey, P.M.; Gaze, W.H.; Wellington, E.M.H. The widespread dissemination of integrons throughout bacterial communities in a riverine system. ISME J. 2018, 12, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Long, S.C.; Das, D.; Dorner, S.M. Are microbial indicators and pathogens correlated? A statistical analysis of 40 years of research. J. Water Health 2011, 9, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.B.; Tchamba, G.T.; Bagré, T.S.; Bouda, S.C.; Fody, A.M.; Kagambèga, A.; Bonkoungou, I.J.O.; Tiendrebéogo, F.; Zongo, C.; Savadogo, A.; et al. Characterization of diarrheagenic Escherichia coli isolated from raw beef, mutton, and intestines sold in Ouagadougou, Burkina Faso. J. Microbiol. Biotechnol. Food Sci. 2016, 5, 470–474. [Google Scholar] [CrossRef]
- Bonkoungou, I.J.O.; Lienemann, T.; Martikainen, O.; Dembelé, R.; Sanou, I.; Traoré, A.S.; Siitonen, A.; Barro, N.; Haukka, K. Diarrhoeagenic Escherichia coli detected by 16-plex PCR in children with and without diarrhoea in Burkina Faso. Clin. Microbiol. Infect. 2011, 18, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Shetty, V.A.; Kumar, S.H.; Shetty, A.K.; Karunasagar, I.; Karunasagar, I. Prevalence and characterization of diarrheagenic Escherichia coli isolated from adults and children in Mangalore, India. J. Lab. Physicians 2012, 4, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Shabana, I.I.; Zaraket, H.; Suzuk, H. Molecular studies on diarrhea-associated Escherichia coli isolated from humans and animals in Egypt. Vet. Microbiol. 2013, 167, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Igbeneghu, O.A.; Lamikanra, A. Multiple-Resistant commensal Escherichia Coli from Nigerian Children: Potential Opportunistic Pathogens. Trop. J. Pharm. Res. 2014, 13, 423–428. [Google Scholar] [CrossRef]
- Saeed, A.; Abd, H.; Sandstrom, G. Microbial aetiology of acute diarrhoea in children under five years of age in Khartoum. Sudan J. Med. Microbiol. 2015, 64, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Buven, G.; Bogaerts, P.; Glupczynski, Y.; Lauwer, S.; Piérard, D. Antimicrobial resistance testing of verocytotocin-producing Escherichia coli and first description of TEM-52 extended-spectrum β-lactamase in serogroup 026. Antimicrob. Agents Chem. 2010, 54, 4907–4909. [Google Scholar] [CrossRef] [PubMed]
- Friedman, N.D.; Temkin, E.; Carmeli, Y. The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 2016, 22, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Loho, T.; Dharmayanti, A. Colistin: An antibiotic and its role in multiresistant Gram-negative infections. Acta Med. Indones. 2015, 47, 157–168. [Google Scholar] [PubMed]
- Da Silva, G.J.; Domingues, S. Interplay between colistin resistance, virulence and fitness in Acinetobacter baumannii. Antibiotics 2017, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Hedden, S.; Cilliers, J. Parched Prospects: The Emerging Water Crisis in South Africa; African Futures Paper 11; Institute for Security Studies: Pretoria, South Africa, 2014. [Google Scholar]
- Turton, A. Sitting on the Horns of a Dilemma: Water as a Strategic Resource in South Africa; South African Institute of Race Relations: Johannesburg, South Africa, 2015. [Google Scholar]
- Van Duijkeren, E.; Wannet, W.J.; Houwers, D.J.; Van Pelt, W. Antimicrobial susceptibility of Salmonella strains isolated from humans, cattle, pigs, and chickens in the Netherlands from 1984 to 2001. J. Clin. Microbiol. 2003, 41, 3574–3578. [Google Scholar] [CrossRef] [PubMed]
- Momtaz, H.; Rahimi, E.; Moshkelani, S. Molecular detection of antimicrobial resistance genes in E. coli isolated from slaughtered commercial chickens in Iran. Vet. Med. 2012, 57, 193–197. [Google Scholar] [CrossRef]
- Janezic, K.J.; Ferry, B.; Hendricks, E.W.; Janiga, B.A.; Johnson, T.; Murphy, S.; Roberts, M.E.; Scott, S.M.; Theisen, A.N.; Hung, K.F.; et al. Phenotypic and genotypic characterisation of Escherichia coli isolated from untreated surface waters. Open Microbiol. J. 2013, 7, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Bej, A.K.; Cesare, D.J.L.; Haff, L.; Atlas, R.M. Detection of Escherichia coli and Shigella spp. in water by using the polymerase chain reaction and gene probes for uidA. Appl. Environ. Microbiol. 1991, 57, 1013–1017. [Google Scholar] [PubMed]
- Rajendran, P.; Ajjampur, S.S.R.; Chidambaram, D.; Chandrabose, G.; Thangaraj, B.; Sarkar, R.; Samuel, P.; Rajan, D.P.; Kang, G. Pathotypes of diarrheagenic Escherichia coli in children attending tertiary care hospital in South India. Diagn. Microbiol. Infect. Dis. 2010, 68, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement; CLSI: Wayne, PA, USA, 2014. [Google Scholar]
- Lina, G.; Quaglia, A.; Reverdy, M.-E.; Leclercq, R.; Vandenesch, F.; Etienne, A. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among Staphylococci. Antimicrob. Agents Chemother. 1999, 43, 1062–1066. [Google Scholar] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Amos, G.C.; Gozzard, E.; Carter, C.E.; Mead, A.; Bowes, M.J.; Hawkey, P.M.; Zhang, L.; Singer, A.C.; Gaze, W.H.; Wellington, E.M. Validated predictive modelling of the environmental resistome. ISME J. 2015, 9, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Turolla, A.; Cattaneo, M.; Marazzi, F.; Mezzanotte, V.; Antonelli, M. Antibiotic resistant bacteria in urban sewage: Role of full-scale wastewater treatment plants on environmental spreading. Chemosphere 2018, 191, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Osińska, A.; Korzeniewska, E.; Harnisz, M.; Niestępski, S. The prevalence and characterization of antibiotic-resistant and virulent Escherichia coli strains in the municipal wastewater system and their environmental fate. Sci. Total Environ. 2017, 577, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Amaya, E.; Reyes, D.; Paniagua, M.; Calderón, S.; Rashid, M.U.; Colque, P.; Kühn, I.; Möllby, R.; Weintraub, A.; Nord, C.E. Antibiotic resistance patterns of Escherichia coli isolates from different aquatic environmental sources in León, Nicaragua. Clin. Microbiol. Infect. 2012, 18, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Canizalez-Roman, A.; Flores-Villaseñor, H.M.; Gonzalez-Nuñez, E.; Velazquez-Roman, J.; Vidal, J.E.; Muro-Amador, S.; Alapizco-Castro, G.; Díaz-Quiñonez, J.A.; León-Sicairos, N. Surveillance of diarrheagenic Escherichia coli strains isolated from diarrhea cases from children, adults and elderly at Northwest of Mexico. Front. Microbiol. 2016, 7, 1924. [Google Scholar] [CrossRef] [PubMed]
- Bryce, J.; Boschi-Pinto, C.; Shibuya, K.; Black, R.E. WHO Child Health Epidemiology Reference Group. WHO estimates of the causes of death in children. Lancet 2005, 365, 1147–1152. [Google Scholar] [CrossRef]
- Mokcracka, J.; Koczura, R.; Jablonska, L.; Kaznowsski, A. Phylogenetic groups, virulence genes and quinolone resistance of intergron-bearing Escherichia coli isolated from a wastewater treatment plant. Antonie van Leeuwen. 2011, 99, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Omar, K.B.; Barnard, T.G. The occurrence of pathogenic Escherichia coli in South African wastewater treatment plants as detected by multiplex PCR. Water SA 2010, 36, 172–176. [Google Scholar]
- Sidhu, J.P.; Ahmed, W.; Hodgers, L.; Toze, S. Occurrence of virulence genes associated with diarrheagenic pathotypes in Escherichia coli isolates from surface water. Appl. Environ. Microbiol. 2013, 79, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Sadowsky, M.J. Escherichia coli in the environment: Implications for water quality and human health. Microb. Environ. 2008, 23, 101–108. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Mora, A.; Blanco, E.J.; Blanco, M.; Alonso, M.P.; Dhabi, G.; Echeita, A.; González, E.A.; Bernárdez, M.I.; Blanco, J. Antimicrobial resistance of Shiga toxin (verotoxin) producing Escherichia coli O157:H7 and non-O157 strains isolated from humans, cattle, sheep and food in Spain. Res. Microbiol. 2005, 156, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wu, D.; Liu, K.; Suolang, S.; He, T.; Liu, X.; Wu, C.; Wang, Y.; Lin, D. Investigation of antimicrobial resistance in Escherichia coli and enterococci isolated from Tibetan pigs. PLoS ONE 2014, 9, e95623. [Google Scholar] [CrossRef] [PubMed]
- Adwan, K.; Jarrar, N.; Abu-Hijleh, A.; Adwan, G.; Awwad, E. Molecular characterization of Escherichia coli isolates from patients with urinary tract infections in Palestine. J. Med. Microbiol. 2014, 63, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Czekalski, N.; Sigdel, R.; Birtel, J.; Matthews, B.; Bürgmann, H. Does human activity impact the natural antibiotic resistance background? Abundance of antibiotic resistance genes in 21 Swiss lakes. Environ. Int. 2015, 81, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Wang, W.; Regev-Yochay, G.; Lipsitch, M.; Hanage, W.P. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 2015, 8, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.H.; Qiao, M.; Lv, Z.E.; Guo, G.X.; Jia, Y.; Su, Y.H.; Zhu, Y.G. Impact of reclaimed water irrigation on antibiotic resistance in public parks, Beijing, China. Env. Pollut. 2014, 184, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Flórez, A.B.; Alegría, Á.; Rossi, F.; Delgado, S.; Felis, G.E.; Torriani, S.; Mayo, B. Molecular identification and quantification of tetracycline and erythromycin resistance genes in Spanish and Italian retail cheeses. BioMed Res. Int. 2014, 2014, 746859. [Google Scholar] [CrossRef] [PubMed]
- Ziembińska-Buczyńska, A.; Felis, E.; Folkert, J.; Meresta, A.; Stawicka, D.; Gnida, A.; Surmacz-Górska, J. Detection of antibiotic resistance genes in wastewater treatment plant–molecular and classical approach. Arch. Environ. Protect. 2015, 41, 23–32. [Google Scholar] [CrossRef]
- Yuan, Q.B.; Guo, M.T.; Yang, J. Fate of antibiotic resistant bacteria and genes during wastewater chlorination: Implication for antibiotic resistance control. PLoS ONE 2015, 10, e119403. [Google Scholar] [CrossRef] [PubMed]
- Curcioa, L.; Luppi, A.; Bonilauri, P.; Gherpelli, Y.; Pezzotti, G.; Pesciaroli, M.; Magistrali, C.F. Detection of the colistin resistance gene mcr-1 in pathogenic Escherichia coli from pigs affected by post-weaning diarrhoea in Italy. J. Glob. Antimicrob. Resist. 2017, 10, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed]
- Quesada, A.; Porrero, M.C.; Téllez, S.; Palomo, G.; García, M.; Domínguez, L. Polymorphism of genes encoding PmrAB in colistin-resistant strains of Escherichia coli and Salmonella enteric isolated from poultry and swine. J. Antimicrob. Chem. 2015, 70, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.; Rahman, Z.; Monira, S.; Rahman, M.A.; Camilli, A.; George, C.M.; Ahmed, N.; Alam, M. Colistin resistant Escherichia coli carrying mcr-1 in urban sludge samples: Dhaka, Bangladesh. Gut Pathog. 2017, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Caniaux, I.; van Belkum, A.; Zambardi, G.; Poirel, L.; Gros, M.F. MCR: Modern colistin resistance. Eur. J. Clin.Microbiol. Infect. Dis. 2017, 36, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Luangtongkum, T.; Jeon, B.; Han, J.; Plummer, P.; Logue, C.M.; Zhang, Q. Antibiotic resistance in Campylobacter: Emergence, transmission and persistence. Future Microbiol. 2009, 4, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kumar, A. Prevalence and antibiotic resistance pattern of Campylobacter species in foods of animal origin. Vet. World 2014, 7, 681–684. [Google Scholar]
- Iweriebor, B.C.; Iwu, C.J.; Obi, L.C.; Nwodo, U.U.; Okoh, A.I. Multiple antibiotic resistances among Shiga toxin producing Escherichia coli O157 in feces of dairy cattle farms in Eastern Cape of South Africa. BMC Microbiol. 2015, 15, 213. [Google Scholar] [CrossRef] [PubMed]
| Primer Sequence (5′–3′) | E. coli and Pathotypes | Targeted Genes | Base Pair | References |
|---|---|---|---|---|
| F-GAACGTTGGTTAATGTGGGGTAA | E. coli | uidA | 147 | [25] |
| R-ACGCGTGGTTACAGTCTTGCG | ||||
| F-GAACGTTGGTTAATGTGGGGTAA | DAEC | daaE | 542 | [26] |
| R-TATTCACCGGTCGGTTATCAGT | ||||
| F-AGCTCAGGCAATGAAACTTTGAC | EIEC | virF | 618 | [26] |
| R-TGGGCTTGATATTCCGATAAGTC | ||||
| F-CACAGGCAACTGAAATAAGTCTGG | EAEC | aafII | 378 | [26] |
| R-ATTCCCATGATGTCAAGCACTTC | ||||
| F-TCAATGCAGTTCCGT TATCAGTT | EPEC | eae | 482 | [26] |
| R-GTAAAGTCCGTTACCCCAACCTG | ||||
| F-CAGTTAATGTGGTGGCGAAGG | EHEC | stx1 | 348 | [26] |
| R-CACCAGACAATGTAACCGCTG | ||||
| F-GCACACGGAGCTCCTCAGTC | ETEC | stII | 129 | [26] |
| Primer Sequence (5′–3′) | Targeted Genes | Base Pair (bp) | Reference |
|---|---|---|---|
| F-GTTCAAGAACAATCAATACA GAG | ermA | 421 | [28] |
| R-GGATCAGGAAAAGGACATTT TAC | |||
| F-CGGTCAGTCCGTTTGTTC | |||
| R-CTTGGTCGGTCTGTAGGG | Mcr-1 | 309 | [29] |
| E. coli Pathotypes | DAEC | EIEC | EPEC | EAEC | ETEC | EHEC |
|---|---|---|---|---|---|---|
| Targeted genes | daaE | virF | eae | aafII | stII | stx1 |
| No of positive isolates | 9 | 0 | 0 | 0 | 0 | 0 |
| Targeted Genes Screened | Total No of Isolates Screened | No of Isolates That Showed Phenotypic Resistance to the Text Antibiotic | No of Confirmed Isolates |
|---|---|---|---|
| ermA | 111 | 111 | 9 |
| mcr-1 | 111 | 65 | 31 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Igwaran, A.; Iweriebor, B.C.; Okoh, A.I. Molecular Characterization and Antimicrobial Resistance Pattern of Escherichia coli Recovered from Wastewater Treatment Plants in Eastern Cape South Africa. Int. J. Environ. Res. Public Health 2018, 15, 1237. https://doi.org/10.3390/ijerph15061237
Igwaran A, Iweriebor BC, Okoh AI. Molecular Characterization and Antimicrobial Resistance Pattern of Escherichia coli Recovered from Wastewater Treatment Plants in Eastern Cape South Africa. International Journal of Environmental Research and Public Health. 2018; 15(6):1237. https://doi.org/10.3390/ijerph15061237
Chicago/Turabian StyleIgwaran, Aboi, Benson Chuks Iweriebor, and Anthony Ifeanyi Okoh. 2018. "Molecular Characterization and Antimicrobial Resistance Pattern of Escherichia coli Recovered from Wastewater Treatment Plants in Eastern Cape South Africa" International Journal of Environmental Research and Public Health 15, no. 6: 1237. https://doi.org/10.3390/ijerph15061237
APA StyleIgwaran, A., Iweriebor, B. C., & Okoh, A. I. (2018). Molecular Characterization and Antimicrobial Resistance Pattern of Escherichia coli Recovered from Wastewater Treatment Plants in Eastern Cape South Africa. International Journal of Environmental Research and Public Health, 15(6), 1237. https://doi.org/10.3390/ijerph15061237





