Bioactive (3Z,5E)-11,20-Epoxybriara-3,5-dien-7,18-olide Diterpenoids from the South China Sea Gorgonian Dichotella gemmacea
Abstract
:1. Introduction
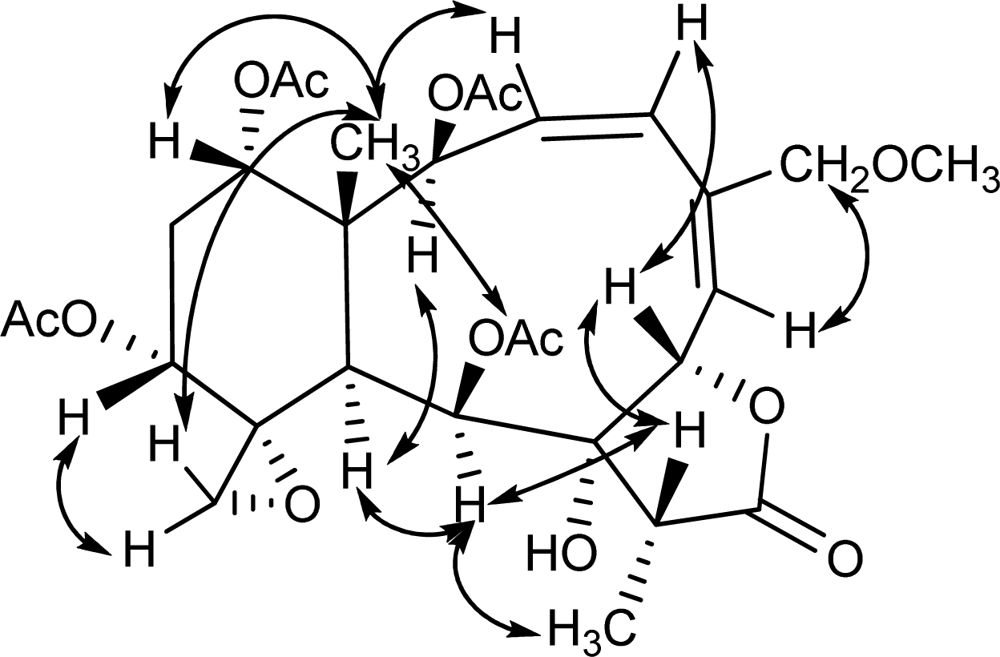
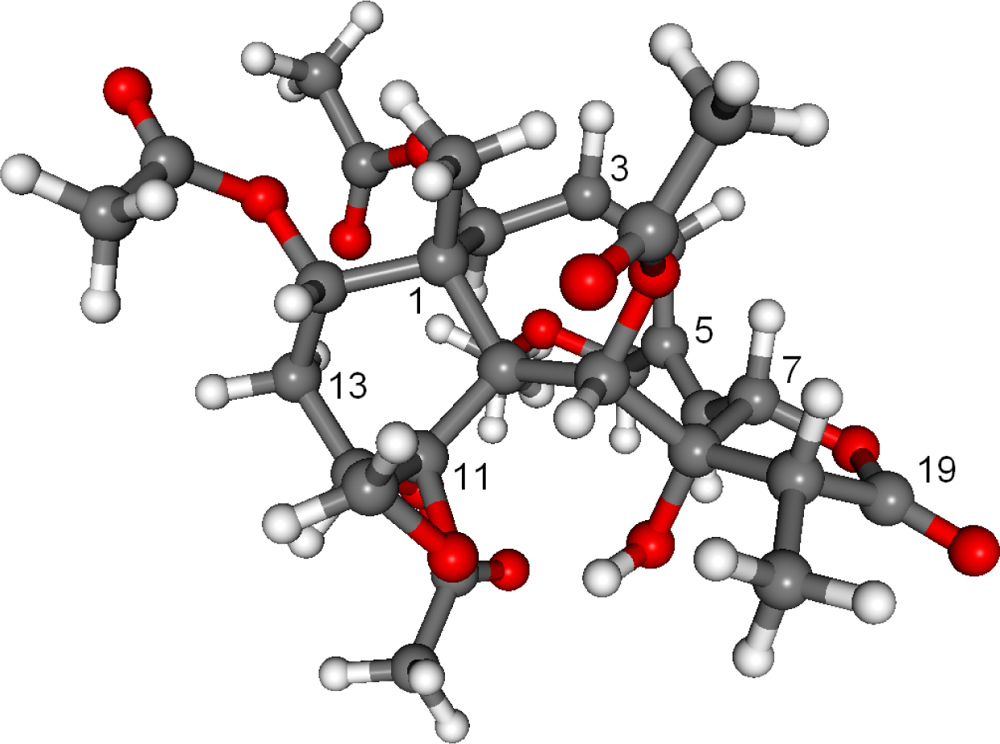
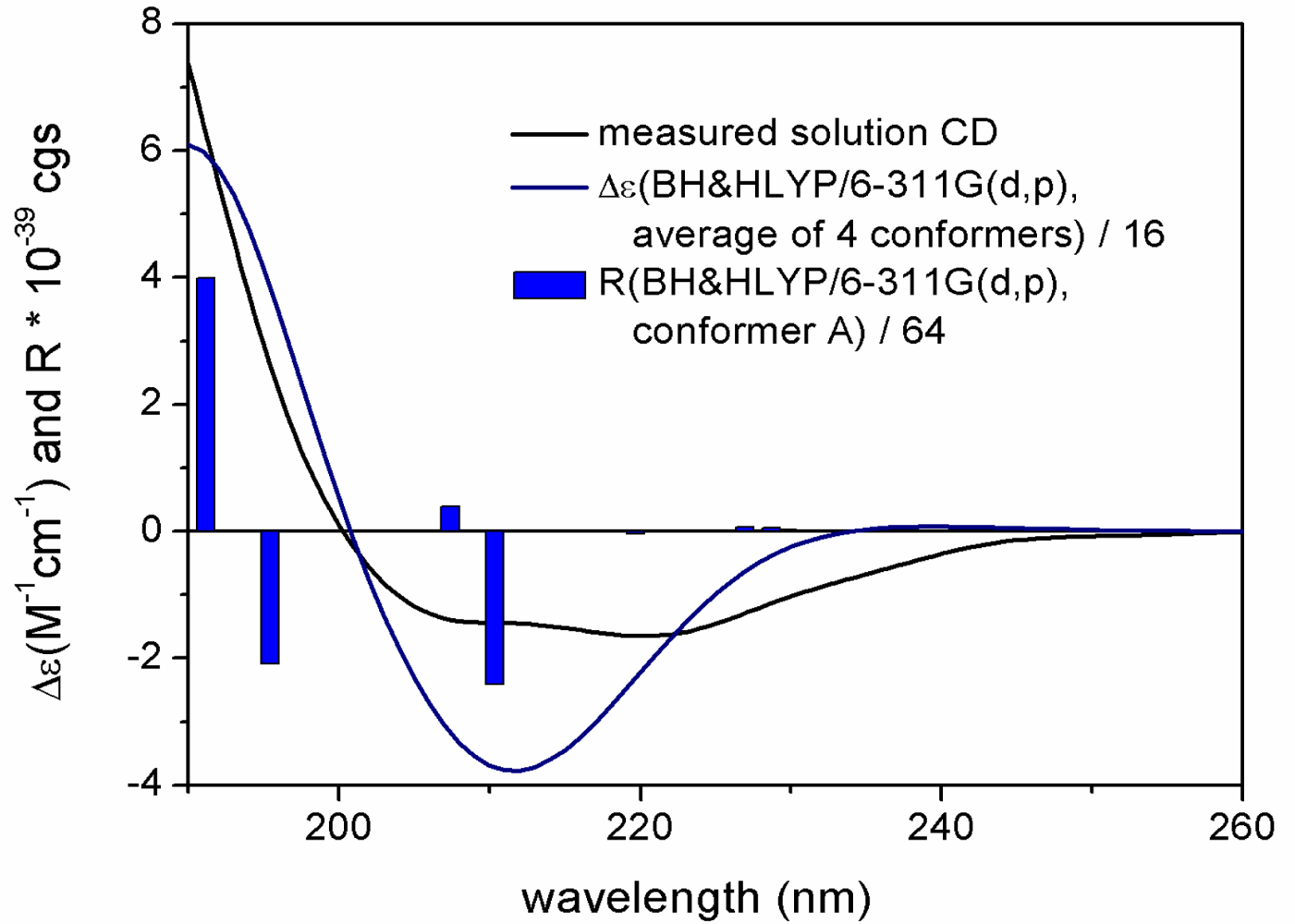
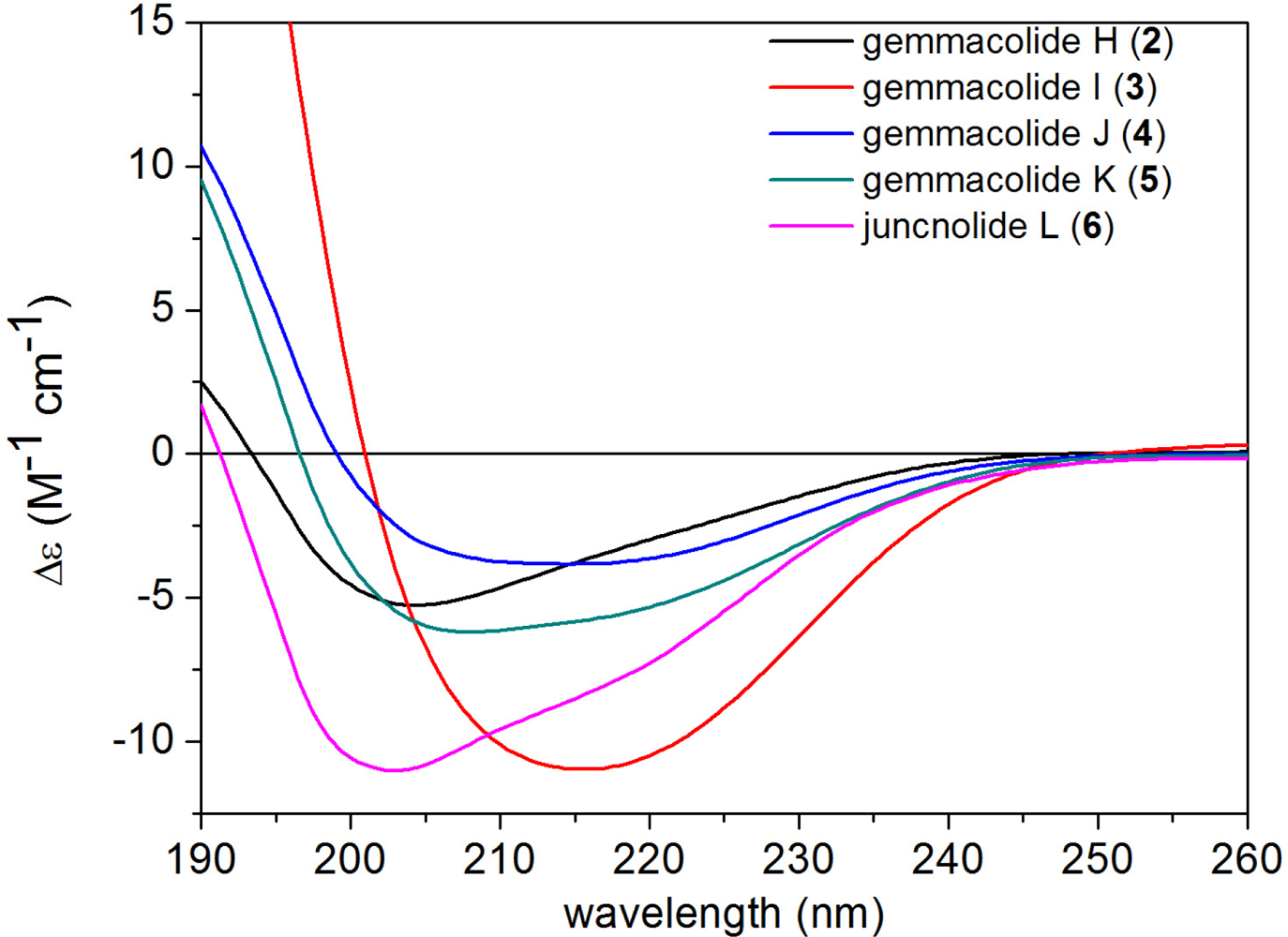
2. Results and Discussion
3. Experimental Section
3.1. General Experimental Procedures
3.2. Animal Material
3.3. Extraction and Isolation
3.4. Computational Section
3.5. Cytotoxicity Assay
3.6. Agar Diffusion Test for Biological Activity
Supporting Information
marinedrugs-09-01403-001.pdfAcknowledgments
- Samples Availability: Not available.
References
- Blunt, JW; Copp, BR; Munro, MHG; Northcote, PT; Prinsep, MR. Marine natural products. Nat. Prod. Rep. 2011, 28, 196–268. [Google Scholar]
- Zhang, W; Guo, YW; Gu, Y. Secondary metabolites from the South China Sea invertebrates: Chemistry and biological activity. Curr. Med. Chem. 2006, 13, 2041–2090. [Google Scholar]
- Burks, JE; van der Helm, D; Chang, CY; Ciereszko, LS. The crystal and molecular structure of briarein A, a diterpenoid from the gorgonian Briareum abestinum. Acta Crystallogr. 1977, B33, 704–709. [Google Scholar]
- Rodriguez, J; Nieto, RM; Jiménez, C. New briarane stecholide diterpenes from the Indonesian gorgonian Briareum sp. J. Nat. Prod. 1998, 61, 313–317. [Google Scholar]
- Sheu, JH; Sung, PJ; Su, JH; Wang, GH; Duh, CY; Shen, YC; Chiang, MY; Chen, IT. Excavatolides U–Z, new briarane diterpenes from the gorgonian Briareum excavatum. J. Nat. Prod. 1999, 62, 1415–1420. [Google Scholar]
- Sung, PJ; Su, JH; Wang, GH; Lin, SF; Duh, CY; Sheu, JH. Excavatolides F–M, new briarane diterpenes from the gorgonian Briareum excavatum. J. Nat. Prod. 1999, 62, 457–463. [Google Scholar]
- Wu, SL; Sung, PJ; Chiang, MY; Wu, JY; Sheu, JH. New polyoxygenated briarane diterpenoids, briaexcavatolides O–R, from the gorgonian Briareum excavatum. J. Nat. Prod. 2001, 64, 1415–1420. [Google Scholar]
- Hoshino, A; Mitome, H; Tamai, S; Takiyama, H; Miyaoka, H. 8,17-Epoxybriarane diterpenoids, briaranolides A–J, from an Okinawan gorgonian Briareum sp. J. Nat. Prod. 2005, 68, 1328–1335. [Google Scholar]
- Shin, J; Park, M; Fenical, W. The junceelloiides new anti-inflammatory diterpenoids of the briarane class from the Chinese gorgonian Junceella fragilis. Tetrahedron 1989, 45, 1633–1638. [Google Scholar]
- Pordesimo, EO; Schmitz, FJ; Ciereszko, LS; Hossain, MB; van der Helm, D. New briarein diterpenes from the Caribbean gorgonians Erythropodium caribaeorum and Briareum sp. J. Org. Chem. 1991, 56, 2344–2357. [Google Scholar]
- Kobayashi, J; Cheng, JF; Nakamura, H; Ohizumi, Y; Tomotake, Y; Matsuzaki, T; Grace, KJS; Jacobs, RS; Kato, Y; Brinen, LS; Clardy, J. Structure and stereochemistry of brianolide, a new antiinflammatory diterpenoid from the Okinawan gorgonian Briareum sp. Experientia 1991, 47, 501–502. [Google Scholar]
- Coval, SJ; Cross, S; Bernardinelli, G; Jefford, CW. Brianthein V, a new cytotoxic and antiviral diterpene isolated from Briareum asbestinum. J. Nat. Prod. 1988, 51, 981–984. [Google Scholar]
- Rodríguez, AD. The natural products chemistry of west Indian gorgonian octocorals. Tetrahedron 1995, 51, 4571–4618. [Google Scholar]
- Qi, SH; Zhang, S; Qian, BY; Xiao, ZH; Li, MY. Ten new antifouling briarane diterpenoids from the South China Sea gorgonian Junceella juncea. Tetrahedron 2006, 62, 9123–9130. [Google Scholar]
- Keifer, PA; Rinehart, KL; Hooper, IR. Renillafoulins, antifouling diterpenes from the sea pansy Renilla reniformis (Octocorallia). J. Org. Chem. 1986, 51, 4450–4454. [Google Scholar]
- Hendrickson, RL; Cardellina, JH, II. Structure and stereochemistry of insecticidal diterpenes from the sea pen Ptilosarcus gurneyi. Tetrahedron 1986, 42, 6565–6570. [Google Scholar]
- El Sayed, KA; Dunbar, DC; Perry, TL; Wilkins, SP; Hamann, MT; Greenplate, JT; Wideman, MA. Marine natural products as prototype insecticidal agents. J. Agric. Food Chem. 1997, 45, 2735–2739. [Google Scholar]
- Hamann, MT; Harrison, KN; Carroll, AR; Scheuer, PJ. Briarane diterpenes from Micronesian gorgonians. Heterocycles 1996, 42, 325–331. [Google Scholar]
- Grasshoff, M. The shallow water gorgonians of New Caledonia and adjacent islands (Coelenterata: Octocorallia). Senckenbergiana Biol. 1999, 78, 1–121. [Google Scholar]
- He, HY; Faulkner, DJ. New chlorinated diterpenes from the gorgonian Junceella gemmacea. Tetrahedron 1991, 47, 3271–3280. [Google Scholar]
- Anjaneyulu, ASR; Rao, VL; Sastry, VG; Venugopal, MJRV; Schmitz, FJ. Juncins I–M, five new briarane diterpenoids from the Indian Ocean gorgonian Junceella juncea Pallas. J. Nat. Prod. 2003, 66, 507–510. [Google Scholar]
- Qi, SH; Zhang, S; Huang, H; Xiao, ZH; Huang, JS; Li, QX. New briaranes from the South China Sea gorgonian Junceella juncea. J. Nat. Prod. 2004, 67, 1907–1910. [Google Scholar]
- Shen, YC; Lin, YC; Ko, CL; Wang, LT. New briaranes from the Taiwanese gorgonian Junceella juncea. J. Nat. Prod. 2003, 66, 302–305. [Google Scholar]
- Han, H; Zhang, W; Yi, YH; Liu, BS; Pan, MX; Wang, XH. A novel sulfated holostane glycoside from sea cucumber Holothuria leucospilota. Chem. Biodivers. 2010, 7, 1764–1769. [Google Scholar]
- Zhang, W; Krohn, K; Ding, J; Miao, ZH; Zhou, XH; Chen, SH; Pescitelli, G; Salvadori, P; Kurtan, T; Guo, YW. Structural and stereochemical studies of α-methylene-γ-lactone-bearing cembrane diterpenoids from a South China Sea soft coral Lobophytum crassum. J. Nat. Prod. 2008, 71, 961–966. [Google Scholar]
- Zhang, W; Gavagnin, M; Guo, Y-W; Mollo, E; Geiselin, M; Cimino, G. Terpenoid metabolites of the nudibranch Hexabranchus sanguineus from the South China Sea. Tetrahedron 2007, 63, 4725–4729. [Google Scholar]
- Zhang, W; Huang, H; Ding, Y; Gavagnin, M; Mollo, E; Cimino, G; Guo, YW. Three new ployoxygenated steroids from two species of the South China Sea gorgonian Muricella flexuosa and Menella verrucosa BRUNDIN. Helv. Chim. Acta 2006, 89, 813–820. [Google Scholar]
- Sun, JF; Huang, H; Chai, XY; Yang, XW; Meng, L; Huang, CG; Zhou, XF; Yang, B; Hu, J; Chen, XQ; Lei, H; Wang, LS; Liu, YH. Dichotellides A–E, five new iodine-containing briarane type diterpenoids from. Dichotella gemmacea. Tetrahedron 2011, 67, 1245–1250. [Google Scholar]
- Bowden, BF; Coll, JC; König, GM. Studies of Australian soft corals. XLVIII New briaran diterpenoids from the gorgonian coral Junceela gemmacea. Aust. J. Chem 1990, 43, 151–159. [Google Scholar]
- Li, C; La, MP; Li, L; Li, XB; Tang, H; Liu, BS; Krohn, K; Sun, P; Yi, YH; Zhang, W. Bioactive 11,20-epoxy-3,5(16)-diene briarane diterpenoids from the South China Sea gorgonian Dichotella gemmacea. J. Nat. Prod. 2011, 74, 1658–1662. [Google Scholar]
- Shen, YC; Chen, YH; Hwang, TL; Guh, JH; Khalil, AT. Four new briarane diterpenoids from the gorgonian coral Junceella fragilis. Helv. Chim. Acta 2007, 90, 1391–1398. [Google Scholar]
- Rodriguez, AD; Ramirez, C; Cobar, OM. Briarein C–L, 10 new briarane diterpenoids from the common Caribbean gorgonian Briareum asbestinum. J. Nat. Prod. 1996, 59, 15–22. [Google Scholar]
- Schrodinger, LLC. Products—MacroModel. Available online: http://www.schrodinger.com/Products/macromodel.html (accessed on 13 August 2009).
- Frisch, MJ; Trucks, GW; Schlegel, HB; Scuseria, GE; Robb, MA; Cheeseman, JR; Montgomery, JJA; Vreven, T; Kudin, KN; Burant, JC; et al. Gaussian 03, Revision C.02; Gaussian Inc.: Wallingford, CT, USA, 2004. [Google Scholar]
- Stephens, PJ; Harada, N. ECD cotton effect approximated by the Gaussian curve and other methods. Chirality 2010, 22, 229–233. [Google Scholar]
- Flukiger, P; Luthi, HP; Portmann, S; Weber, J. MOLEKEL 5.4.; Swiss Center for Scientific Computing: Manno, Switzerland, 2000–2002. [Google Scholar]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar]

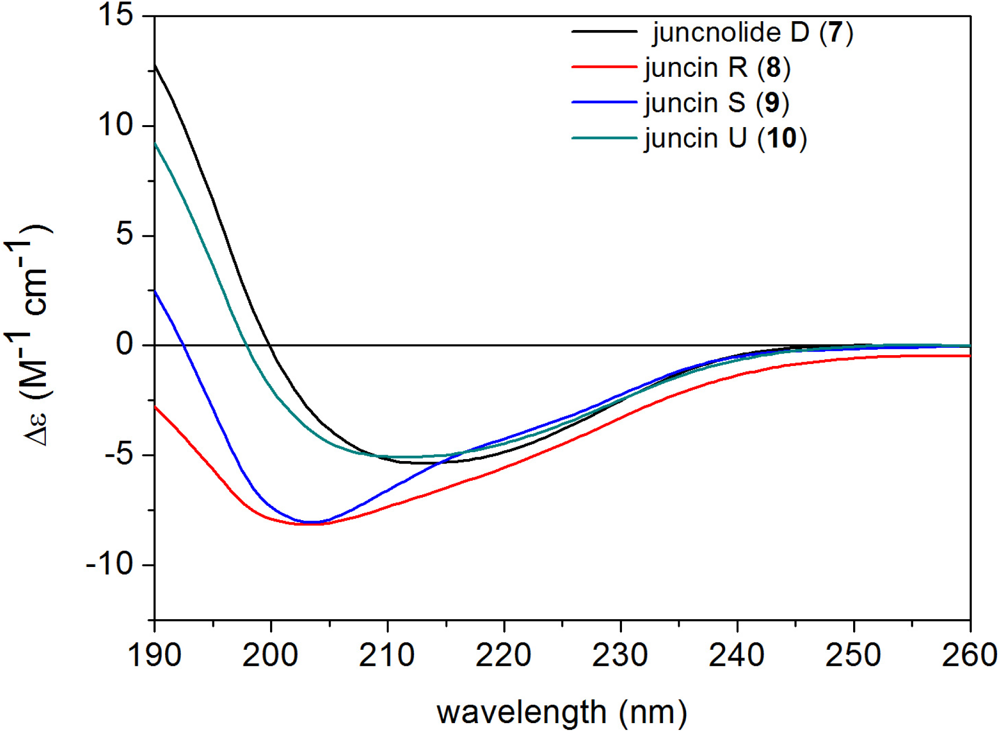
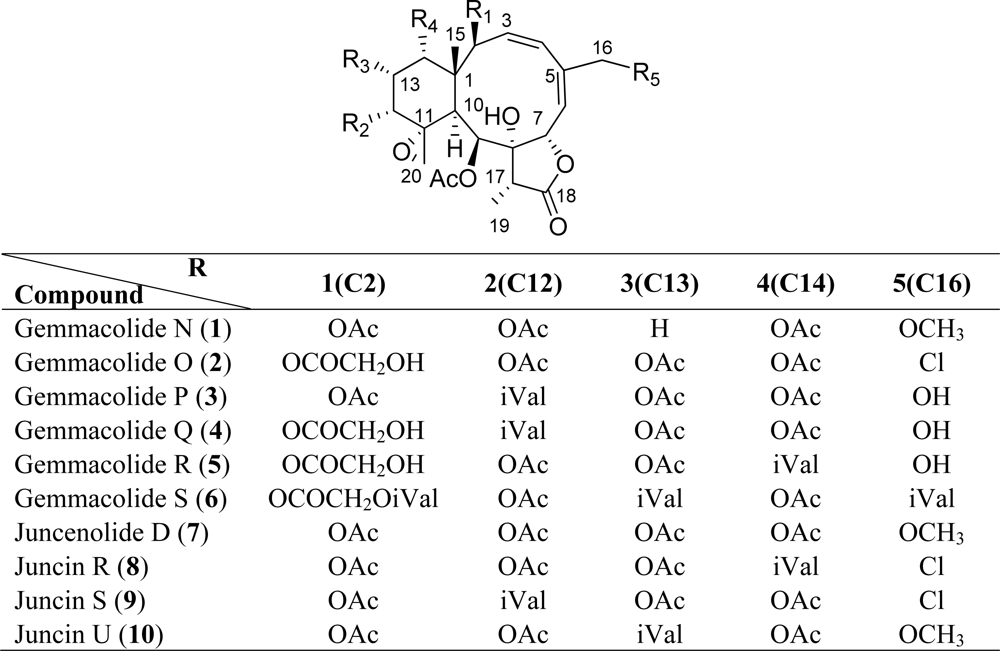
| Carbon | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 47.1 s | 46.5 s | 46.5 s | 46.6 s | 46.6 s | 46.4 s |
| 2 | 74.3 d | 75.2 d | 75.6 d | 77.2 d | 76.7 d | 75.5 d |
| 3 | 131.6 d | 131.1 d | 131.2 d | 130.7 d | 130.6 d | 131.4 d |
| 4 | 128.3 d | 129.1 d | 129.6 d | 130.1 d | 130.1 d | 128.5 d |
| 5 | 141.8 s | 139.7 s | 144.7 s | 144.2 s | 144.3 s | 139.7 s |
| 6 | 122.4 d | 126.3 d | 123.7 d | 123.9 d | 123.6 d | 122.4 d |
| 7 | 79.0 d | 78.5 d | 78.7 d | 78.7 d | 78.3 d | 78.6 d |
| 8 | 81.1 s | 81.0 s | 81.0 s | 80.1 s | 80.1 s | 81.1 s |
| 9 | 64.0 d | 63.6 d | 63.8 d | 63.8 d | 63.8 d | 63.8 d |
| 10 | 32.8 d | 32.6 d | 32.8 d | 32.7 d | 32.6 d | 32.6 d |
| 11 | 59.0 s | 58.1 s | 58.4 s | 58.2 s | 58.3 s | 58.4 s |
| 12 | 73.1 d | 73.0 d | 72.8 d | 72.7 d | 73.1 d | 73.2 d |
| 13 | 28.5 t | 66.4 d | 66.4 d | 66.3 d | 66.4 d | 66.4 d |
| 14 | 73.6 d | 73.8 d | 73.7 d | 73.8 d | 73.5 d | 73.8 d |
| 15 | 14.0 q | 14.3 q | 14.5 q | 14.4 q | 14.5 q | 14.5 q |
| 16 | 72.3 t | 44.5 t | 63.9 t | 63.8 t | 63.8 t | 62.7 t |
| 17 | 44.2 d | 44.0 d | 44.1 d | 44.1 d | 44.1 d | 44.1 d |
| 18 | 175.6 s | 174.9 s | 175.2 s | 175.2 s | 175.1 s | 175.2 s |
| 19 | 6.3 q | 6.3 q | 6.3 q | 6.3 q | 6.3 q | 6.3 q |
| 20 | 49.0 t | 48.9 t | 49.0 t | 49.1 t | 49.1 t | 48.8 t |
| 9-OAc | 170.2 s | 170.1 s | 170.2 s | 170.2 s | 170.2 s | 170.2 s |
| 21.5 q | 21.5 q | 21.6 q | 21.6 q | 21.1 q | 21.5 q | |
| R1 | 169.2 s | 172.2 s | 170.9 s | 172.9 s | 172.4 s | 166.6 s |
| 21.1 q | 61.1 t | 20.5 q | 61.2 t | 61.3 t | 60.9 t | |
| 172.4 s | ||||||
| 42.7 t | ||||||
| 25.6 d | ||||||
| 22.4 q (×2) | ||||||
| R2 | 170.0 s | 169.6 s | see 1′–5′ | see 1′–5′ | 169.8 s | 169.7 s |
| 21.1 q | 20.9 q | 21.6 q | 20.7 q | |||
| R3 | n.o. | 169.7 s | 169.7 s | 169.7 s | 169.7 s | see 1′–5′ |
| 20.5 q | 21.5 q | 20.5 q | 20.6 q | |||
| R4 | 169.8 s | 170.6 s | 170.1 s | 170.7 s | see 1′–5′ | 170.5 s |
| 21.1 q | 20.9 q | 20.8 q | 20.9 q | 20.7 q | ||
| R5 | 58.4 q | see 1″–5″ | ||||
| 1′ | 172.0 s | 171.9 s | 172.9 s | 171.7 s | ||
| 2′ | 43.5 t | 43.4 t | 43.1 t | 42.6 t | ||
| 3′ | 25.7 d | 25.7 d | 25.2 d | 25.0 d | ||
| 4′ | 22.3 q (×2) | 22.4 q (×2) | 22.5 q | 22.3 q (×2) | ||
| 5′ | 22.4 q | |||||
| 1″ | 172.0 s | |||||
| 2″ | 43.3 t | |||||
| 3″ | 25.7 d | |||||
| 4″ and 5″ | 22.4 q (×2) |
| Proton | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 2 | 5.71 (d, 9.7) | 5.65 (ov) | 5.63 (ov) | 5.76 (d, 9.4) | 5.69 (d, 9.6) | 5.69 (ov) |
| 3 | 5.60 (t, 10.5, 9.7) | 5.64 (ov) | 5.60 (ov) | 5.63 (t, 9.4, 10.7) | 5.63 (t, 9.6, 10.5) | 5.63 (ov) |
| 4 | 6.27 (d, 10.5) | 6.42 (d, 9.7) | 6.35 (d, 9.6) | 6.39 (d, 10.7) | 6.39 (d, 10.5) | 6.34 (d, 10.6) |
| 6 | 5.90 (d, 8.5) | 6.07 (d, 9.0) | 5.82 (d, 7.4) | 5.86 (d, 8.9) | 5.86 (d, 8.3) | 5.69 (ov) |
| 7 | 5.01 (d, 8.5) | 4.95 (d, 9.0) | 4.96 (d, 7.4) | 4.95 (d, 8.9) | 4.95 (d, 8.3) | 4.96 (d, 8.6) |
| 9 | 4.81 (br d, 4.6) | 4.76 (br d, 4.5) | 4.74 (br d, 4.8) | 4.75 (br d, 4.5) | 4.78 (br d, 4.7) | 4.73 (br d, 4.9) |
| 10 | 3.69 (br d, 4.6) | 3.62 (ov) | 3.61 (br d, 4.8) | 3.63 (ov) | 3.62 (ov) | 3.63 (ov) |
| 12 | 4.51 (ov) | 4.88 (br d, 2.6) | 4.92 (br d, 3.5) | 4.93 (br d, 2.9) | 4.90 (br d, 2.2) | 4.88 (br s) |
| 13β | 1.92 (ov) | 5.06 (dd, 2.6, 2.6) | 5.09 (dd, 3.5, 3.5) | 5.08 (dd, 2.9, 3.9) | 5.09 (dd, 2.2, 2.2) | 5.09 (br s) |
| 13α | 2.31 (ov) | |||||
| 14 | 4.90 (br s) | 5.17 (br d, 2.6) | 5.23 (br d, 3.5) | 5.19 (br d, 2.9) | 5.24 (br d, 2.2) | 5.20 (br s) |
| 15 | 1.04 (s) | 1.14 (s) | 1.14 (s) | 1.15 (s) | 1.15 (s) | 1.13 (s) |
| 16a | 4.48 (d, 15.2) | 4.66 (d, 13.8) | 4.51 (br s) | 4.55 (d, 15.6) | 4.54 (d, 15.0) | 5.46 (d, 16.3) |
| 16b | 4.24 (d, 15.2) | 4.56 (d, 13.8) | 4.45 (d, 15.6) | 4.47 (d, 15.0) | 4.56 (d, 16.3) | |
| 17 | 2.32 (ov) | 2.31 (q, 7.1) | 2.30 (ov) | 2.31 (ov) | 2.31 (ov) | 2.29 (ov) |
| 19 | 1.16 (d, 7.1) | 1.15 (ov) | 1.13 (d, 7.2) | 1.14 (d, 7.2) | 1.16 (ov) | 1.14 (d, 6.9) |
| 20a | 3.55 (br d, 2.4) | 3.60 (ov) | 3.63 (br d, 2.5) | 3.62 (ov) | 3.62 (br s) | 3.61 (br s) |
| 20b | 2.78 (br d, 2.4) | 2.93 (br s) | 2.93 (br d, 2.5) | 2.94 (br s) | 2.93 (br s) | 2.93 (br s) |
| 9-OAc | 2.19 (s) | 2.19 (s) | 2.19 (s) | 2.20 (s) | 2.19 (s) | 2.18 (s) |
| R1 | 1.96 (s) | 4.15 (d, 16.9) | 1.99 (s) | 4.15 (d, 17.2) | 4.16 (d, 16.4) | 4.52 (d, 15.5) |
| 4.03 (d, 16.9) | 4.04 (d, 17.2) | 4.03 (d, 16.4) | 4.42 (d, 15.5) | |||
| 2.28 (ov) | ||||||
| 2.13 (ov) | ||||||
| 0.97 (d, 6.5) (×2 Me) | ||||||
| R2 | 2.10 (s) | 2.16 (s) | see 2′–5′ | see 2′–5′ | 2.17 (s) | 2.16 (s) |
| R3 | n.o. | 1.95 (s) | 1.94 (s) | 1.94 (s) | 1.94 (s) | see 2′–5′ |
| R4 | 2.02 (s) | 2.09 (s) | 2.09 (s) | 2.11 (s) | see 2′–5′ | 2.10 (s) |
| R5 | 3.45 (s) | see 2″–5″ | ||||
| 2′a | 2.35 (ov) | 2.34 (m) | 2.31 (m) | 2.08 (ov) (×2) | ||
| 2′b | 2.27 (ov) | 2.23 (m) | 2.20 (m) | |||
| 3′ | 2.17 (m) | 2.16 (m) | 2.05 (m) | 1.99 (m) | ||
| 4′ | 0.99 (d, 6.3) | 0.99 (d, 6.5) | 0.98 (d, 7.1) | 0.92 (d, 6.5) (×2 Me) | ||
| 5′ | 1.01 (d, 6.3) | 1.01 (d, 6.5) | 1.00 (d, 7.1) | |||
| 2″ | 2.28 (ov) | |||||
| 3″ | 2.13 (ov) | |||||
| 4″ and 5″ | 0.98 (d, 6.1) (×2 Me) |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Adriamycin a | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A549 | >50.5 | >44.6 | >44.1 | 21.6 ± 1.8 | 27.2 ± 2.4 | 16.4 ± 2.3 | 37.1 ± 4.2 | 13.9 ± 2.5 | 20.2 ± 2.3 | >43.2 | 2.8 ± 0.32 |
| MG63 | >50.5 | >44.6 | >44.1 | 20.5 ± 2.1 | 23.7 ± 2.8 | 18.8 ± 3.9 | >46.0 | 5.6 ± 1.2 | 16.5 ± 2.4 | >43.2 | 3.2 ± 0.37 |
| M. violaceum | S. tritici | E. coli | B. megaterium | |
|---|---|---|---|---|
| 1 | 0 | 7.5 | 12.5 | 0 |
| 2 | 6.0 | 6.5 | 13.0 | 6.0 |
| 3 | 6.0 | 6.5 | 7.5 | 5.5 |
| 4 | 6.5 | 7.5 | 10.0 | 5.5 |
| 5 | 0 | 5.5 | 5.5 | 6.0 |
| 6 | 7.0 | 0 | 6.0 | 0 |
| 7 | 6.0 | 7.5 | 12.5 | 6.0 |
| 8 | 7.5 | 7.5 | 14.0 | 6.0 |
| 9 | 5.5 | 7.0 | 10.0 | 0 |
| 10 | 0 | 7.5 | 11.0 | 8.0 |
| penicillin | 7.0 | 6.0 | 15.0 | 8.0 |
| streptomycin | 8.0 | 5.5 | 9.0 | 5.5 |
| ketoconazole | 15.0 | 12.5 | 9.0 | 11.0 |
| acetone | 0 | 0 | 0 | 0 |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, C.; La, M.-P.; Sun, P.; Kurtan, T.; Mandi, A.; Tang, H.; Liu, B.-S.; Yi, Y.-H.; Li, L.; Zhang, W. Bioactive (3Z,5E)-11,20-Epoxybriara-3,5-dien-7,18-olide Diterpenoids from the South China Sea Gorgonian Dichotella gemmacea. Mar. Drugs 2011, 9, 1403-1418. https://doi.org/10.3390/md9081403
Li C, La M-P, Sun P, Kurtan T, Mandi A, Tang H, Liu B-S, Yi Y-H, Li L, Zhang W. Bioactive (3Z,5E)-11,20-Epoxybriara-3,5-dien-7,18-olide Diterpenoids from the South China Sea Gorgonian Dichotella gemmacea. Marine Drugs. 2011; 9(8):1403-1418. https://doi.org/10.3390/md9081403
Chicago/Turabian StyleLi, Cui, Ming-Ping La, Peng Sun, Tibor Kurtan, Attila Mandi, Hua Tang, Bao-Shu Liu, Yang-Hua Yi, Ling Li, and Wen Zhang. 2011. "Bioactive (3Z,5E)-11,20-Epoxybriara-3,5-dien-7,18-olide Diterpenoids from the South China Sea Gorgonian Dichotella gemmacea" Marine Drugs 9, no. 8: 1403-1418. https://doi.org/10.3390/md9081403
APA StyleLi, C., La, M.-P., Sun, P., Kurtan, T., Mandi, A., Tang, H., Liu, B.-S., Yi, Y.-H., Li, L., & Zhang, W. (2011). Bioactive (3Z,5E)-11,20-Epoxybriara-3,5-dien-7,18-olide Diterpenoids from the South China Sea Gorgonian Dichotella gemmacea. Marine Drugs, 9(8), 1403-1418. https://doi.org/10.3390/md9081403





