Diverse Bacterial PKS Sequences Derived From Okadaic Acid-Producing Dinoflagellates
Abstract
1. Introduction
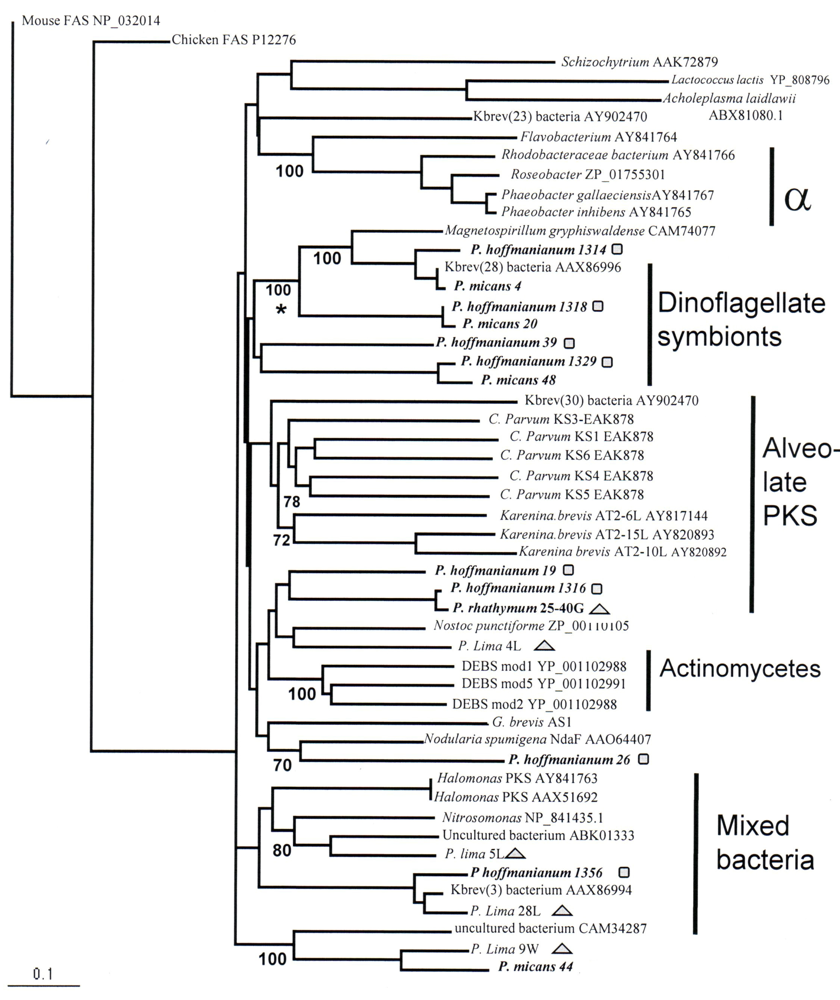
2. Results and Discussion
3. Conclusion
4. Experimental Section
4.1 Culture Conditions
4.2 Amplification, cloning and sequencing of PKS and 16S genes
4.2.1 DNA Extraction from dinoflagellates
4.2.2 Isolation of bacteria from P. lima
4.2.3 Polymerase Chain Reaction (PCR)
4.2.4 Amplicon Purification and Cloning
4.2.5 Sequencing Reactions
4.3 Phylogenetic analysis
4.4 Protein Phophatase (PP2A) inhibition assay
4.5 OA/ DTX ELISA
Acknowledgements
- Samples Availability: Dinoflagellate strains used in this study are available from the Provasoli-Guillard Center for the Culture of Marine Phytoplankton (CCMP). Sixteen nucleotide sequences for PKS amplicons have been deposited to GenBank; Accession numbers EU698032 - EU698047
References and Notes
- Hernandez-Becerril, DU; Cortes Altamirano, R; Alonso, R. The dinoflagellate genus Prorocentrum of the Mexican Pacific. Hydrobiol 2000, 418, 111–121. [Google Scholar]
- Gestal-Otero, JJ. Seafood and freshwater toxins. Food Science and Technology 2000, 103, 45–64. [Google Scholar]
- Balmer-Hanchey, EL; Jaykus, L; Green, DP; McClellan-Green, PJ. Marine biotoxins of algal origin and seafood safety. Aquat Food Prod Technol 2003, 12, 29–53. [Google Scholar]
- Tachibana, K; Scheuer, PJ; Tsukitani, Y; Kikuchi, H; Van, Engen D; Clardy, J; Gopichand, Y; Schmitz, FJ. Okadaic acid, a cytotoxic polyether from two marine sponges of the genus Halichondria. J Am Chem Soc 1981, 103, 2469–2471. [Google Scholar]
- Yasumoto, T; Seino, N; Murakami, Y; Murata, M. Toxins produced by benthic dinoflagellates. Biological Bulletin (Woods Hole, MA, United States) 1987, 172, 128–131. [Google Scholar]
- Murakami, Y; Oshima, Y; Yasumoto, T. Identification of okadaic acid as a toxic component of a marine dinoflagellateProrocentrum lima. Nippon Suisan Gakkaishi 1982, 48, 69–72. [Google Scholar]
- Dickey, RW; Bobzin, SC; Faulkner, DJ; Bencsath, FA; Andrzejewski, D. Identification of okadaic acid from a Caribbean dinoflagellateProrocentrum concavum. Toxicon 1990, 28, 371–377. [Google Scholar]
- Zhou, J; Fritz, L. Ultrastructure of two toxic marine dinoflagellates, Prorocentrum lima and Prorocentrum maculosum. Phycologia 1993, 32, 444–450. [Google Scholar]
- Morton, SL; Moeller, PD; Young, KA; Lanoue, B. Okadaic acid production from the marine dinoflagellate Prorocentrum belizeanum Faust isolated from the Belizean coral reef ecosystem. Toxicon : official journal of the International Society on Toxinology 1998, 36, 201–206. [Google Scholar]
- Morton, SL. Morphology and toxicology of Prorocentrum fausitae sp. nov., a toxic species of non-planktonic dinoflagellate from Heron Island Australia. Bot Mar 1998, 41, 565–569. [Google Scholar]
- Ten-Hage, L; Delaunay, N; Pichon, V; Coute, A; Puiseux-Dao, S; Turquet, J. Okadaic acid production from the marine benthic dinoflagellate Prorocentrum arenarium Faust (Dinophyceae) isolated from Europa Island coral reef ecosystem. Toxicon 2000, 38, 1043–1054. [Google Scholar]
- Draisci, R; Giannetti, L; Lucentini, L; Marchiafava, C; James, KJ; Bishop, AG; Healy, BM; Kelly, SS. Isolation of a new okadaic acid analog from phytoplankton implicated in diarrhetic shellfish poisoning. J Chromatogr A 1998, 798, 137–145. [Google Scholar]
- Murata, M; Shimatani, M; Sugitani, H; Oshima, Y; Yasumoto, T. Isolation and structure elucidation of the causative toxin of the diarrhetic shellfish poisoning. Nippon Suisan Gakkaishi 1982, 48, 549–552. [Google Scholar]
- Murata, M; Murakami, Y; Kumagai, M; Yanagi, T; Yasumoto, T; Iwashita, T; Naoki, H. okadaic acid and its derivatives as diarrhetic shellfish toxins. Tennen Yuki Kagobutsu Toronkai Koen Yoshishu 1986, 28, 192–199. [Google Scholar]
- Schmitz, FJ; Prasad, RS; Gopichand, Y; Hossain, MB; Van, der Helm D; Schmidt, P. Acanthafolicin, a new episulfide-containing polyether carboxylic acid from extracts of the marine sponge Pandaros acanthifolium. J Am Chem Soc 1981, 103, 2467–2469. [Google Scholar]
- Suarez-Gomez, B; Souto, ML; Norte, M; Fernandez, JJ. Isolation and structural determination of DTX-6, a new okadaic acid derivative. J Nat Prod 2001, 64, 1363–1364. [Google Scholar]
- Suarez-Gomez, B; Souto, ML; Cruz, PG; Fernandez, JJ; Norte, M. New targets in diarrhetic shellfish poisoning control. J Nat Prod 2005, 68, 596–599. [Google Scholar]
- Suzuki, T; Beuzenberg, V; Mackenzie, L; Quilliam, MA. Discovery of okadaic acid esters in the toxic dinoflagellate Dinophysis acuta from New Zealand using liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom 2004, 18, 1131–1138. [Google Scholar]
- Hu, T; Marr, J; DeFreitas, ASW; Quilliam, MA; Walter, JA; Wright, JLC; Pleasance, S. New diol esters isolated from cultures of the dinoflagellates Prorocentrum lima and Prorocentrum concavum. J Nat Prod 1992, 55, 1631–1637. [Google Scholar]
- Hu, T; deFreitas, ASW; Doyle, J; Jackson, D; Marr, J; Nixon, E; Pleasance, S; Quilliam, MA; Walter, JA; Wright, JLC. New DSP toxin derivatives isolated from toxic mussels and the dinoflagellates, Prorocentrum lima and Prorocentrum concavum. Developments in Marine Biology 1993, 3, 507–512. [Google Scholar]
- Hu, T; deFreitas, ASW; Doyle, J; Jackson, D; Marr, J; Nixon, E; Pleasance, S; Quilliam, MA; Walter, JA; Wright, JLC. Developments in Marine Biology 1993, 3, 507–512.
- Fernandez, JJ; Suarez-Gomez, B; Souto, ML; Norte, M. Identification of new okadaic acid derivatives from laboratory cultures of Prorocentrum lima. J Nat Prod 2003, 66, 1294–1296. [Google Scholar]
- Dounay, AB; Forsyth, CJ. Okadaic acid: the archetypal serine/threonine protein phosphatase inhibitor. Curr Med Chem 2002, 9, 1939–1980. [Google Scholar]
- Needham, J; McLachlan, JL; Walter, JA; Wright, JLC. Biosynthetic origin of C-37 and C-38 in the polyether toxins okadaic acid and dinophysistoxin-1. J Chem Soc, Chem Comm 1994, 2599–2600. [Google Scholar]
- Wright, JLC; Hu, T; McLachlan, JL; Needham, J; Walter, JA. Biosynthesis of DTX-4: Confirmation of a polyketide pathway, proof of a Baeyer-Villiger oxidation step, and evidence for an unusual carbon deletion process. J Am Chem Soc 1996, 118, 8757–8758. [Google Scholar]
- Macpherson, GR; Burton, IW; LeBlanc, P; Walter, JA; Wright, JLC. Studies of the biosynthesis of DTX-5a and DTX-5b by the dinoflagellate Prorocentrum maculosum: regiospecificity of the putative Baeyer-Villigerase and insertion of a single amino acid in a polyketide chain. J Org Chem 2003, 68, 1659–1664. [Google Scholar]
- Van Lanen, SG; Shen, B. Advances in polyketide synthase structure and function. Curr Op Drug Dis & Dev 2008, 11, 186–195. [Google Scholar]
- Seshime, Y; Juvvadi, PR; Fujii, I; Kitamoto, K. Discovery of a novel superfamily of type III polyketide synthases in Aspergillus oryzae. Biochem Biophys Res Commun 2005, 331, 253–260. [Google Scholar]
- Snyder, RV; Gibbs, PDL; Palacios, A; Abiy, L; Dickey, R; Lopez, JV; Rein, KS. Polyketide synthase genes from marine dinoflagellates. Marine Biotechnology 2003, 5, 1–12. [Google Scholar]
- Zhu, G; LaGier, MJ; Stejskal, F; Millership, JJ; Cai, X; Keithly, JS. Cryptosporidium parvum: the first protist known to encode a putative polyketide synthase. Gene 2002, 298, 79–89. [Google Scholar]
- De, Traubenberg CR; Geraud, ML; Soyer-Gobillard, MO; Emdadi, D. The toxic dinoflagellate Prorocentrum lima and its associated bacteria. I. An ultrastructural study. Eur J Protistol 1995, 31, 318–326. [Google Scholar]
- De, Traubenberg CR; Soyer-Gobillard, MO. Bacteria associated with a photosynthetic dinoflagellate in culture. Symbiosis 1990, 8, 117–133. [Google Scholar]
- Biegala, IC; Kennaway, G; Alverca, E; Lennon, J; Vaulot, D; Simon, N. Identification of bacteria associated with dinoflagellates (Dinophyceae) Alexandrium spp. using tyramide signal amplification-fluorescent in situ hybridization and confocal microscopy. J Phycol 2002, 38, 404–411. [Google Scholar]
- Maki, T; Imai, I. Relationship between intracellular bacteria and the bivalve killer dinoflagellate Heterocapsa circularisquama (dinophyceae). Fisheries Science 2001, 67, 794–803. [Google Scholar]
- Lucas, AN; Vesk, M. The fine structure of two photosynthetic species of Dinophysis (Dinophyceae). J Phycol 1990, 26, 345–357. [Google Scholar]
- LaFay, B; Ruimy, R; De, Traubenberg CR; Breittmayer, V; Gauthier, MJ; Christen, R. Roseobacter algicola sp. nov., a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellateProrocentrum lima. Int J Sys Bacterio 1995, 45, 290–296. [Google Scholar]
- Prokic, I; Brummer, F; Brigge, T; Gortz, HD; Gerdts, G; Schutt, C; Elbrachter, M; Muller, WEG. Bacteria of the genus Roseobacter associated with the toxic dinoflagellate Prorocentrum lima. Protist 1998, 149, 347–357. [Google Scholar]
- Scorzetti, G; Brand, LE; Hitchcock, GL; Rein, KS; Sinigalliano, C; Fell, JW. Multiple simultaneous detection of Harmful Algal Bloom (HABs) through a high throughput bead array technology, with potential use in phytoplankton community analysis. Harmful Algae 2008, in press. [Google Scholar]
- Loeblich, AR. Spector, DL, Ed.; Dinoflagellate evolution. In Dinoflagellates; Academic Press Inc: Orlando, FL, 1984; pp. 482–516. [Google Scholar]
- Faust, MA. Morphologic details of six benthic species of Prorocentrum (Pyrrophyta) from a mangrove island, Twin Cays, Belize, including two new species. J Phycol 1990, 26, 548–558. [Google Scholar]
- Steidinger, KA. A re-evaluation of the toxic dinoflagellate biology and ecology. Progr Phycol Res 1983, 2, 147–188. [Google Scholar]
- Cortes-Altamirano, R; Sierra-Beltran, AP. Morphology and taxonomy of Prorocentrum mexicanum and reinstatement of Prorocentrum rhathymum (Dinophyceae). J Phycol 2003, 38, 221–225. [Google Scholar]
- De, Traubenberg CR. Interaction entre un dinoflagelle et sa microflore bacterienne associee: role des bacteries dans la toxicite de Prorocentrum lima Ehrenberg (Dodge); Universite de Nantes: Nantes, France, 1993. [Google Scholar]
- Martens, T; Gram, L; Grossart, H; Kessler, D; Mueller, R; Simon, M; Wenzel, SC; Brinkhoff, T. Bacteria of the Roseobacter clade show potential for secondary metabolite production. Microb Ecol 2007, 54, 31–42. [Google Scholar]
- Snyder, RV; Guerrero, MA; Sinigalliano, CD; Winshell, J; Perez, R; Lopez, JV; Rein, KS. Localization of polyketide synthase encoding genes to the toxic dinoflagellate Karenia brevis. Phytochemistry 2005, 66(15), 1767–80. [Google Scholar]
- Guillard, RRL. In Culture of phytoplankton for feeding marine invertebrates. In Cult Mar Invertebr Anim 1975, 29–60. [Google Scholar]
- McLachlan, J. Some consideration of the growth of marine algae in artificial media. Can J Microbiol 1964, 10, 769–782. [Google Scholar]
- Guillard, RRL. Hallegraeff, GM, Anderson, DM, Cembella, AD, Eds.; 1995; Culture methods. In Manual on Harmful Marine Microalgae; UNESCO Publishing: New York, 1995; pp. 45–62.
- Sambrook, JD; Russell, W; Maniatis, T. Molecular cloning : a laboratory manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, N.Y, 2001. [Google Scholar]
- Altschul, SF; Madden, TL; Schaffer, AA; Zhang, J; Zhang, Z; Miller, W; Lipman, DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–3402. [Google Scholar]
- Thompson, JD; Gibson, TJ; Plewniak, F; Jeanmougin, F; Higgins, DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res 1997, 25, 4876–4882. [Google Scholar]
- Sfanos, KAS; Harmody, DK; McCarthy, PJ; Dang, P; Pomponi, SA; Lopez, JV. Systematic survey of cultured microbial associates of deep water marine invertebrates. Syst Appl Microbiol 2005, 28, 242–264. [Google Scholar]
- Swofford, DL. 2002; PAUP* Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer; Sunderland Mass. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenetics: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar]
- Nei, M; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University: Oxford, 2000. [Google Scholar]
- Posada, D; Crandall, KA. MODELTEST: testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar]
- Simon, JF; Vernoux, JP. Highly sensitive assay of okadaic acid using protein phosphatase and paranitrophenyl phosphate. Nat Tox 1994, 2, 293–301. [Google Scholar]
- Tubaro, A; Florio, C; Luxich, E; Sosa, S; Loggia, RD; Yasumoto, T. A protein phosphatase 2A inhibition assay for a fast and sensitive assessment of okadaic acid and contamination in mussels. Toxicin 1996, 34, 743–652. [Google Scholar]
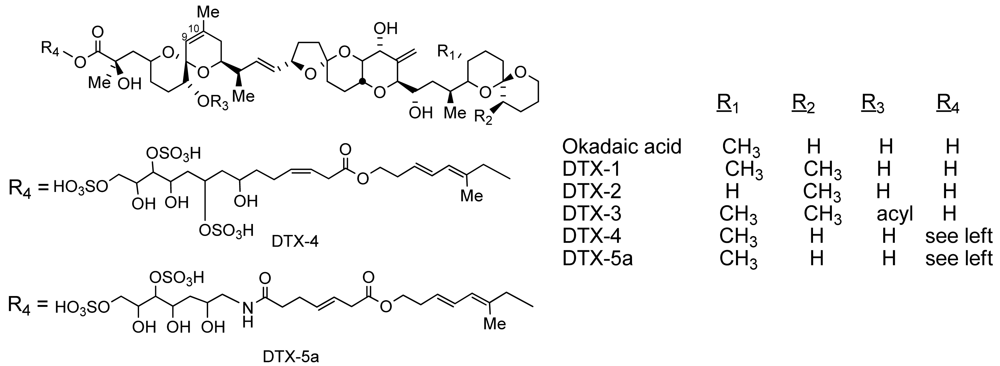
| OA equivalents by PP2A (ppb±sd) | OA equivalents by ELISA(range in ppb) | |
|---|---|---|
| P. lima | 235 ± 5 | 328 ± 6 |
| P. hoffmanianum | 97 ± 3 | 81 ± 1 |
| P. rhathymum | 0.45 ± 0.004 | 0.57 ± 0.003 |
| P. micans | nda | nda |
| P. donghaiense | nda | nda |
| Name | Sequence (5’→3’) | Product Size |
|---|---|---|
| PKS4U | MGIGARGCIYTICARATGGAYCCICARCARMG | 700bp |
| PKS5L | GGRTCNCCIARYTGIGTICCIGTICCRTGIGC | |
| 16SF | GGAGAGTTTGATCATGGCT | 1.3kbp |
| 16SR | ACGGYTACCTTGTTACGACTT |
| Accession number | Nearest BLASTn Match Description | PL | TB | PH1 | PH2 | PR | PD | PM |
|---|---|---|---|---|---|---|---|---|
| DQ167249 | Roseobacter prionitis | 28.5 | 17 | 60 | ||||
| AJ968652 | Roseobacter pelophilus | 28.5 | 37 | |||||
| DQ120726 | Roseobacter sp. 812 | 6 | 17 | |||||
| DQ104407 | Roseobacter sp. JL-351 | 49 | ||||||
| DQ659415.1 | Roseobacter sp. COL2P | 8.7 | ||||||
| AJ878874 | Thalassobius mediterraneus | 6 | 9 | |||||
| AM420114.1 | Uncultured Alpha-Proteobacterium | 6 | 9 | |||||
| AY162118 | Planctomycete GMD16E07 | 6 | 22. | |||||
| AJ889010 | Stappia alba strain 5OM30 | 6 | 5 | |||||
| AY163576 | Croceibacter atlanticus | 3 | 51 | |||||
| EF414083 | Uncultured Bacteroidetes bacterium | 3 | 3 | |||||
| Y15341 | Rhizobium sp. | 17.5 | 3 | |||||
| AY654759 | Mucus bacterium 110 from O. patagonica | 4.5 | ||||||
| AY258089 | Mesorhizobium sp. DG943 | 3 | ||||||
| AF441991.1 | Uncultured CFB group bacterium clone | 3 | ||||||
| EF12346.1 | Uncultured Gamma-Proteobacterium clone | 3 | ||||||
| DQ446117 | Uncultured spirochete clone | 3 | ||||||
| AJ227758 | Caulobacter henricii strain ATCC 15253 | 9 | ||||||
| AY136121 | Marinobacter sp. MED106 | 6 | ||||||
| AY701447 | Uncultured Bacteroidetes bacterium | 6 | ||||||
| AM176885.1 | Uncultured bacterium clone SZB60 | 6 | ||||||
| AY917783 | Uncultured bacterium clone 1971b-30 | 6 | ||||||
| DQ811828 | Uncultured Delta-Proteobacterium clone | 5.8 | ||||||
| AY162122 | MSB | 4.3 | ||||||
| AY960750 | Planctomycete GMD14H10 | 3 | ||||||
| AY345437 | Maribacter dokdonensis strain DSW-9 | 3 | ||||||
| AY517632 | Bacterium K2–12 | 3 | ||||||
| DQ513013 | Marinobacter flavimaris strain SW-145 | 3 | ||||||
| DQ486493 | Uncultured bacterium clone FS140-15B-02 | 3 | ||||||
| EU196324 | Gamma-Proteobacterium DG1253 | 3 | ||||||
| EU148878 | Sphingomonas sp. NP31 | 12 | ||||||
| EF106349 | Uncultured bacterium clone PP6–13 | 9 | ||||||
| EF658677 | Uncultured planctomycete | 9 | ||||||
| AY038570 | Uncultured bacterium clone YHSS3 | 9 | ||||||
| AY682384 | Uncultured alpha-Proteobacterium | 12 | ||||||
| CP000264 | Kordiimonas gwangyangensis strain | 6 | ||||||
| EF658677.1 | GW14–5 | 6 | ||||||
| AY539822 | Jannaschia sp. CCS1 | |||||||
| AY258095 | Uncultured bacterium clone YHSS3 | 8.5 | 34 | |||||
| EU249979 | Gamma-Proteobacterium BT-P-1 | 4.5 | ||||||
| AY664364 | Sulfitobacter sp. DG1020 | 4.5 | ||||||
| AY562560 | Uncultured Pseudomonas sp | 3 | ||||||
| AM697073 | Pseudoalteromonas sp. PM02 | 3 | ||||||
| AY344411.1 | Alpha proteobacterium CRA 4C | 41 | ||||||
| EF123623.1 | Uncultured bacterium | 17.5 | ||||||
| EU107173.1 | Unidentified bacterium clone K2-S-32 | 10.5 | ||||||
| EF512127.1 | Uncultured Bacteroidetes bacterium | 8.5 | ||||||
| AB255368.1 | Pseudomonas sp. N9-1 | 4.5 | ||||||
| DQ822527.1 | Stappia alba strain | 4.5 | ||||||
| EU196324.1 | Gilvibacter sediminis | 3 | ||||||
| EU005335.1 | Bacterium QM28 | 3 | ||||||
| AB073564 | Sphingomonas sp. NP31 | 3 | ||||||
| Uncultured alpha proteobacterium clone | ||||||||
| G7–25 | ||||||||
| Cytophaga sp. MBIC04693 |
| PKS sequence name | Total Occurrence Frequency (%) | Organism Libraries |
|---|---|---|
| PL 28L | 24 | PL, PD, PR, PM, |
| PH 19 | 18 | CCMP2804 |
| PL 5L | 18 | CCMP683, TB |
| PL 4L | 11 | PL, CCMP683, TB |
| PL 9W | 1 | PL, CCMP683, TB |
| PH 26 | 0.5 | PL, TBCCMP683 |
| PH 39 | 0.5 | CCMP683 |
| PR 25 | 1 | PR, CCMP2804 |
| PM 44 | 6 | PM |
| PM 4 | 3 | PM |
| PM 48 | 2 | PM |
| PM 20 | 0.5 | PM, CCMP2804 |
| PH1316 | 10 | CCMP2804 |
| PH1329 | 2 | CCMP2804 |
| PH1356 | 1 | CCMP2804 |
| PH1314 | 1 | CCMP2804 |
Share and Cite
Perez, R.; Liu, L.; Lopez, J.; An, T.; Rein, K.S. Diverse Bacterial PKS Sequences Derived From Okadaic Acid-Producing Dinoflagellates. Mar. Drugs 2008, 6, 164-179. https://doi.org/10.3390/md6020164
Perez R, Liu L, Lopez J, An T, Rein KS. Diverse Bacterial PKS Sequences Derived From Okadaic Acid-Producing Dinoflagellates. Marine Drugs. 2008; 6(2):164-179. https://doi.org/10.3390/md6020164
Chicago/Turabian StylePerez, Roberto, Li Liu, Jose Lopez, Tianying An, and Kathleen S. Rein. 2008. "Diverse Bacterial PKS Sequences Derived From Okadaic Acid-Producing Dinoflagellates" Marine Drugs 6, no. 2: 164-179. https://doi.org/10.3390/md6020164
APA StylePerez, R., Liu, L., Lopez, J., An, T., & Rein, K. S. (2008). Diverse Bacterial PKS Sequences Derived From Okadaic Acid-Producing Dinoflagellates. Marine Drugs, 6(2), 164-179. https://doi.org/10.3390/md6020164




