Fine-Scale Spatial Heterogeneity in the Distribution of Waterborne Protozoa in a Drinking Water Reservoir
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Selection and Sampling
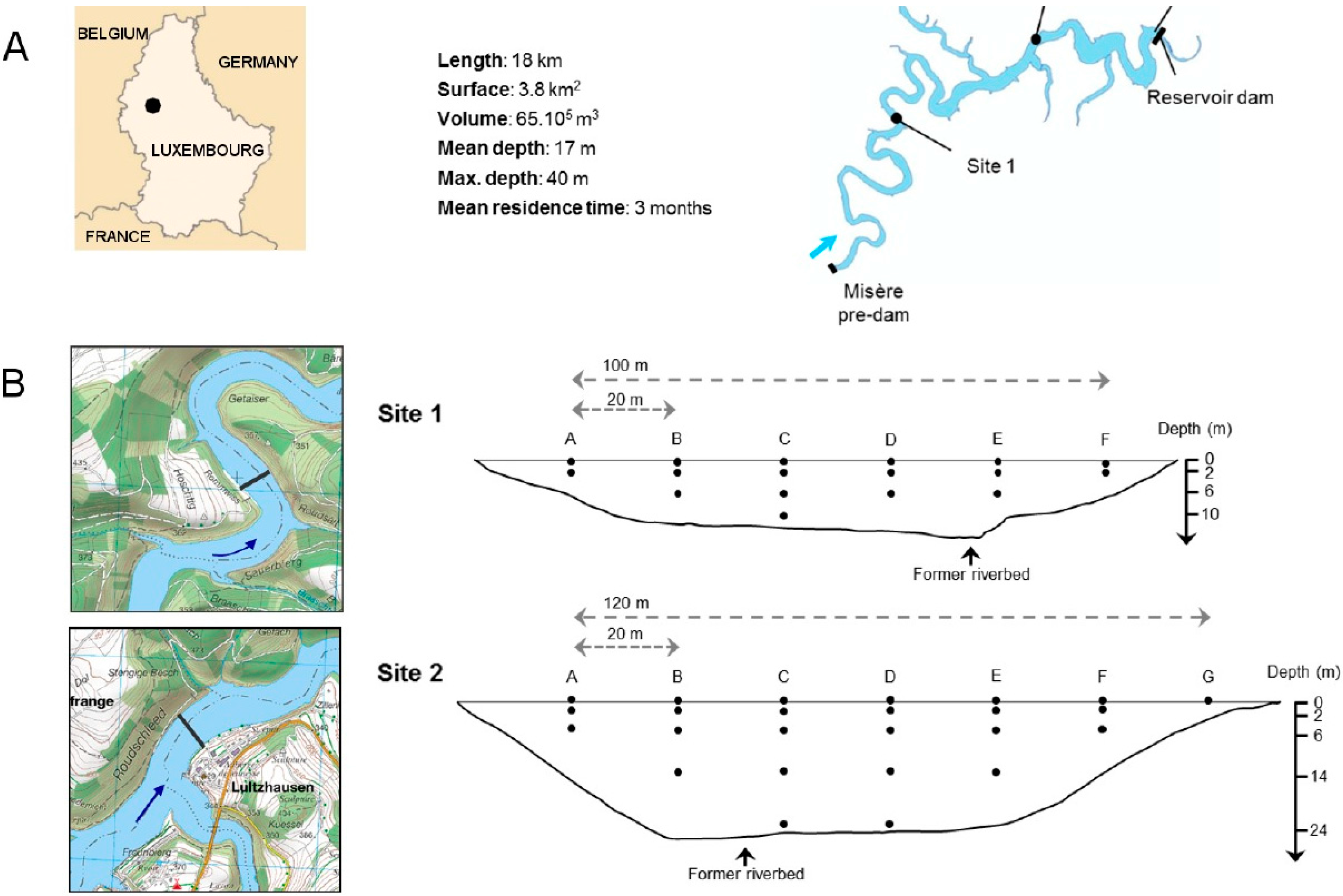
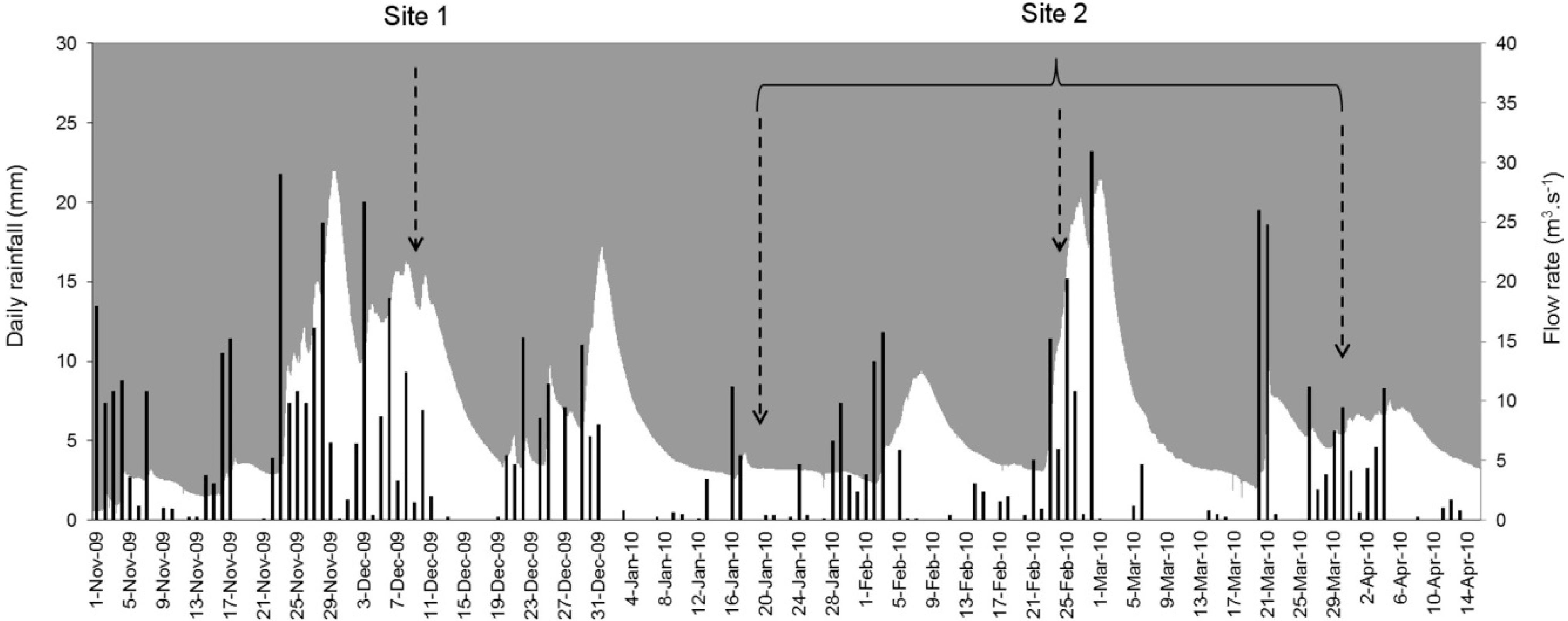
2.2. Sample Processing
2.3. Reproducibility Assay
2.4. Drifting Buoy Assays
2.5. Statistical Analyses
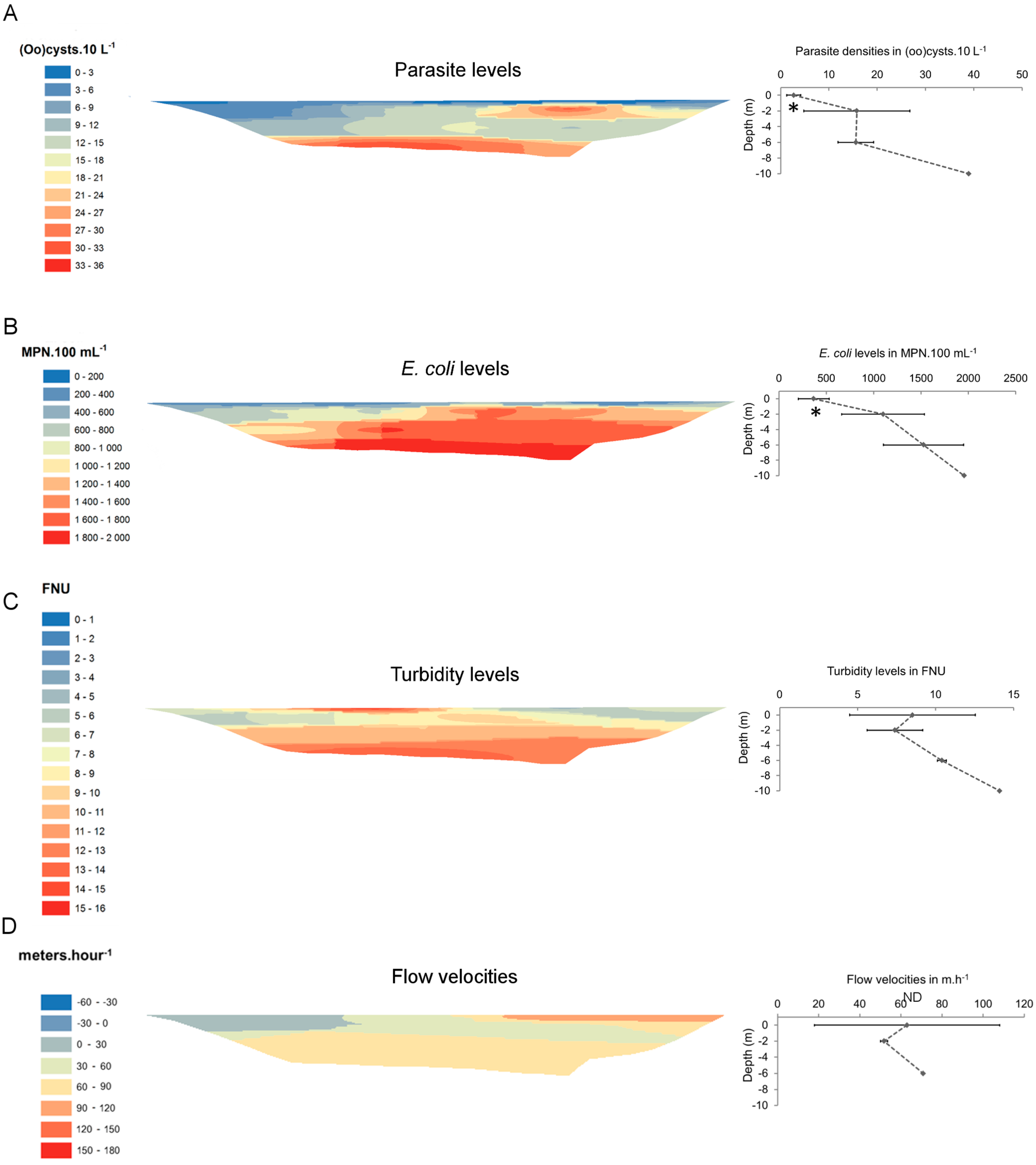
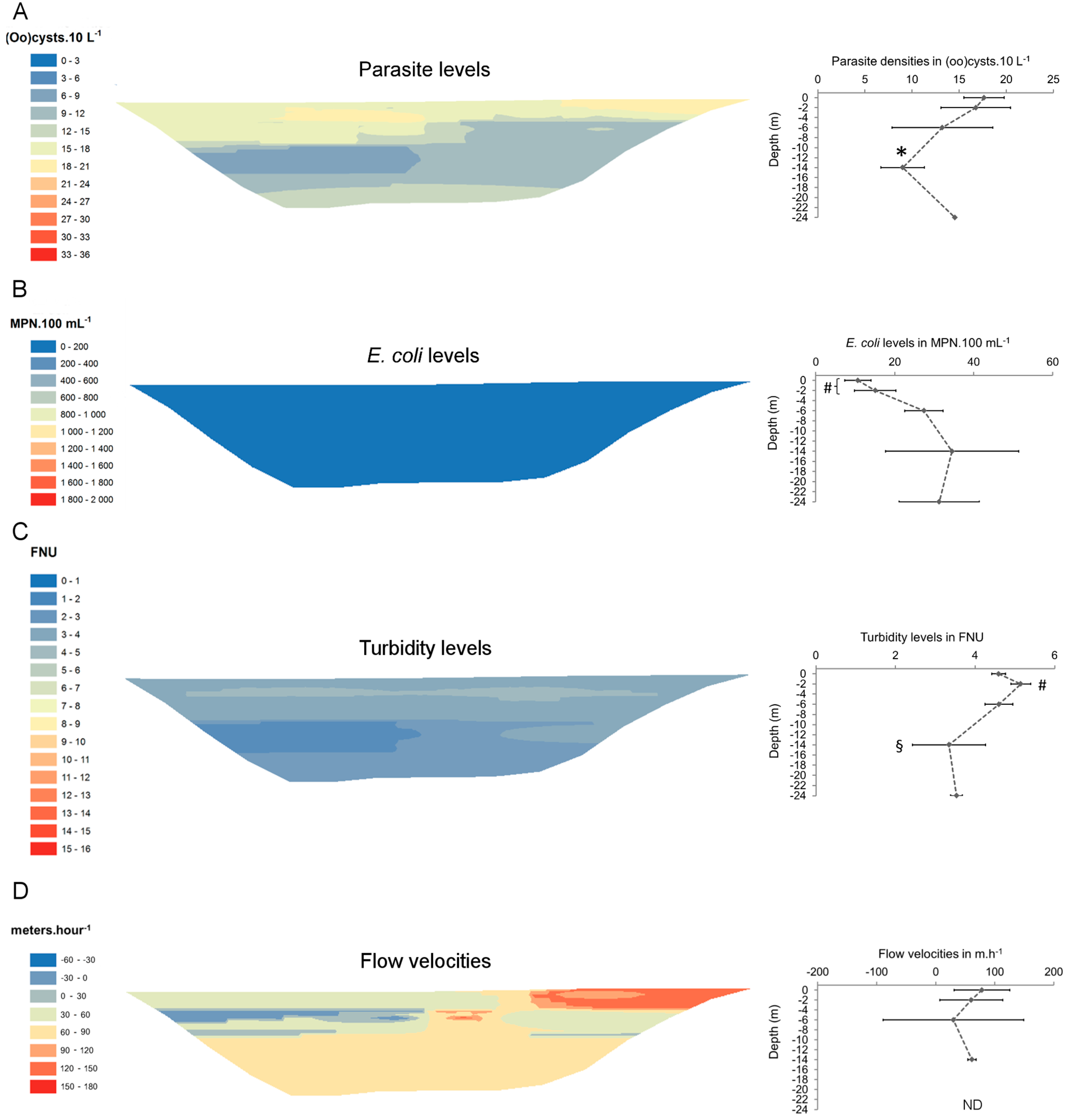
3. Results
| Sampling Site/Date | Cryptosporidium (oocysts·10L−1) | Giardia (cysts·10 L−1) | E. coli * (MPN·100 mL−1) | Total Coliforms * (MPN·100mL−1) | Turbidity (FNU) | TSS (mg·L−1) | Temperature (°C) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Cross-section | Pos./Tot. | Average (min–max) | Pos./Tot. | Average (min–max) | Average (min–max) | Average (min–max) | Average (min–max) | Average (min–max) | Average (min–max) | |
| Site 1 | 09-Dec-09 | 14/17 | 1.9 (0.3–5.1) | 17/17 | 10.6 (1.2–34.6) | 992 (256–1956) | 3588 (650–9208) | 8.9 (4.6–15.7) | 4.7 (2.3–8.3) | 7.8 (7.1–8.4) |
| Site 2 | 19-Jan-10 | 17/24 | 0.6 (0.2–2.1) | 22/24 | 2.9 (0.3–6.0) | 172 (10–657) | 406 (20–1624) | 3.4 (2.3–4.4) | 1.7 (0.9–2.4) | 2.8 (2.3–3.8) |
| 24-Feb-10 | 25/25 | 0.7 (0.1–2.4) | 25/25 | 5.7 (0.4–9.8) | 299 (20–2987) | 542 (115–3609) | 3.3 (1.8–18.2) | 2.0 (0.9–9.4) | 3.2 (2.2–4.6) | |
| 30-Mar-10 | 23/23 | 2.1 (0.3–5.0) | 23/23 | 12.9 (4.0–19.1) | 21 (6–53) | 106 (55–225) | 4.4 (2.7–5.6) | 3.0 (1.8–4.0) | 8.2 (5.3–10.5) | |
| Cross-Section | Rho Value | Combination of Variables | |
|---|---|---|---|
| Site 1 | 09-Dec-09 | 0.784 | Depth, E. coli and TSS |
| Site 2 | 19-Jan-10 | 0.066 | TSS |
| 24-Feb-10 | 0.031 | Conductivity | |
| 30-Mar-10 | 0.425 | Turbidity, TSS | |
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Appendix

| Variable | Sampling Point | Mean Standard Deviation (Mean CV) * | |||||
|---|---|---|---|---|---|---|---|
| C0 | C2 | C6 | D0 | D2 | D6 | ||
| Cryptosporidium (oocysts·10 L−1) | 0.3 ± 0.2 (54%) | 0.4 ± 0.1 (17%) | 0.2 ± 0.1 (40%) | 0.4 ± 0.4 (89%) | 5.9 ± 0.8 (14%) | 2.1 ± 2.2 (104%) | 0.6 (52.8%) |
| Giardia (cysts·10 L−1) | 11.5 ± 1.2 (11%) | 14.6 ± 1.8 (12%) | 12.8 ± 1.8 (14%) | 10.0 ± 0.9 (9%) | 14.3 ± 1.6 (11%) | 14.3 ± 1.7 (12%) | 1.5 (11.5%) |
| E. coli (MPN·100 mL−1) | 17.0 ± 4.5 (26%) | 16.4 ± 2.7 (17%) | 14.9 ± 2.8 (19%) | 17.3 ± 4.2 (24%) | 18.9 ± 6.7 (35%) | 15.4 ± 9.1 (59%) | 5.0 (30.1%) |
| Total coliforms (MPN·100 mL−1) | 41.0 ± 8.8 (22%) | 38.3 ± 6.8 (18%) | 33.8 ± 6.6 (20%) | 100.1 ± 92.7 (93%) | 42.9 ± 13.5 (31%) | 37.9 ± 12.3 (32%) | 23.4 (35.9%) |
| Turbidity (FNU) | 3.4 ± 0.3 (8.4%) | 3.0 ± 0.2 (5.5%) | 3.1 ± 0.2 (7.0%) | 3.1 ± 0.1 (4.8%) | 3.1 ± 0.1 (3.6%) | 3.3 ± 0.5 (14.1%) | 0.2 (7.2%) |
| TSS (mg·L−1) | 30.7 ± 0.3 (0.9%) | 30.6 ± 0.3 (0.9%) | 30.6 ± 0.3 (1.1%) | 34.4 ± 6.7 (19.4%) | 41.8 ± 3.2 (7.7%) | 36.9 ± 7.3 (19.8%) | 3.0 (8.3%) |
| Temperature (°C) | 3.9 ± 0.2 (5.5%) | 4.4 ± 0.1 (3.2%) | 4.4 ± 0.1 (2.9%) | 4.0 ± 0.2 (4.1%) | 4.2 ± 0.2 (4.5%) | 4.5 ± 0.1 (2.1%) | 0.2 (3.7%) |
| pH | 6.8 ± 0.1 (2.1%) | 6.7 ± 0.6 (8.8%) | 7.0 ± 0.1 (1.2%) | 6.8 ± 0.1 (2.1%) | 6.8 ± 0.3 (5.1%) | 7.1 ± 0.4 (5.1%) | 0.3 (4.0%) |
| Conductivity (µS·cm−1) | 154.3 ± 0.7 (0.5%) | 153.5 ± 0.2 (0.1%) | 153.2 ± 0.7 (0.4%) | 154.6 ± 0.7 (0.5%) | 153.5 ± 0.7 (0.5%) | 153.4 ± 0.4 (0.2%) | 0.6 (0.4%) |
| Dissolved oxygen (mg·L−1) | 9.6 ± 0.2 (1.9%) | 9.1 ± 0.2 (2.0%) | 9.3 ± 0.2 (1.9%) | 9.5 ± 0.5 (5.3%) | 9.0 ± 0.1 (1.7%) | 9.6 ± 0.2 (1.7%) | 0.2 (2.4%) |
Conflicts of Interest
References
- White, W.R. World Water: Resources, Usage and the Role of Man-Made Reservoirs; Foundation for Water Research: Bucks, UK, 2010; pp. 1–60. [Google Scholar]
- Brookes, J.D.; Antenucci, J.; Hipsey, M.; Burch, M.D.; Ashbolt, N.J.; Ferguson, C. Fate and transport of pathogens in lakes and reservoirs. Environ. Int. 2004, 30, 741–759. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.B.; Gerba, C.P.; Jakubowski, W. Survey of potable water supplies for Cryptosporidium and Giardia. Environ. Sci. Technol. 1991, 25, 1393–1400. [Google Scholar] [CrossRef]
- Onichandran, S.; Kumar, T.; Lim, Y.A.; Sawangjaroen, N.; Andiappan, H.; Salibay, C.C.; Chye, T.T.; Ithoi, I.; Dungca, J.Z.; Sulaiman, W.Y.; et al. Waterborne parasites and physico-chemical assessment of selected lakes in Malaysia. Parasitol. Res. 2013, 112, 4185–4191. [Google Scholar] [CrossRef] [PubMed]
- Baldursson, S.; Karanis, P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—An update 2004–2010. Water Res. 2011, 45, 6603–6614. [Google Scholar] [CrossRef]
- Mackenzie, W.R.; Hoxie, N.J.; Proctor, M.E.; Gradus, M.S.; Blair, K.A.; Peterson, D.E.; Kazmierczak, J.J.; Addiss, D.G.; Fox, K.R.; Rose, J.B.; et al. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. New Eng. J. Med. 1994, 331, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Nygard, K.; Schimmer, B.; Sobstad, O.; Walde, A.; Tveit, I.; Langeland, N.; Hausken, T.; Aavitsland, P. A large community outbreak of waterborne giardiasis-delayed detection in a non-endemic urban area. BMC Public Health 2006, 6, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Widerstrom, M.; Schonning, C.; Lilja, M.; Lebbad, M.; Ljung, T.; Allestam, G.; Ferm, M.; Bjorkholm, B.; Hansen, A.; Hiltula, J.I.; et al. Large outbreak of Cryptosporidium hominis infection transmitted through the public water supply, Sweden. Emerg. Infect. Dis. 2014, 20, 581–589. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation (WHO). Water Safety Plan Manual (WSP Manual): Step-by-step Risk Management for Drinking-water Suppliers. Available online: http://www.who.int/water_sanitation_health/publication_9789241562638/en/ (accessed on 27 July 2015).
- Dreelin, E.A.; Ives, R.L.; Molloy, S.; Rose, J.B. Cryptosporidium and Giardia in surface water: A case study from Michigan, USA to inform management of rural surface water systems. Int. J. Environ. Res. Public Health 2014, 11, 10480–10503. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.; Moorehead, W.; Ross, A.; Isaac-Renton, J. Studies of Giardia spp. and Cryptosporidium spp. in two adjacent watersheds. Appl. Environ. Microbiol. 1996, 62, 2798–2805. [Google Scholar] [PubMed]
- Wilkes, G.; Ruecker, N.J.; Neumann, N.F.; Gannon, V.P.J.; Jokinen, C.; Sunohara, M.; Topp, E.; Pintar, K.D.M.; Edge, T.A.; Lapen, D.R. Spatiotemporal analysis of Cryptosporidium species/genotypes and relationships with other zoonotic pathogens in surface water from mixed-use watersheds. Appl. Environ. Microbiol. 2013, 79, 434–448. [Google Scholar] [CrossRef] [PubMed]
- Dorner, S.M.; Anderson, W.B.; Gaulin, T.; Candon, H.L.; Slawson, R.M.; Payment, P.; Huck, P.M. Pathogen and indicator variability in a heavily impacted watershed. J. Water Health 2007, 5, 241–257. [Google Scholar] [PubMed]
- Keeley, A.; Faulkner, B.R. Influence of land use and watershed characteristics on protozoa contamination in a potential drinking water resources reservoir. Water Res. 2008, 42, 2803–2813. [Google Scholar] [CrossRef] [PubMed]
- Helmi, K.; Skraber, S.; Burnet, J.B.; Leblanc, L.; Hoffmann, L.; Cauchie, H.M. Two-year monitoring of Cryptosporidium parvum and Giardia lamblia occurrence in a recreational and drinking water reservoir using standard microscopic and molecular biology techniques. Environ. Monit. Assess. 2011, 179, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Brookes, J.D.; Hipsey, M.R.; Burch, M.D.; Regel, R.H.; Linden, L.G.; Ferguson, C.M.; Antenucci, J.P. Relative value of surrogate indicators for detecting pathogens in lakes and reservoirs. Environ. Sci. Technol. 2005, 39, 8614–8621. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, V.; Lair, N.; Hoffmann, L.; Cauchie, H.M. Spatio-temporal patterns of protozoan communities in a meso-eutrophic reservoir (Esch-sur-Sûre, Luxembourg). Hydrobiologia 2005, 551, 49–60. [Google Scholar] [CrossRef]
- Douglas-Helders, G.M.; O'Brien, D.P.; McCorkell, B.E.; Zilberg, D.; Gross, A.; Carson, J.; Nowak, B.F. Temporal and spatial distribution of paramoebae in the water column—A pilot study. J. Fish. Dis. 2003, 26, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Bettarel, Y.; Amblard, C.; Sime-Ngando, T.; Carrias, J.F.; Sargos, D.; Garabétian, F.; Lavandier, P. Viral lysis, flagellate grazing potential, and bacterial production in Lake Pavin. Microb. Ecol. 2003, 45, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Goddard, V.J.; Baker, A.C.; Davy, J.E.; Adams, D.G.; De Ville, M.M.; Thackeray, S.J.; Maberly, S.C.; Wilson, W.H. Temporal distribution of viruses, bacteria and phytoplankton throughout the water column in a freshwater hypereutrophic lake. Aquat. Microb. Ecol. 2005, 39, 211–223. [Google Scholar] [CrossRef]
- De Wever, A.; Muylaert, K.; Van Der Gucht, K.; Pirlot, S.; Cocquyt, C.; Descy, J.P.; Plisnier, P.D.; Vyverman, W. Bacterial community composition in Lake Tanganyika: Vertical and horizontal heterogeneity. Appl. Environ. Microbiol. 2005, 71, 5029–5037. [Google Scholar] [CrossRef] [PubMed]
- Ongerth, J.E.; Saeed, F.M.A. Distribution of Cryptosporidium oocysts and Giardia cysts in water above and below the normal limit of detection. Parasitol. Res. 2013, 112, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Alderisio, K.A.; Jiang, J. Detection of Cryptosporidium oocysts in water: effect of the number of samples and analytic replicates on test results. Appl. Environ. Microbiol. 2006, 72, 5942–5947. [Google Scholar] [CrossRef] [PubMed]
- LeChevallier, M.W.; Di Giovanni, G.D.; Clancy, J.L.; Bukhari, Z.; Bukhari, S.; Rosen, J.S.; Sobrinho, J.; Frey, M.M. Comparison of method 1623 and cell culture-PCR for detection of Cryptosporidium spp. in source waters. Appl. Environ. Microbiol. 2003, 69, 971–979. [Google Scholar] [CrossRef] [PubMed]
- King, B.; Fanok, S.; Phillips, R.; Swaffer, B.; Monis, P. Integrated Cryptosporidium assay to determine oocyst density, infectivity and genotype for risk assessment of source and reuse water. Appl. Environ. Microbiol. 2015, 81, 3471–3481. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Long, S.C.; Das, D.; Dorner, S.M. Are microbial indicators and pathogens correlated? A statistical analysis of 40 years of research. J. Water Health 2011, 9, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Burnet, J.B.; Penny, C.; Ogorzaly, L.; Cauchie, H.M. Spatial and temporal distribution of Cryptosporidium and Giardia in a drinking water resource: Implications for monitoring and risk assessment. Sci. Total Environ. 2014, 472, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, R.G. Water movements. In Limnology: Lake and River Ecosystems, 3rd ed.; Academic Press: New York, NY, USA, 2001; pp. 93–128. [Google Scholar]
- CRHS. État Des Lieux De La Haute-Sûre (in French). Available online: http://www.crhs.eu/index.php?id=8;lang=fr (accessed on 27 July 2015).
- Agrometeorological Measurement Network of Luxembourg. Available online: www.agrimeteo.lu (accessed on 27 July 2015).
- Hydro-Climatological Observatory of Luxembourg. Available online: www.hydroclimato.lu (accessed on 27 July 2015).
- Burnet, J.B. Spatial and Temporal Dynamics of Cryptosporidium and Giardia in a Drinking Water Reservoir and its Catchment: The Upper-Sûre Reservoir Complex. PhD Thesis, University of Liège, Arlon, Belgium, 16 October 2012. [Google Scholar]
- Clarke, K.R.; Warwick, R.M. Quantifying structural redundancy in ecological communities. Oecologia 1998, 113, 278–289. [Google Scholar] [CrossRef]
- Van Breemen, L.W.C.A.; Ketelaars, H.; Hoogenboezem, W.; Medema, G. Storage reservoirs—A first barrier for pathogenic micro-organisms in the Netherlands. Water Sci. Technol. 1998, 37, 253–260. [Google Scholar] [CrossRef]
- Gallas-Lindemann, C.; Sotiriadou, I.; Plutzer, J.; Karanis, P. Prevalence and distribution of Cryptosporidium and Giardia in wastewater and the surface, drinking and ground waters in the Lower Rhine, Germany. Epidemiol. Infect. 2013, 141, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Antenucci, J.P.; Brookes, J.D.; Hipsey, M.R. A simple model for quantifying Cryptosporidium transport, dilution, and potential risk in reservoirs. J. Am. Water Works Ass. 2005, 97, 86–93. [Google Scholar]
- Emelko, M.B.; Schmidt, P.J.; Reilly, P.M. Particle and microorganism enumeration data: Enabling quantitative rigor and judicious interpretation. Environ. Sci. Technol. 2010, 44, 1720–1727. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, C.; Kaucner, C.; Krogh, M.; Deere, D.; Warnecke, M. Comparison of methods for the concentration of Cryptosporidium oocysts and Giardia cysts from raw waters. Can. J. Microbiol. 2004, 50, 675–682. [Google Scholar] [CrossRef] [PubMed]
- US EPA. Method 1623: Cryptosporidium and Giardia in Water by Filtration/IMS/FA. 2005; EPA 815-R-05-002. Available online: http://water.epa.gov/scitech/methods/cwa/bioindicators/upload/method_1623.pdf (accessed on 27 July 2015). [Google Scholar]
- Zuckerman, U.; Tzipori, S. Portable continuous flow centrifugation and method 1623 for monitoring of waterborne protozoa from large volumes of various water matrices. J. Appl. Microbiol. 2006, 100, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Edzwald, J.K.; Kelley, M.B. Control of Cryptosporidium: From reservoirs to clarifiers to filters. Water Sci. Technol. 1998, 37, 1–8. [Google Scholar] [CrossRef]
- Quilliam, R.S.; Clements, K.; Duce, C.; Cottrill, S.B.; Malham, S.K.; Jones, D.L. Spatial variation of waterborne Escherichia coli—Implications for routine water quality monitoring. J. Water Health 2011, 9, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Hawley, N.; Wang, X.; Brownawell, B.; Flood, R. Resuspension of bottom sediments in Lake Ontario during the unstratified period, 1992–1993. J. Great Lakes Res. 1996, 22, 707–721. [Google Scholar] [CrossRef]
- Chung, G.E.; Bonbardelli, F.A.; Shladow, S.G. Sediment resuspension in a shallow lake. Water Resour. Res. 2009, 45. [Google Scholar] [CrossRef]
- Molloy, S.L.; Montgomery, A.E.; Huffman, D.E.; Rose, J.B. Detection of Cryptosporidium parvum oocysts in sediment and biosolids by immunomagnetic separation. Water Environ. Res. 2006, 78, 1013–1016. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Reckhow, K.H.; Lukasik, J.; Harwood, V.J. Assessment of sources of human pathogens and faecal contamination in a Florida freshwater lake. Water Res. 2012, 46, 5799–5812. [Google Scholar] [CrossRef] [PubMed]
- Nasser, A.M.; Zaruk, N.; Tenenbaum, L.; Netzan, Y. Comparative survival of Cryptosporidium, coxsackievirus A9 and Escherichia coli in stream, brackish and sea waters. Water Sci. Tech. 2003, 47, 91–96. [Google Scholar]
- Young, I.; Smith, B.A.; Fazil, A. A systematic review and meta-analysis of the effects of extreme weather events and other weather-related variables on Cryptosporidium and Giardia in fresh surface waters. J. Water Health 2015, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Atherholt, T.B.; LeChevallier, M.W.; Norton, W.D.; Rosen, J.S. Effect of rainfall on Giardia and Crypto. J. Am. Water Works Ass. 1998, 90, 66–80. [Google Scholar]
- Weintraub, J.M. Improving Cryptosporidium testing methods: A public health perspective. J. Water Health 2006, 4, 23–26. [Google Scholar] [PubMed]
- Aw, T.G.; Rose, J.B. Detection of pathogens in water: from phylochips to qPCR to pyrosequencing. Curr. Opin. Biotechnol. 2012, 23, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Bridle, H.; Miller, B.; Desmulliez, M.P.Y. Application of microfluidics in pathogen monitoring: A review. Water Res. 2014, 55, 256–271. [Google Scholar] [CrossRef] [PubMed]
- Burnet, J.B.; Ogorzaly, L.; Tissier, A.; Penny, C.; Cauchie, H.M. Novel quantitative TaqMan real-time PCR assays for detection of Cryptosporidium at the genus level and genotyping of major human and cattle-infecting species. J. Appl. Microbiol. 2013, 114, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, J.A.; Bonilla, T.D.; Abdelzaher, A.M.; Scott, T.M.; Lukasik, J.; Solo-Gabriele, H.M.; Palmer, C.J. Quantification of protozoa and viruses from small water volumes. Int. J. Environ. Res. Public Health 2015, 12, 7118–7132. [Google Scholar] [CrossRef] [PubMed]
- Hill, V.R.; Polaczyk, A.L.; Kahler, A.M.; Cromeans, T.L.; Hahn, D.; Amburgey, J.E. Comparison of hollow-fiber ultrafiltration to the usepa viradel technique and usepa method 1623. J. Environ. Qual. 2009, 38, 822–825. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, A.B.; Schladow, S.G.; Rueda, F.J. A hydrodynamics-based approach to evaluating the risk of waterborne pathogens entering drinking water intakes in a large, stratified lake. Water Res. 2015, 83, 227–236. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burnet, J.-B.; Ogorzaly, L.; Penny, C.; Cauchie, H.-M. Fine-Scale Spatial Heterogeneity in the Distribution of Waterborne Protozoa in a Drinking Water Reservoir. Int. J. Environ. Res. Public Health 2015, 12, 11910-11928. https://doi.org/10.3390/ijerph120911910
Burnet J-B, Ogorzaly L, Penny C, Cauchie H-M. Fine-Scale Spatial Heterogeneity in the Distribution of Waterborne Protozoa in a Drinking Water Reservoir. International Journal of Environmental Research and Public Health. 2015; 12(9):11910-11928. https://doi.org/10.3390/ijerph120911910
Chicago/Turabian StyleBurnet, Jean-Baptiste, Leslie Ogorzaly, Christian Penny, and Henry-Michel Cauchie. 2015. "Fine-Scale Spatial Heterogeneity in the Distribution of Waterborne Protozoa in a Drinking Water Reservoir" International Journal of Environmental Research and Public Health 12, no. 9: 11910-11928. https://doi.org/10.3390/ijerph120911910
APA StyleBurnet, J.-B., Ogorzaly, L., Penny, C., & Cauchie, H.-M. (2015). Fine-Scale Spatial Heterogeneity in the Distribution of Waterborne Protozoa in a Drinking Water Reservoir. International Journal of Environmental Research and Public Health, 12(9), 11910-11928. https://doi.org/10.3390/ijerph120911910








