Sarcoeleganolides C–G, Five New Cembranes from the South China Sea Soft Coral Sarcophyton elegans
Abstract
:1. Introduction
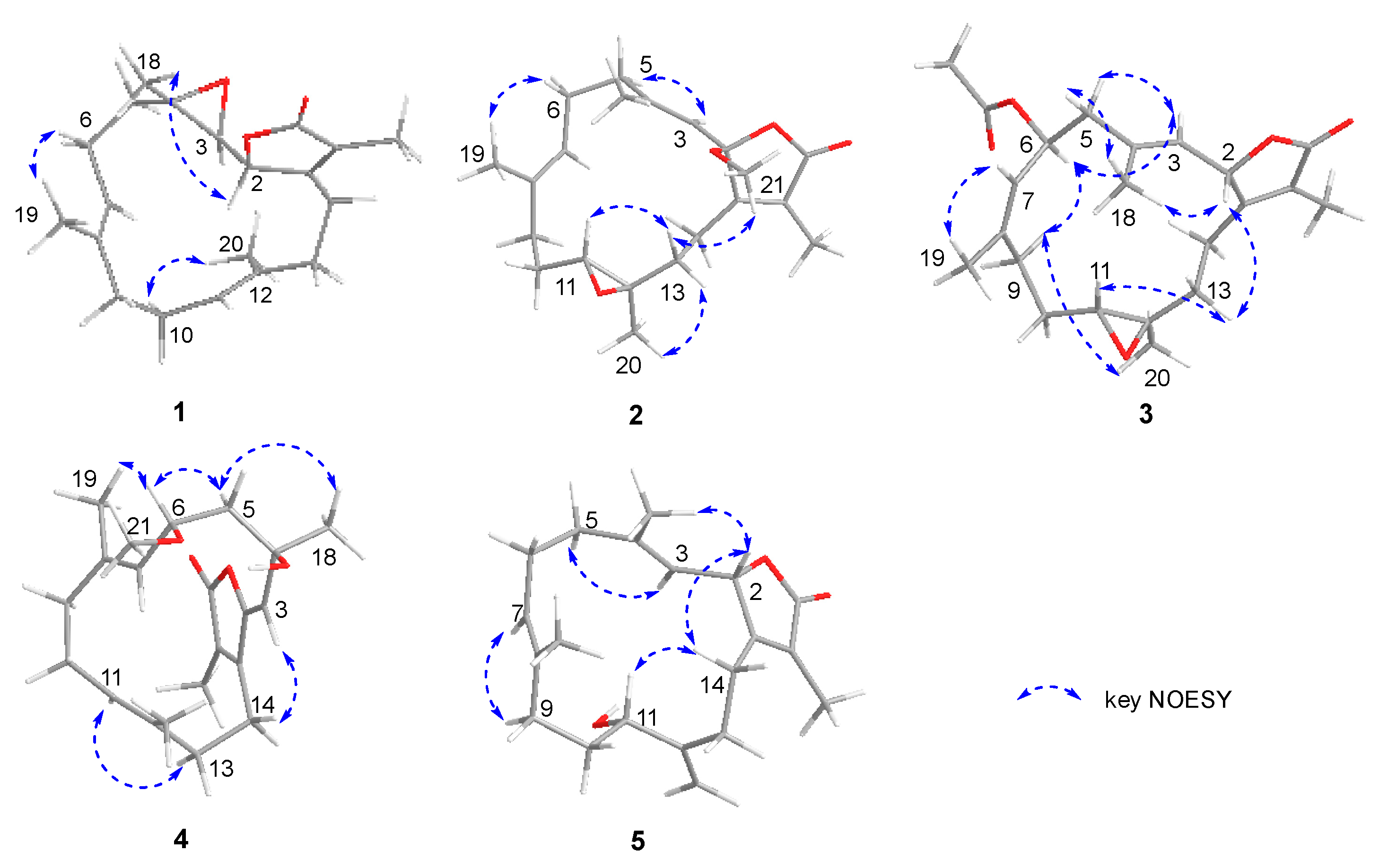
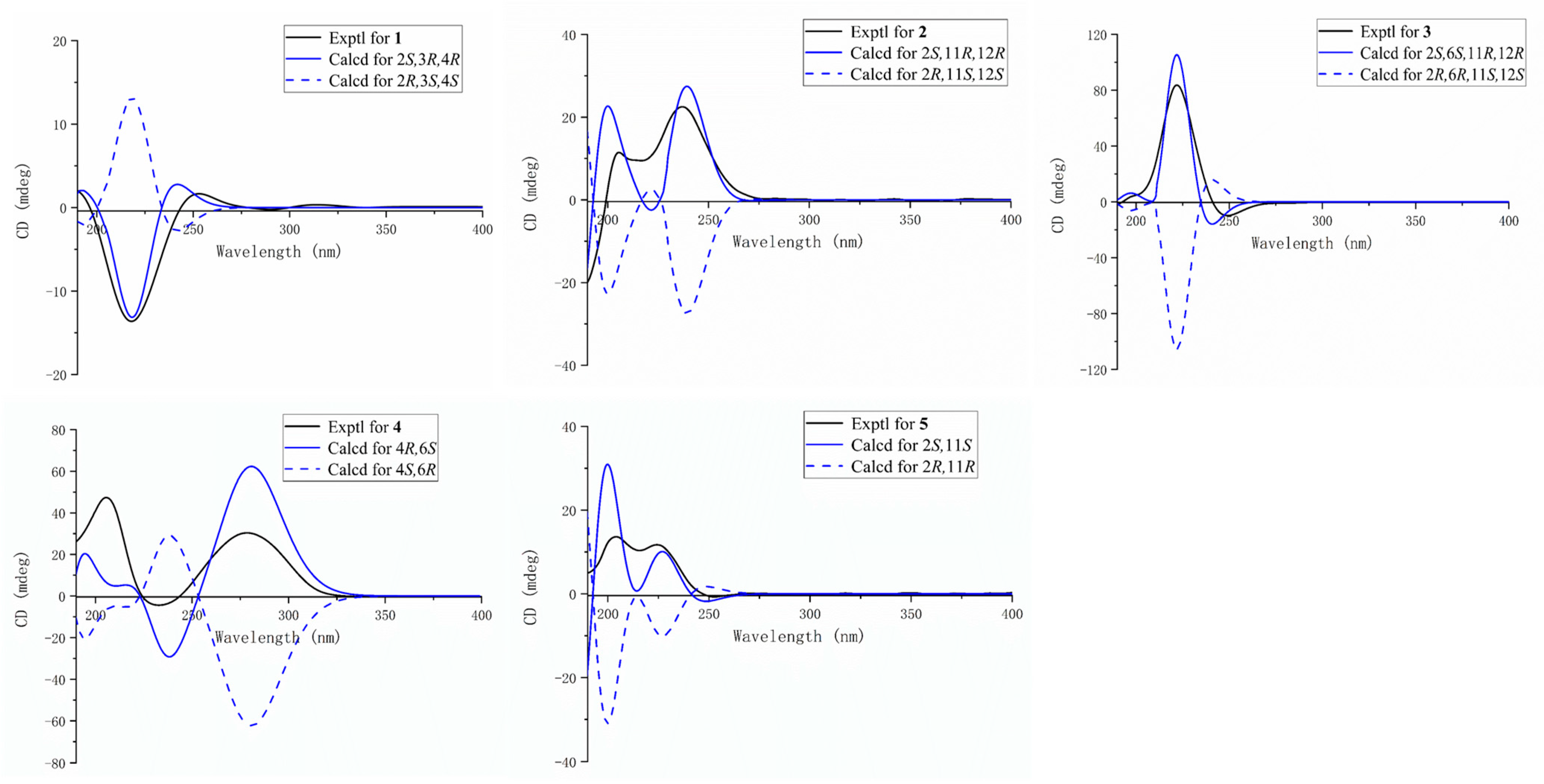
2. Results
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Animal Material
3.3. Extraction and Isolation
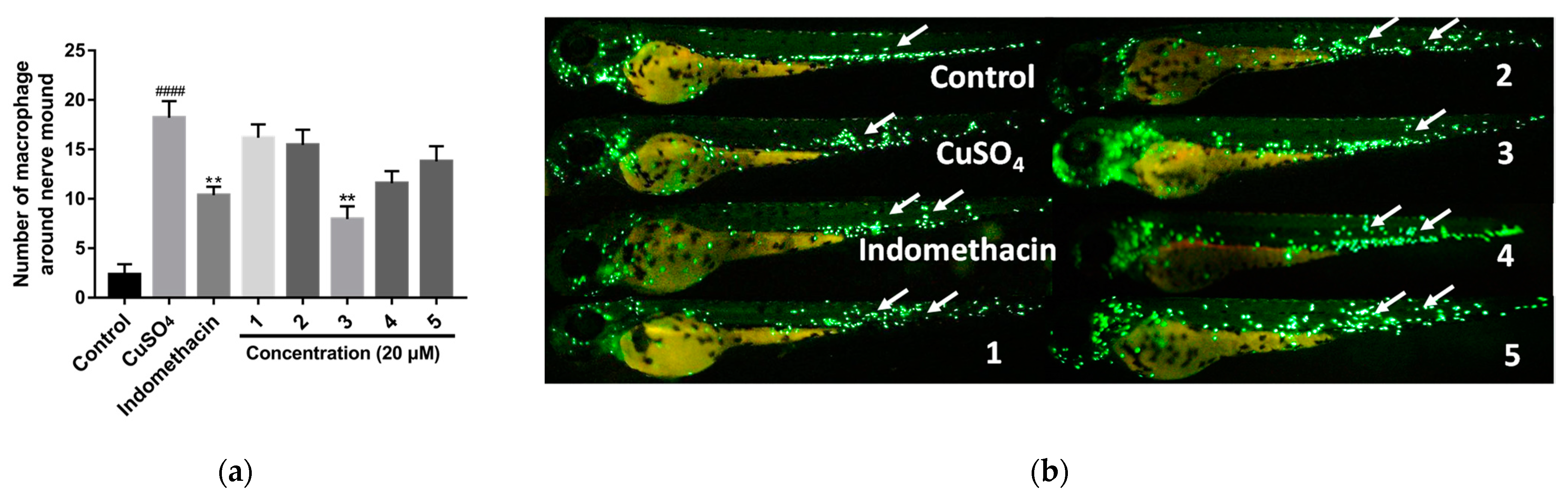
3.4. Anti-Inflammatory Activity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hegazy, M.E.F.; Mohamed, T.A.; Alhammady, M.A.; Shaheen, A.M.; Reda, E.H.; Elshamy, A.I.; Aziz, M.; Pare, P.W. Molecular architecture and biomedical leads of terpenes from red sea marine invertebrates. Mar. Drugs 2015, 13, 3154–3181. [Google Scholar] [CrossRef]
- Nweze, J.A.; Mbaoji, F.N.; Li, Y.M.; Yang, L.Y.; Huang, S.S.; Pan, L.X.; Yang, D.F.; Nweze, J.A.; Chigor, V.N.; Eze, E.A.; et al. Potentials of marine natural products against malaria, leishmaniasis, and trypanosomiasis parasites: A review of recent articles. Infect. Dis. Poverty 2021, 10, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zhou, X.F.; Lin, X.P.; Liu, J.; Peng, Y.; Yang, X.W.; Liu, Y. Cembrane diterpenes chemistry and biological properties. Curr. Org. Chem. 2012, 16, 1512–1539. [Google Scholar] [CrossRef]
- Gross, H.; Kehraus, S.; Nett, M.; Konig, G.M.; Beil, W.; Wright, A.D. New cytotoxic cembrane based diterpenes from the soft corals Sarcophyton cherbonnieri and Nephthea sp. Org. Biomol. Chem. 2003, 1, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.; Tung, P.T.; Ngoc, N.T.; Hanh, T.T.H.; Nguyen, P.T.; Nguyen, V.T.; Nguyen, X.C.; Thao, D.T.; Huong, T.T.; Thung, D.C.; et al. Cytotoxic biscembranoids from the soft coral Sarcophyton pauciplicatum. Chem. Pharm. Bull. 2015, 63, 636–640. [Google Scholar]
- Koenig, G.M.; Wright, A.D. New cembranoid diterpenes from the soft coral Sarcophyton ehrenbergi. J. Nat. Prod. 1998, 61, 494–496. [Google Scholar] [CrossRef]
- Chao, C.H.; Li, W.L.; Huang, C.Y.; Ahmed, A.F.; Dai, C.F.; Wu, Y.C.; Lu, M.C.; Liaw, C.C.; Sheu, J.H. Isoprenoids from the soft coral Sarcophyton glaucum. Mar. Drugs 2017, 15, 202. [Google Scholar] [CrossRef]
- Ahmed, A.F.; Chen, Y.W.; Huang, C.Y.; Tseng, Y.J.; Lin, C.C.; Dai, C.F.; Wu, Y.C.; Sheu, J.H. Isolation and structure elucidation of cembranoids from a dongsha atoll soft coral Sarcophyton stellatum. Mar. Drugs 2018, 16, 210. [Google Scholar] [CrossRef]
- Ninh Thi, N.; Tran Thi Hong, H.; Tran Hong, Q.; Nguyen Xuan, C.; Nguyen Hoai, N.; Thi Thao, D.; Cuong, P.V.; Do Cong, T.; Phan Van, K.; Van Minh, C. Cembranoids from the Vietnamese soft coral Sarcophyton ehrenbergi. Nat. Prod. Res. 2021, in press. [Google Scholar] [CrossRef]
- Mohamed, T.A.; Elshamy, A.I.; Abdel-Tawab, A.M.; AbdelMohsen, M.M.; Ohta, S.; Pare, P.W.; Hegazy, M.E.F. Oxygenated Cembrene Diterpenes from Sarcophyton convolutum: Cytotoxic Sarcoconvolutum A-E. Mar. Drugs 2021, 19, 519. [Google Scholar] [CrossRef]
- Ren, L.; Leng, X.; Li, T.; Liu, J.; Wang, W.; Yan, X.; Yan, X.; He, S. Four new cembranoids from the South China Sea soft coral Sarcophyton trocheliophorum. Nat. Prod. Res. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tang, X.L.; Han, X.; Feng, D.Q.; Luo, X.C.; Ofwegen, L.V.; Li, P.L.; Li, G.Q. Sarcoglaucins A-I, new antifouling cembrane-type diterpenes from the South China Sea soft coral Sarcophyton glaucum. Org. Chem. Front. 2019, 6, 2004–2013. [Google Scholar] [CrossRef]
- Badria, F.A.; Guirguis, A.N.; Perovic, S.; Steffen, R.; Muller, W.E.G.; Schroder, H.C. Sarcophytolide: A new neuroprotective compound from the soft coral Sarcophyton glaucum. Toxicology 1998, 131, 133–143. [Google Scholar] [CrossRef]
- Yang, M.; Li, X.L.; Wang, J.R.; Lei, X.; Tang, W.; Li, X.W.; Sun, H.; Guo, Y.W. Sarcomililate A, an Unusual Diterpenoid with Tricyclo[11.3.0.02,16]hexadecane Carbon Skeleton, and Its Potential Biogenetic Precursors from the Hainan Soft Coral Sarcophyton mililatensis. J. Org. Chem. 2019, 84, 2568–2576. [Google Scholar] [CrossRef]
- Shen, S.M.; Li, W.S.; Ding, X.; Luo, H.; Zhang, H.Y.; Guo, Y.W. Ximaoglaucumins A–F, new cembranoids with anti-inflammatory activities from the South China Sea soft coral Sarcophyton glaucum. Bioorg. Med. Chem. 2021, 38, 116139. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Cuadrado, C.; Gao, C.; Wu, Q.; Li, X.; Pang, T.; Daranas, A.H.; Guo, Y.; Li, X. Polyoxygenated anti-inflammatory biscembranoids from the soft coral Sarcophyton tortuosum and their stereochemistry. Chin. Chem. Lett. 2021, 32, 271–276. [Google Scholar] [CrossRef]
- Liu, Z.; Cheng, W.; Liu, D.; van Ofwegen, L.; Proksch, P.; Lin, W. Capnosane-type cembranoids from the soft coral Sarcophyton trocheliophorum with antibacterial effects. Tetrahedron 2014, 70, 8703–8713. [Google Scholar] [CrossRef]
- Liu, K.M.; Lan, Y.H.; Su, C.C.; Sung, P.J. Trocheliolide B, a New Cembranoidal Diterpene from the Octocoral Sarcophyton Trocheliophorum. Nat. Prod. Commun. 2016, 11, 21–22. [Google Scholar] [CrossRef]
- Su, J.Y.; Zhong, Y.L.; Lou, G.X.; Li, X.Q.; Zeng, L.M.; Huang, Y.-Q.; Hu, S.Z. Studies on the stereochemistry of (-)-sartrochine. Acta Chim. Sin. 1994, 52, 813–816. [Google Scholar]
- El Sayed, K.A.; Hamann, M.T.; Waddling, C.A.; Jensen, C.; Lee, S.K.; Dunstan, C.A.; Pezzuto, J.M. Structurally novel bioconversion products of the marine natural product sarcophine effectively inhibit JB6 cell transformation. J. Org. Chem. 1998, 63, 7449–7455. [Google Scholar] [CrossRef]
- Wang, S.K.; Duh, C.Y. New cytotoxic cembranolides from the soft coral Lobophytum michaelae. Mar. Drugs 2012, 10, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.T.; Wang, S.K.; Soong, K.; Duh, C.Y. New cytotoxic cembranolides from the soft coral Lobophytum michaelae. Chem. Pharm. Bull. 2007, 55, 766–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, F.L.P.; de Albuquerque, A.C.F.; Fiorot, R.G.; Lião, L.M.; Martorano, L.H.; Mota, G.V.S.; Valverde, A.L.; Carneiro, J.W.M.; dos Santos Junior, F.M. Structural characterisation of natural products by means of quantum chemical calculations of NMR parameters: New insights. Org. Chem. Front. 2021, 8, 2019–2058. [Google Scholar] [CrossRef]
- Han, X.; Luo, X.C.; Xue, L.; van Ofwegen, L.V.; Zhang, W.J.; Liu, K.C.; Zhang, Y.; Tang, X.L.; Li, P.L.; Li, G.Q. Dolabellane Diterpenes and Elemane Alkaloids from the Soft Coral Clavularia inflata Collected in the South China Sea. J. Nat. Prod. 2022, 85, 276–283. [Google Scholar] [CrossRef]
- Li, J.J.; Zhang, Y.; Han, L.W.; Tian, Q.P.; He, Q.X.; Wang, X.M.; Sun, C.; Han, J.; Liu, K.C. Tenacissoside H exerts an anti-inflammatory effect by regulating the nf-κb and p38 pathways in zebrafish. Fish Shellfish Immunol. 2018, 83, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.H.; Jiao, W.H.; Zhou, M.; Zhang, Y.; Zeng, D.Q.; Zhu, H.R.; Liu, K.C.; Sun, F.; Chen, H.F.; Lin, H.W. Septosones A-C, in Vivo Anti-inflammatory Meroterpenoids with Rearranged Carbon Skeletons from the Marine Sponge Dysidea septosa. Org. Lett. 2019, 21, 767–770. [Google Scholar] [CrossRef]
- Pereira, T.C.B.; Campos, M.M.; Bogo, M.R. Copper toxicology, oxidative stress and inflammation using zebrafish as experimental model. J. Appl. Toxicol. 2016, 36, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, V.; Vuksanovic, M.; Tomic, N.; Stojanovic, D.; Husovic, T.V.; Uskokovic, P. Improvement in cavitation resistance of poly(vinyl butyral) composite films with silica nanoparticles: A technical note. Polym. Polym. Compos. 2021, 29, S1664–S1669. [Google Scholar] [CrossRef]
- Asri, N.; Nazemalhosseini Mojarad, E.; Mirjalali, H.; Mohebbi, S.R.; Baghaei, K.; Rostami-Nejad, M.; Yadegar, A.; Rezaei-Tavirani, M.; Asadzadeh Aghdaei, H.; Rostami, K.; et al. Toward finding the difference between untreated celiac disease and COVID-19 infected patients in terms of CD4, CD25 (IL-2 Rα), FOXP3 and IL-6 expressions as genes affecting immune homeostasis. BMC Gastroenterol. 2021, 21, 462. [Google Scholar] [CrossRef]
- Schrödinger Release 2019-1: MacroModel; Schrödinger, LLC: New York, NY, USA, 2019.
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J Chem Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef]
- Smith, S.G.; Goodman, J.M. Assigning Stereochemistry to Single Diastereoisomers by GIAO NMR Calculation: The DP4 Probability. J. Am. Chem. Soc. 2010, 132, 12946–12959. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.; Trucks, G.; Schlegel, H.; Scuseria, G.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Petersson, G.; Nakatsuji, H. Gaussian; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Su, L.H.; Geng, C.A.; Li, T.Z.; Ma, Y.B.; Huang, X.Y.; Zhang, X.M.; Chen, J.J. Artatrovirenols A and B: Two Cagelike Sesquiterpenoids from Artemisia atrovirens. J. Org. Chem. 2020, 85, 13466–13471. [Google Scholar] [CrossRef] [PubMed]
- Li, S.W.; Cuadrado, C.; Yao, L.G.; Daranas, A.H.; Guo, Y.W. Quantum Mechanical-NMR-Aided Configuration and Conformation of Two Unreported Macrocycles Isolated from the Soft Coral Lobophytum sp.: Energy Calculations versus Coupling Constants. Org. Lett. 2020, 22, 4093–4096. [Google Scholar] [CrossRef] [PubMed]
- Suramitr, S.; Piriyagagoon, A.; Wolschann, P.; Hannongbua, S. Theoretical study on the structures and electronic properties of oligo(p-phenylenevinylene) carboxylic acid and its derivatives: effects of spacer and anchor groups. Theor. Chem. Acc. 2012, 131, 1–15. [Google Scholar] [CrossRef]


| No. | 1 a | 2 a | 3 b | 4 a | 5 a | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| δHc (J in Hz) | δC d | δH c (J in Hz) | δC d | δH e (J in Hz) | δC f | δH e (J in Hz) | δC f | δH e (J in Hz) | δC f | |
| 1 | 160.7, qC | 158.3, qC | 160.0, qC | 151.9, qC | 161.6, qC | |||||
| 2 | 4.94, m | 79.1, CH | 108.3, qC | 4.91, d, (10.0) | 78.4, CH | 148.2, qC | 5.45, d, (10.5) | 79.9, CH | ||
| 3 | 2.77, d, (4.2) | 61.5, CH | 5.16, s | 120.6, CH | 4.74, d, (10.0) | 125.3, CH | 5.23, s | 117.0, CH | 4.89, d, (10.5) | 120.0, CH |
| 4 | 61.5, qC | 143.8, qC | 139.0, qC | 72.0, qC | 144.7, qC | |||||
| 5a | 1.37, m | 38.8, CH2 | 2.20, m | 40.2, CH2 | 2.38, dd, (10.0, 3.0) | 46.2, CH2 | 2.16, m | 48.5, CH2 | 2.32, m | 39.5, CH2 |
| 5b | 2.08, m | 2.20, m | 2.04, t, (11.5) | 2.03, m | 2.20, m | |||||
| 6a | 2.20, m | 23.7, CH2 | 2.35, m | 24.6, CH2 | 5.36, td, (10.0, 2.0) | 71.1, CH | 4.30, t, (9.0) | 73.4, CH | 2.20, m | 24.2, CH2 |
| 6b | 2.08, m | 2.14, m | 2.35, m | |||||||
| 7 | 5.05, t, (7.2) | 124.3, CH | 5.02, t, (6.6) | 125.7, CH | 5.19, d, (10.0) | 127.3, CH | 4.88, d, (9.0) | 124.8, CH | 4.92, d, (5.0) | 123.0, CH |
| 8 | 135.2, qC | 134.3, qC | 141.9, qC | 141.1, qC | 135.5, qC | |||||
| 9a | 2.11, m | 38.8, CH2 | 2.29, m | 37.0, CH2 | 2.61, td, (14.0, 2.5) | 29.1, CH2 | 2.10, m | 38.7, CH2 | 2.03, m | 33.9, CH2 |
| 9b | 2.18, m | 2.03, m | 1.76, m | 2.10, m | 2.03, m | |||||
| 10a | 2.26, m | 24.4, CH2 | 2.06, m | 24.2, CH2 | 1.24, m | 24.3, CH2 | 2.22, m | 24.3, CH2 | 1.71, m | 34.4, CH2 |
| 10b | 2.20, m | 1.34, m | 1.86, m | 2.10, m | 1.71, m | |||||
| 11 | 5.09, t, (6.6) | 126.4, CH | 2.69, dd, (9.6, 3.3) | 61.5, CH | 2.30, dd, (10.5, 2.5) | 58.8, CH | 4.86, d, (4.0) | 125.9, CH | 3.98, t, (6.5) | 72.1, CH |
| 12 | 133.6, qC | 61.6, qC | 59.9, qC | 131.7, qC | 151.9, qC | |||||
| 13a | 2.04, m | 36.6, CH2 | 1.68, m | 34.0, CH2 | 1.09, m | 35.2, CH2 | 2.33, m | 36.2, CH2 | 2.24, m | 32.1, CH2 |
| 13b | 2.45, m | 1.89, m | 1.63, m | 2.33, m | 2.17, m | |||||
| 14a | 2.74, m | 24.9, CH2 | 2.45, m | 23.4, CH2 | 1.74, m | 22.0, CH2 | 2.54, m | 22.5, CH2 | 2.26, m | 27.0, CH2 |
| 14b | 2.39, m | 2.14, m | 1.59, m | 2.54, m | 2.46, m | |||||
| 15 | 124.0, qC | 126.4, qC | 124.0, qC | 123.2, qC | 124.2, qC | |||||
| 16 | 174.4, qC | 172.2, qC | 173.9, qC | 170.0, qC | 174.9, qC | |||||
| 17 | 1.83, s | 8.8, CH3 | 1.90, s | 8.8, CH3 | 1.61, s | 8.8, CH3 | 1.93, s | 9.3, CH3 | 1.87, s | 9.0, CH3 |
| 18 | 1.53, s | 17.9, CH3 | 1.57, s | 15.9, CH3 | 1.34, s | 18.3, CH3 | 1.45, s | 32.9, CH3 | 1.78, s | 15.9, CH3 |
| 19 | 1.58, s | 16.1, CH3 | 1.66, s | 15.0, CH3 | 1.46, s | 22.4, CH3 | 1.66, s | 17.2, CH3 | 1.64, s | 17.1, CH3 |
| 20 | 1.68, s | 17.0, CH3 | 1.29, s | 16.6, CH3 | 1.11, s | 17.3, CH3 | 1.60, s | 17.1, CH3 | 5.19, s; 5.02, s | 110.9, CH2 |
| 21 | 3.14, s | 50.2, CH3 | 169.4, qC | 3.21, s | 55.1, CH3 | |||||
| 22 | 1.65, s | 20.9, CH3 | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Zhang, J.; Shi, X.; Li, K.; Li, F.; Tang, X.; Li, G.; Li, P. Sarcoeleganolides C–G, Five New Cembranes from the South China Sea Soft Coral Sarcophyton elegans. Mar. Drugs 2022, 20, 574. https://doi.org/10.3390/md20090574
Wang C, Zhang J, Shi X, Li K, Li F, Tang X, Li G, Li P. Sarcoeleganolides C–G, Five New Cembranes from the South China Sea Soft Coral Sarcophyton elegans. Marine Drugs. 2022; 20(9):574. https://doi.org/10.3390/md20090574
Chicago/Turabian StyleWang, Cili, Jiarui Zhang, Xing Shi, Kai Li, Fengling Li, Xuli Tang, Guoqiang Li, and Pinglin Li. 2022. "Sarcoeleganolides C–G, Five New Cembranes from the South China Sea Soft Coral Sarcophyton elegans" Marine Drugs 20, no. 9: 574. https://doi.org/10.3390/md20090574
APA StyleWang, C., Zhang, J., Shi, X., Li, K., Li, F., Tang, X., Li, G., & Li, P. (2022). Sarcoeleganolides C–G, Five New Cembranes from the South China Sea Soft Coral Sarcophyton elegans. Marine Drugs, 20(9), 574. https://doi.org/10.3390/md20090574






