Resistance Markers and Genetic Diversity in Acinetobacter baumannii Strains Recovered from Nosocomial Bloodstream Infections
Abstract
:1. Introduction
2. Experimental Section
2.1. Bacterial Strains, Identification and Susceptibility Testing
2.2. Phenotypic Detection of Class B (Metallo-β-Lactamase) Enzymes Related to Resistance
2.2.1. Disk Approximation (DA) Test
2.2.2. MΒLE-Test ®
2.3. Polymerase Chain Reaction for the Detection of β-Lactamase Genes
| Genes | Primers | Product (pb) | Reference |
|---|---|---|---|
| blaSPM-1 | 5’-AAAATCTGGGTACGCAAACG-3’ 5’-ACATTATCCGCTGGAACAGG-3’ | 271 pb | [15] |
| blaIMP-1 | 5’-GAATAGAGTGGCTTAACTCTC-3’ 5’-AGATAACCTAGTGGTTTGG-3’ | 189 pb | [15] |
| blaVIM-1 | 5’-CGAATGCGCAGCACCAG-3’ 5’-CTGGTGCTGCGCATTCG-3’ | 390 pb | [15] |
| blaGIM-1 | 5’-TCGACACACCTTGGTCTGAA-3’ 5’-AACTTCCAACTTTGCCATGC-3’ | 477 pb | [15] |
| blaSIM-1 | 5’-TACAAGGGATTCGGCATCG-3’ 5’-TAATGGCCTGTTCCCATGTG-3’ | 571 pb | [16] |
| blaAmpC | 5’-CAGTAGCGAGACTGCGCA-3’ 5’-ATAACCACCCAGTCACGC-3’ | 631 pb | [17,18] |
| blaOXA23 | 5’-GATCGGATTGGAGAACCAGA-3’ 5’-ATTTCTGACCGCATTTCCAT-3’ | 501 pb | [6] |
| blaOXA24 | 5’-GGTTAGTTGGCCCCCTTAAA-3’ 5’-AGTTGAGCGAAAAGGGGATT-3’ | 246pb | [6] |
| blaOXA51 | 5’-TAATGCTTTGATCGGCCTTG-3’ 5’-ATTTCTGACCGCATTTCCAT-3’ | 353 pb | [6] |
| blaOXA58 | 5’-AAGTATTGGGGCTTGTGCTG-3’ 5’-CCCCTCTGCGCTCTACATAC-3’ | 599 pb | [6] |
PCR Product Analysis
2.4. Characterization of the Genetic Profiles of the Strains
2.5. Ethical Considerations
3. Results and Discussion
3.1. Antimicrobial Susceptibility Profiles
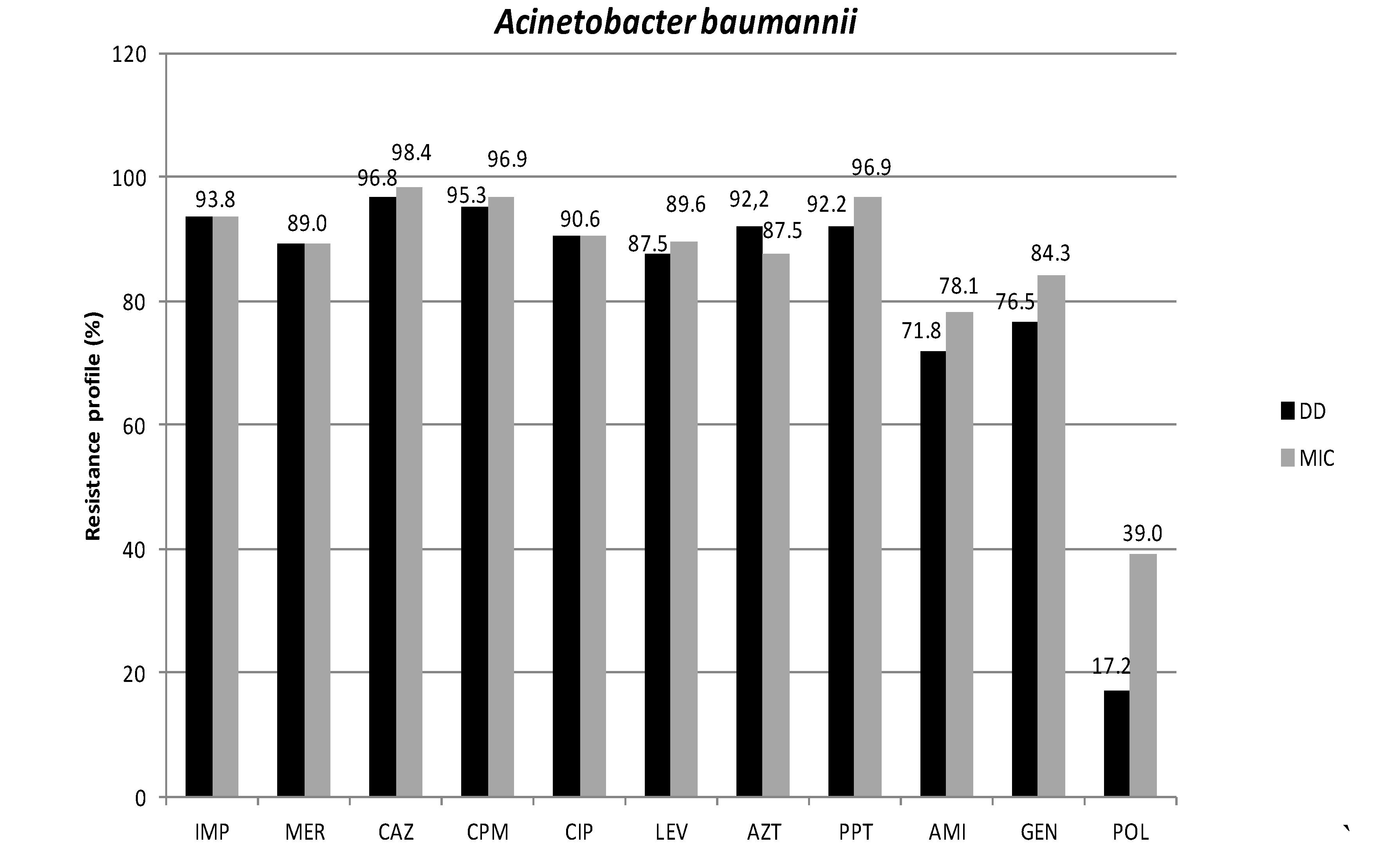
3.2. Phenotypic Detection of Enzymes Related to Resistance
3.3. Detection of β-Lactamase Genes
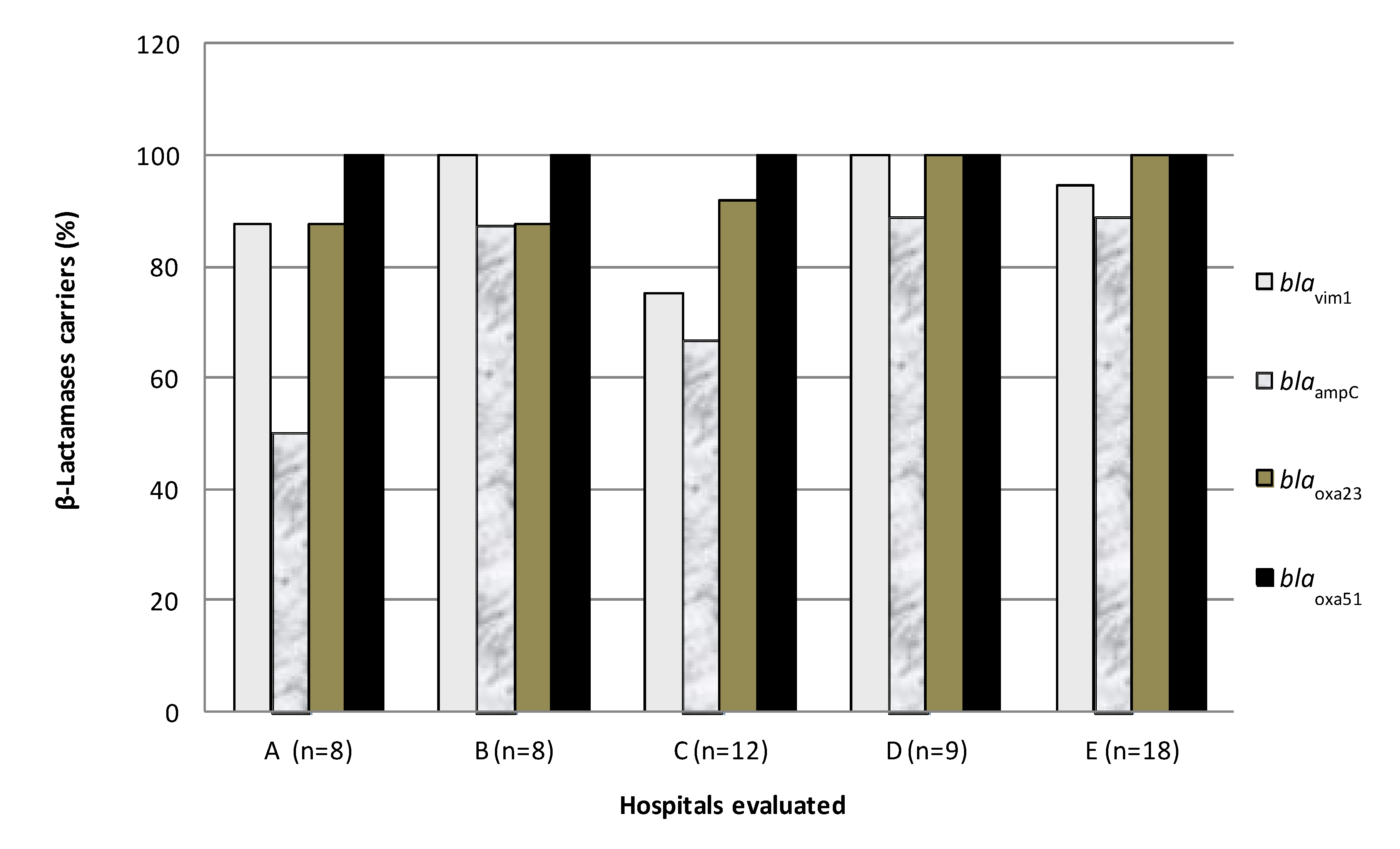
| Genes | Frequency | % |
|---|---|---|
| blaVIM-1-blaAmpC-blaOXA23-blaOXA51 | 37 | 67.3 |
| blaVIM-1-blaOXA23-blaOXA51 | 11 | 20.0 |
| blaAmpC-blaOXA23-blaOXA51 | 3 | 5.5 |
| blaVIM-1-blaAmpC-blaOXA51 | 2 | 3.6 |
| blaOXA23-blaOXA51 | 1 | 1.8 |
| blaVIM-1- blaOxa51 | 1 | 1.8 |
3.4. Genotypic Characterization of A. Baumannii Strains
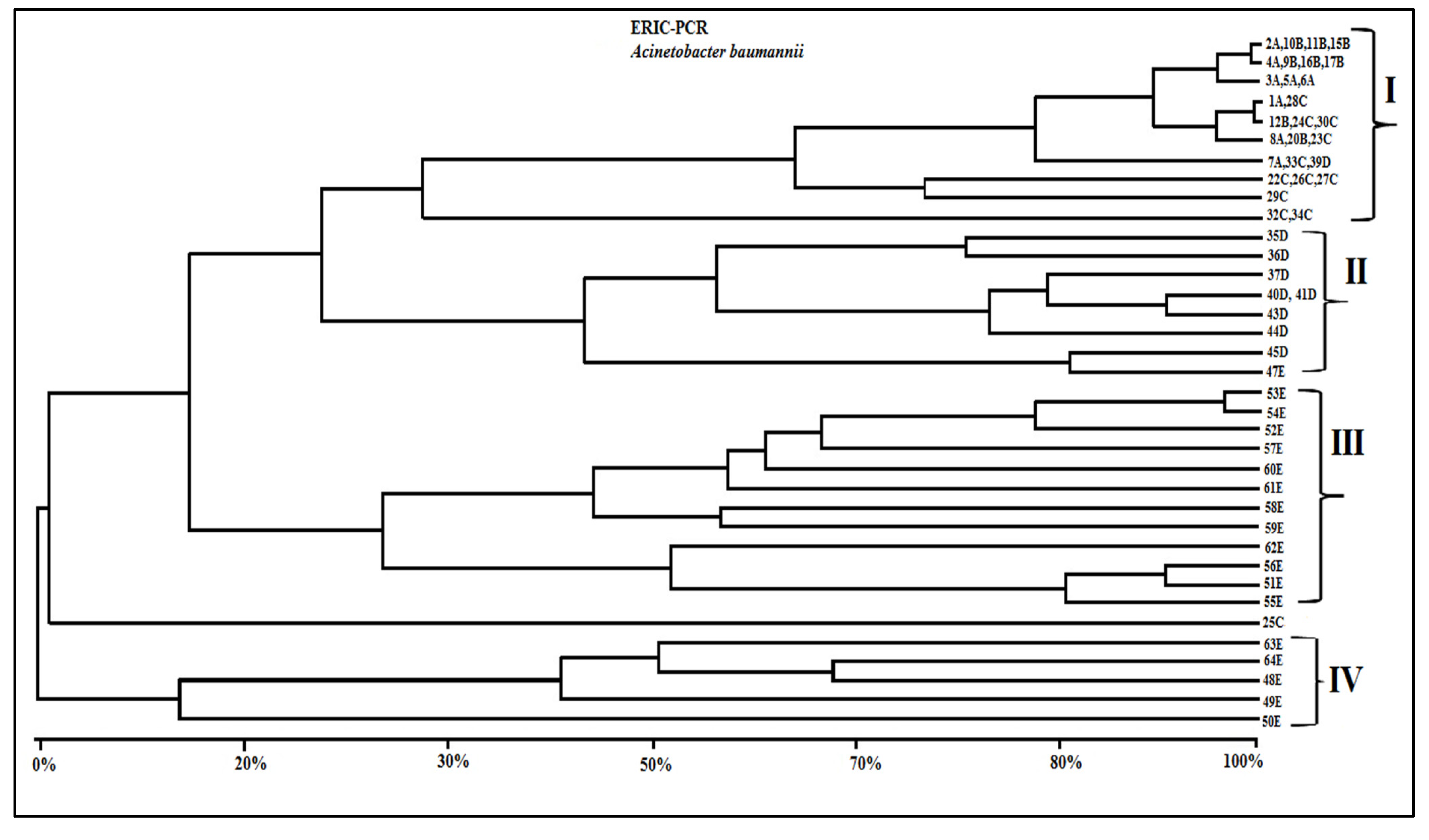
| Isolate/ Hospital | ERIC-PCR Groups | Resistance Genes | Etest MBL | Phenotypic Profile Resistance |
|---|---|---|---|---|
| 1A | I | blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 2A | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 3A | I | blaVIM-1, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, AZ, PT, AM, GE, PO |
| 4A | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, PT |
| 5A | I | blaVIM-1, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 6A | I | blaVIM-1, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, AZ, PT, AM, GE, PO |
| 7A | I | blaVIM-1, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 8A | I | blaVIM-1, blaAmpC, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 9B | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 10B | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 11B | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 12B | I | blaVIM-1, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 15B | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 16B | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 17B | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 20B | I | blaVIM-1, blaAmpC, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 22C | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 23C | I | blaVIM-1, blaAmpC, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 24C | I | blaVIM-1, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 25C | Ungrouped | blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 26C | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, AZ, PT, AM, GE, PO |
| 27C | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, LE, AZ, PT, AM |
| 28C | I | blaAmpC, blaOxa23, blaOxa51 | − | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 29C | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 30C | I | blaVIM-1, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 32C | I | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 33C | I | blaVIM-1, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 34C | I | blaVIM-1,blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 35D | II | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 36D | II | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 37D | II | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 39D | I | blaVIM-1, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 40D | II | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 41D | II | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 43D | II | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 44D | II | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 45D | II | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, AZ, PT, AM, GE |
| 47E | II | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 48E | IV | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 49E | IV | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE, PO |
| 50E | IV | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 50E | IV | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 51E | III | blaVIM-1, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 52E | III | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 53E | III | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 54E | III | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 55E | III | blaAmpc, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 56E | III | blaVIM-1, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 57E | III | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, CA, CP, CI, LE, AZ, PT |
| 58E | III | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM |
| 59E | III | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, |
| 60E | III | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 61E | III | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT, AM, GE |
| 62E | III | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT |
| 63E | IV | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | + | IM, ME, CA, CP, CI, LE, AZ, PT |
| 64E | IV | blaVIM-1, blaAmpC, blaOxa23, blaOxa51 | − | IM, CA, CP, CI, LE, AZ, PT, AM, GE |
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wisplinghoff, H.H.; Paulus, T.T.; Lugenheim, M.M.; Stefanik, D.D.; Higgins, P.G.; Edmond, M.B.; Wenzel, R.P.; Seifert, H. Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J. Infect. 2012, 64, 282–290. [Google Scholar] [CrossRef]
- Cisneros-Herreros, J.M.; Garnacho-Montero, J.; Pachon-Ibanez, M.E. Nosocomial pneumonia due to Acinetobacter baumannii. Enferm. Infecc. Microbiol. Clin. 2005, 3, 46–51. [Google Scholar]
- Marra, A.R.; Sampaio Camargo, T.Z.; Goncalves, P.; Sogayar, A.M.; Moura, D.F., Jr.; Guastelli, L.R.; Alves Rosa, C.A.; da Silva Victor, E.; Pavao dos Santos, O.F.; Edmond, M.B. Preventing catheter-associated urinary tract infection in the zero-tolerance era. Am. J. Infect. Control. 2011, 39, 817–822. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Suscetibility Testing; Seventeenth Informational Supplement; M100-S17. CLSI: Pennsylvania, PA, USA, 2011. Available online: http://www.microbiolab-bg.com/CLSI.pdf (accessed on 28 October 2013).
- Cloete, T.E. Resistance mechanisms of bacteria to antimicrobial compounds. Int. Biodeterior. Biodegr. 2003, 51, 277–282. [Google Scholar] [CrossRef]
- Turton, J.F.; Ward, M.E.; Woodford, N.; Kaufmann, M.E.; Pike, R.; Livermore, D.M.; Pitt, T.L. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 2006, 258, 72–77. [Google Scholar] [CrossRef]
- Woodford, N.; Ellington, M.J.; Coelho, J.M.; Turton, J.F.; Ward, M.E.; Brown, S.; Amyes, S.G.B.; Livermore, D.M. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Ag. 2006, 27, 351–353. [Google Scholar] [CrossRef]
- Vila, J.; Marti, S.; Sanchez-Cespedes, J. Porins, efflux pumps and multidrug resistance in Acinetobacter Baumannii. J. Antimicrob. Chemother. 2007, 59, 1210–1215. [Google Scholar] [CrossRef]
- Unal, S.; Garcia-Rodriguez, J.A. Activity of meropenem and comparators against Pseudomonas aeruginosa and Acinetobacter spp. isolated in the MYSTIC Program, 2002–2004. Diagn. Microbiol. Infect. Dis. 2005, 53, 265–271. [Google Scholar] [CrossRef]
- Perez, F.; Hujer, A.M.; Hujer, K.M.; Decker, B.K.; Rather, P.N.; Bonomo, R.A. Global challenge of multidrug-resistant Acinetobacter Baumannii. Antimicrob. Agents Ch. 2007, 51, 3471–3484. [Google Scholar] [CrossRef]
- Arakawa, Y. Convenient test for screening metallo-βlactamase-producing gram-negative bacteria by using thiol compounds. J. Clin. Microbiol. 2000, 38, 40–43. [Google Scholar]
- Yong, D.; Lee, K.; Yum, J.H.; Shin, H.B.; Rossolini, G.M.; Chong, Y. Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 2002, 40, 3798–3801. [Google Scholar] [CrossRef]
- Mendes, R.E.; Castanheira, M.; Pignatari, A.C.C.; Gales, A.C. Metalo-β-lactamases. J. Bras. Patol. Med. Lab. 2006, 42, 103–113. [Google Scholar] [CrossRef]
- Hammami, S.; Boutiba-Ben Boubaker, I.; Ghozzi, R.; Saidani, M.; Amine, S.; Ben Redjeb, S. Nosocomial outbreak of imipenem-resistant Pseudomonas aeruginosa producing VIM-2 metallo-beta-lactamase in a kidney transplantation unit. Diagn. Pathol. 2011, 6. [Google Scholar] [CrossRef]
- Walsh, T.R.; Toleman, M.A.; Hryniewicz, W.; Bennett, P.M.; Jones, R.N. Evolution of an integron carrying blaVIM-2 in Eastern Europe: report from the SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 2003, 52, 116–119. [Google Scholar] [CrossRef]
- Yan, J.J.; Hsueh, P.R.; Ko, W.C.; Luh, K.T.; Tsai, S.H.; Wu, H.M.; Wu, J.J. Metallo-beta-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Ch. 2001, 45, 2224–2228. [Google Scholar] [CrossRef]
- Lee, K.; Yum, J.H.; Yong, D.; Lee, H.M.; Kim, H.D.; Docquier, J.D.; Rossolini, G.M.; Chong, Y. Novel acquired metallo-beta-lactamase gene, bla(SIM-1), in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Ch. 2005, 49, 4485–4491. [Google Scholar] [CrossRef]
- Sompolinsky, D.; Nitzan, Y.; Tetry, S.; Wolk, M.; Vulikh, I.; Kerrn, M.B.; Sandvang, D.; Hershkovit, G.; Katcoff, D.J. Integron-mediated ESBL resistance in rare serotypes of Escherichia coli causing infections in an elderly population of Israel. J. Antimicrob. Chemother. 2005, 55, 119–122. [Google Scholar]
- Coudron, P.E.; Moland, E.S.; Thomson, K.S. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J. Clin. Microbiol. 2000, 38, 1791–1796. [Google Scholar]
- Silbert, S.; Pfaller, M.A.; Hollis, R.J.; Barth, A.L.; Sader, H.S. Evaluation of three molecular typing techniques for nonfermentative Gram-negative bacilli. Infect. Control. Hosp. Epidemiol. 2004, 25, 847–851. [Google Scholar]
- Antonio, C.S.; Neves, P.R.; Medeiros, M.; Mamizuka, E.M. High Prevalence of Carbapenem-Resistant Acinetobacter. baumannii carrying the blaOXA-143 Gene in Brazilian Hospitals. Antimicrob. Agents Ch. 2011, 55, 1322–1323. [Google Scholar] [CrossRef]
- Werneck, J.S.; Picao, R.C.; Girardello, R.; Cayo, R.; Marguti, V.; Dalla-Costa, L.; Gales, A.C.; Antonio, C.S.; Neves, P.R.; Medeiros, M.; Mamizuka, E.M.; Elmor de Araujo, M.R.; Lincopan, N. Low prevalence of blaOXA-143 in private hospitals in Brazil. Antimicrob. Agents Ch. 2011, 55, 4494–4495. [Google Scholar] [CrossRef]
- Ferreira, A.E.; Marchetti, D.P.; da Cunha, G.R.; de Oliveira, L.M.; Fuentefria, D.B.; Dall Bello, A.G.; Corcao, G. Molecular characterization of clinical multiresistant isolates of Acinetobacter. sp. from hospitals in Porto Alegre, State of Rio Grande do Sul, Brazil. Rev. Soc. Bras. Med. Trop. 2011, 44, 725–730. [Google Scholar] [CrossRef]
- Ambler, R.P. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B, Biol. Sci. 1980, 16, 321–331. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A.; Medeiros, A.A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Ch. 1995, 39, 1211–1233. [Google Scholar] [CrossRef]
- Figueiredo, D.Q.; Castro, L.F.S.; Santos, K.R.N.; Teixeira, L.M.; Mondino, S.S.B. Detection of metallo-beta-lactamases in hospital strains of Pseudomonas aeruginosa and Acinetobacter baumannii. J. Bras. Patol. Med. Lab. 2009, 45, 177–184. [Google Scholar]
- Arda, B.; Sipahi, O.R.; Yamazhan, T.; Tasbakan, M.; Pullukcu, H.; Tunger, A.; Buke, C.; Ulusoy, S. Short-term effect of antibiotic control policy on the usage patterns and cost of antimicrobials, mortality, nosocomial infection rates and antibacterial resistance. J. Infect. 2007, 55, 41–48. [Google Scholar] [CrossRef]
- Laranjeiras, V.S.; Marchetti, D.P.; Steyer, J.C.; Corcao, G.; Picoli, S.U. Investigation of metallo-β-lactamase producing Acinetobacter sp. and Pseudomonas aeruginosa at an emergency hospital in Porto Alegre, State of Rio Grande do Sul, Brazil. Rev. Soc. Bras. Med. Trop. 2010, 43, 462–464. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC beta-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYS Numerical Taxonomy and Multivariate Analysis System, version 1.60; Exeter Software: New York, NY, USA, 1990. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Martins, H.S.I.; Bomfim, M.R.Q.; França, R.O.; Farias, L.M.; Carvalho, M.A.R.; Serufo, J.C.; Santos, S.G. Resistance Markers and Genetic Diversity in Acinetobacter baumannii Strains Recovered from Nosocomial Bloodstream Infections. Int. J. Environ. Res. Public Health 2014, 11, 1465-1478. https://doi.org/10.3390/ijerph110201465
Martins HSI, Bomfim MRQ, França RO, Farias LM, Carvalho MAR, Serufo JC, Santos SG. Resistance Markers and Genetic Diversity in Acinetobacter baumannii Strains Recovered from Nosocomial Bloodstream Infections. International Journal of Environmental Research and Public Health. 2014; 11(2):1465-1478. https://doi.org/10.3390/ijerph110201465
Chicago/Turabian StyleMartins, Hanoch S. I., Maria Rosa Q. Bomfim, Rafaela O. França, Luiz M. Farias, Maria Auxiliadora R. Carvalho, José Carlos Serufo, and Simone G. Santos. 2014. "Resistance Markers and Genetic Diversity in Acinetobacter baumannii Strains Recovered from Nosocomial Bloodstream Infections" International Journal of Environmental Research and Public Health 11, no. 2: 1465-1478. https://doi.org/10.3390/ijerph110201465
APA StyleMartins, H. S. I., Bomfim, M. R. Q., França, R. O., Farias, L. M., Carvalho, M. A. R., Serufo, J. C., & Santos, S. G. (2014). Resistance Markers and Genetic Diversity in Acinetobacter baumannii Strains Recovered from Nosocomial Bloodstream Infections. International Journal of Environmental Research and Public Health, 11(2), 1465-1478. https://doi.org/10.3390/ijerph110201465





