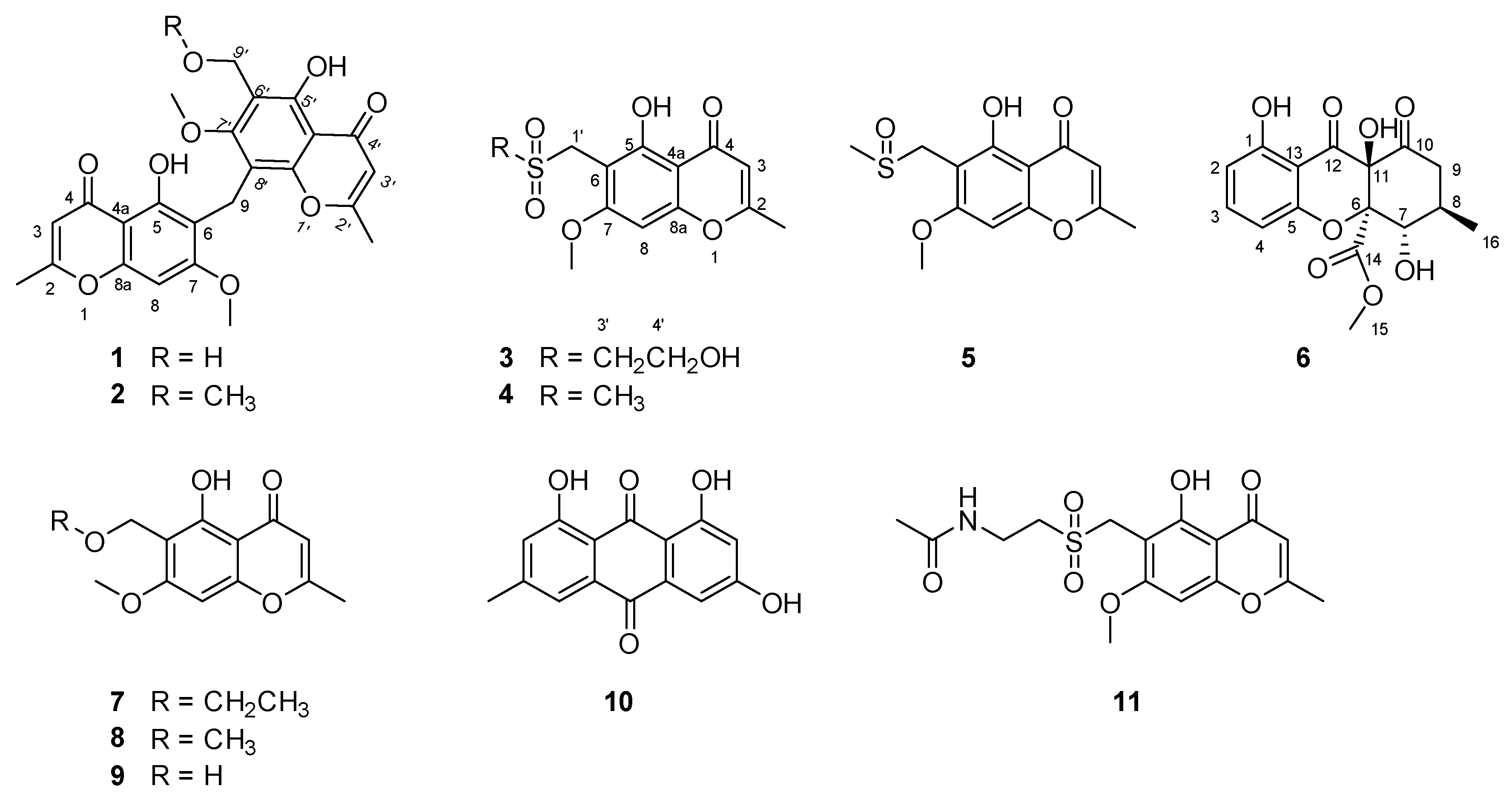
2. Results and Discussion
Amycolachromone A (
1) displayed HRESIMS peak at
m/
z 477.1172 [M + Na]
+ (calcd 477.1162) corresponding to the molecular formula C
24H
22O
9, indicating fourteen degrees of unsaturation. Analysis of the NMR data of
1 (
Table 1, see
Supplementary Materials) revealed three aromatic protons at
δH 6.64 (1H, s, H-8), 6.22 (1H, s, H-3′), 6.20 (1H, s, H-3), two methoxy groups at
δH 3.83 (3H, s, CH
3O-7) and 3.75 (3H, s, CH
3O-7′), two methyl groups at
δH 2.35 (3H, s, CH
3-2) and 3.21 (3H, s, CH
3-2′), two methylenes at
δH 4.45 (2H, d,
J = 4.8 Hz, H-9′) and 3.98 (2H, s, H-9), two phenolic hydroxyl groups at
δH 13.13 (1H s, OH-5) and 13.10 (1H s, OH-5′), a hydroxyl group at
δH 4.78 (1H, t,
J = 5.2 Hz, OH-9′). The
13C NMR (
Table 1) revealed 24 carbon signals: the two carbonyls C-4 (
δC 183.2) and C-4′ (
δC 182.5), the three aromatic carbons C-3 (
δC 108.7), C-3′ (
δC 108.4) and C-8 (
δC 90.6), five non-oxygenated quaternary aromatic carbons at C-4a (
δC 110.8), C-6 (
δC 104.4), C-4a’ (
δC 106.7), C-6′ (
δC 117.6), and C-8′ (
δC 112.5), eight oxygenated quaternary aromatic carbons at C-2 (
δC 168.5), C-2′ (
δC 168.3), C-7 (
δC 163.8),C-7′ (
δC 163.5), C-5 (
δC 158.5), C-5′ (
δC 158.3), C-8a (
δC 156.7), and C-8a’ (
δC 154.8), two methoxy groups CH
3O-7 (
δC 63.2) and CH
3O-7′ (
δC 56.7), two methyl groups CH
3-2 (
δC 20.4) and CH
3-2′ (
δC 20.0), and two methylenes C-9′ (
δC 52.1) and C-9 (
δC 16.9). Analysis of the
1H and
13C NMR data of
1 revealed the presence of the same 5-hydroxy-4H-chromen-4-one moiety as found in xanthones [
18,
19], and therefore suggested a compound comprising two xanthone building blocks.
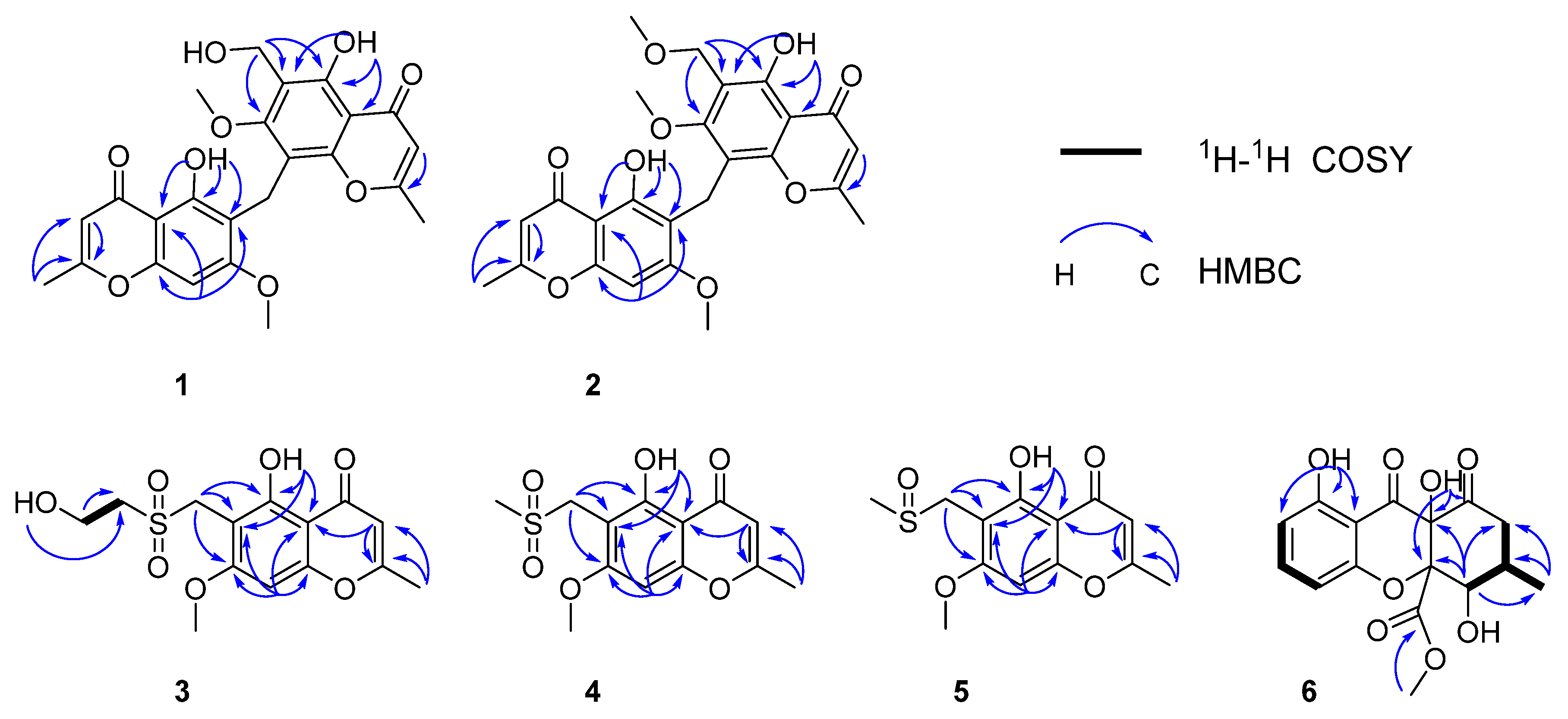
1H-
1H COSY correlations were observed from H-9′ to OH-9′. Further confirmation was found for HMBC correlations of 5-OH to C-5, C-6, 4a; H-8 to C-6, C-4a, C-8a; H-3 to C-2; 2-CH
3 to C-2, C-3, indicating the same Eugenin. HMBC correlations from 5′-OH to C-5′, C-6′, 4a′; H-3′ to C-2′; 2′-CH
3 to C-2′, C-3′; H-9′ to C-5′, C-6′, C-7′, indicated the same 6-Hydroxymethyleugenin (
10) [
20]. HMBC correlations from H-9 to C-7, C-5, C-4a, C8′, C-7, C-8a′, indicated that Eugenin and 6-hydroxymethyleugenin are linked with C-9. Selected key correlations in the observed NMR spectrum are shown in
Figure 3. On the basis of these results, the structure of compound
1 was established as shown.
Amycolachromone B (
2) displayed HRESIMS peak at
m/
z 469.1502 [M + H]
+ (calcd 469.1499),
m/
z 491.1333 [M + Na]
+ (calcd 469.1318), corresponding to the molecular formula C
25H
24O
9 (fourteen degrees of unsaturation). Analysis of the NMR data of
2 (
Table 1) revealed for three aromatic protons at
δH 6.64 (1H, s, H-8), 6.22 (1H, s, H-3′), 6.23 (1H, s, H-3), two methoxy groups at
δH 3.82 (3H, s, CH
3O-7) and 3.76 (3H, s, CH
3O-7′), two methyl groups at
δH 2.22 (3H, s, CH
3-2) and 3.37 (3H, s, CH
3-2′), two methylene at
δH 4.35 (2H, s, H-9′) and 3.98 (2H, s, H-9), two phenolic hydroxyl groups at
δH 13.21 (1H s, OH-5) and 13.11 (1H s, OH-5′). The
13C NMR (
Table 1) revealed 25 carbon signals: the two carbonyl group C-4 (
δC 183.2), C-4′ (
δC 182.4), three aromatic carbon C-3 (
δC 108.7), C-3′ (
δC 108.5) and C-8 (
δC 90.6), five nonoxygenated quaternary aromatic carbons at C-4a (
δC 110.8), C-6 (
δC 100.9), C-4a′ (
δC 106.7), C-6′ (
δC 114.1), and C-8′ (
δC 112.5), eight oxygenated quaternary aromatic carbons at C-2 (
δC 168.8), C-2′ (
δC 168.2), C-7 (
δC 164.2), C-7′ (
δC 163.4), C-5 (
δC 158.9), C-5′ (
δC 158.6),C-8a (
δC 156.7), and C-8a′ (
δC 154.8), three methoxy groups CH
3O-7(
δC 62.5), CH
3O-9′ (
δC 57.7), and CH
3O-7′ (
δC 56.6), two methyl groups CH
3-2 (
δC 20.4) and CH
3-2′ (
δC 20.0), two methylene C-9′ (
δC 63.2) and C-9 (
δC 16.9). Analysis of the
1H and
13C NMR data of
2 revealed the presence of the same 5-hydroxy-4H-chromen-4-one moiety as found in xanthones [
18,
19], and comprised two xanthones. In contrast, the NMR data of
2 showed them to be nearly identical except for a methoxy group linked with C-9′. Further confirmation was found for HMBC correlations of 5-OH to C-5, C-6, 4a; H-8 to C-6, C-4a, C-8a, C-7; H-3 to C-2; 2-CH
3 to C-2, C-3, indicated that same as Eugenin [
20]. HMBC correlations from 5′-OH to C-5′, C-6′, 4a′; H-3′ to C-2′; 2′-CH
3 to C-2′, C-3′; H-9′ to C-5′, C-6′, C-7′, indicated that same as 6-Methoxymethyleugenin (
9) [
21]. HMBC correlations from H-9 to C-7, C-5, C-4a, C8′, C-7, C-8a′, indicated that Eugenin and Methoxymethyleugenin are linked with C-9. Selected key correlations in the observed NMR spectrum are shown in
Figure 3. On the basis of these results, the structure of compound
2 was established as shown.
Amycolachromone C (
3) displayed HRESIMS ion at
m/
z 351.0515 [M + Na]
+ (calcd 351.0514), corresponding to the molecular formula C
14H
16O
7S, indicating nine degrees of unsaturation. Analysis of the NMR data of
3 (
Table 2) revealed two aromatic protons at
δH 6.78 (1H, s, H-8), 6.32 (1H, s, H-3), a methoxy group at
δH 3.90 (3H, s, CH
3O-7), a methyl group at
δH 2.39 (3H, s, CH
3-2), three methylenes at
δH 4.39 (2H, s, H-1′),
δH 3.80 (2H, q,
J =6.1 Hz, H-4′), and 3.21 (2H, t,
J = 6.3, H-3′), a phenolic hydroxyl group at
δH 11.40 (1H s, OH-5), and a hydroxyl group at
δH 5.03 (1H, t,
J = 5.4 Hz, 1H, OH-4′). Examination of the
13C NMR spectrum (
Table 2) revealed 14 carbon signals: a carbonyl group C-4 (
δC 182.4), the two aromatic carbons C-3 (
δC 108.9) and C-8 (
δC 91.3), three non-oxygenated quaternary aromatic carbons at C-8a (
δC 158.2), C-4a (
δC 104.6), and C-6 (
δC 101.0), and three oxygenated quaternary aromatic carbons at C-2 (
δC 168.9), C-5 (
δC 159.9), and C-7 (
δC 163.9) and the methylene C-1′ (
δC 49.3). Analysis of the
1H and
13C NMR data of
2 revealed the presence of the same 5-hydroxy-4H-chromen-4-one moiety as found in xanthones [
18,
19].
In the
1H-
1H COSY spectrum, there were correlations from H-4′ to OH-5′ and H-3′. According to the HMBC, there were correlations from H-1′ to C-6, C-5, and C-7, H-4′ to C-3′, OH-4′ to C-3′. The sulfur atom present in
3 was shown to be attached at C-1′and C-3′, indicated that C-1′was attached at C-6. Further confirmation was found for HMBC correlations of OH-5 to C-4a, C-6, C-5; CH
3-2 to C-3, C-2; CH
3O-7 to C7, H-3 to CH
3-2, C-4a, C-2; H-8 to C-4a, C-6, C-8a, C-7, C-4, a hydroxyl group could be located at C-5, a methoxy groups could be located at C-7, a methyl group could be located at C-2. Selected key correlations in the observed NMR spectrum are shown in
Figure 3. On the basis of these results, the structure of compound
3 was established as shown.
Amycolachromone D (
4) displayed HRESIMS peak at
m/
z 321.0406 [M + Na]
+ (calcd 321.0409), corresponding to the molecular formula C
13H
14O
6S (nine degrees of unsaturation). Analysis of the NMR data of
4 (
Table 2) revealed for two aromatic protons at
δH 6.76 (1H, s, H-8), 6.30 (1H, s, H-3), a methoxy groups at
δH 3.90 (3H, s, CH
3O-7), two methyl groups at
δH 2.91 (3H, s, H-3′) and 2.39 (3H, s, CH
3-2), a methylene group at
δH 4.33 (2H, s, H-1′). The
13C NMR (
Table 2) revealed 13 carbon signals: a carbonyl group C-4 (
δC 182.3), two aromatic carbon C-3 (
δC 108.9) and C-8 (
δC 91.1), three nonoxygenated quaternary aromatic carbons at C-8a (
δC 158.2),C-4a (
δC 104.7), and C-6 (
δC 101.2), three oxygenated quaternary aromatic carbons at C-2 (
δC 168.7), C-5 (
δC 159.1), and C-7 (
δC 163.7), a methoxy groups CH
3O-7 (
δC 57.2), two methyl groups C-3′ (
δC 42.0) and CH
3-2 (
δC 20.4), a methylene group C-1′ (
δC 49.4). Analysis of the
1H and
13C NMR data of
4 revealed the presence of the same 5-hydroxy-4H-chromen-4-one moiety as found in xanthones [
18,
19]. A side-by-side comparison of the NMR spectroscopic data with those of
3 showed them to be nearly identical except for the final hydroxymethyl unit on the side chain.
According to the HMBC correlations from H-1′ to C-6, C-5 and C-7, the sulfur atom present in
4 was shown to be attached at C-1′and C-3′, indicating that C-1′ was attached at C-6, Further confirmation was found for HMBC correlations of CH
3O-7 to C7; H-3 to CH
3-2, C-4a, C-2; H-8 to C-4a, C-6, C-8a, C-7, C-4, a methoxy group could be located at C-7 and a methyl group could be located at C-2. Selected key correlations in the observed NMR spectrum are shown in
Figure 3. On the basis of these results, the structure of compound
4 was established as shown.
Amycolachromone E (
5) displayed HRESIMS peak at
m/
z 305.0462 [M + Na]
+ (calcd 305.0460), corresponding to the molecular formula C
13H
14O
5S (eight degrees of unsaturation). Analysis of the NMR data of
5 (
Table 2) revealed for two aromatic protons at
δH 6.76 (1H, s, H-8), 6.29 (1H, s, H-3), a methoxy groups at
δH 3.90 (3H, s, CH
3O-7), two methyl groups at
δH 2.54 (3H, s, H-3′) and 2.39 (3H, s, CH
3-2) and a methylene group at
δH 3.99 (2H, d,
J = 6.7 Hz, H-1′). The
13C NMR (
Table 2) revealed 13 carbon signals: a carbonyl group C-4 (
δC 182.3), two aromatic carbon C-3 (
δC 108.9) and C-8 (
δC 91.2), three nonoxygenated quaternary aromatic carbons at C-8a (
δC 157.8), C-4a (
δC 104.7), and C-6 (
δC 102.3), three oxygenated quaternary aromatic carbons at C-2 (
δC 168.8), C-5 (
δC 159.6), and C-7 (
δC 163.6), a methoxy groups CH
3O-7 (
δC 57.2), two methyl groups C-3′ (
δC 39.1) and CH
3-2 (
δC 20.4), a methylene group C-1′ (
δC 48.4). Analysis of the
1H and
13C NMR data of
5 revealed the presence of the same 5-hydroxy-4H-chromen-4-one moiety as found in xanthones [
18,
19]. A side-by-side comparison of the NMR spectroscopic data with those of
3 showed them to be nearly identical except for the final sulfur monoxide unit on the side chain.
According to the HMBC correlations from H-1′ to C-6, C-5 and C-7, H-3′ to C-1′, the sulfur atom present in
5 was shown to be attached at C-1′and C-3′, indicating that C-1′ was attached at C-6, Further confirmation was found for HMBC correlations of CH
3O-7 to C7; H-3 to CH
3-2, C-4a, C-2; H-8 to C-4a, C-6, C-8a, C-7 and C-4, a methoxy group could be located at C-7, a methyl group could be located at C-2. Selected key correlations in the observed NMR spectrum are shown in
Figure 3. On the basis of these results, the structure of compound
5 was established as shown.
Amycolachromone F (
6),
−54 (
c 0.1, MeOH), displayed HRESIMS peak at
m/
z 337.0915 [M + H]
+ (calcd 337.0923), corresponding to the molecular formula C
16H
16O
8 (nine degrees of unsaturation). Analysis of the
1H data of
6 (
Table 3) revealed resonance for three aromatic protons at
δH 7.52 (1H, t,
J = 8.3 Hz, H-3), 6.60 (1H, d,
J = 8.3 Hz, H-4), 6.53 (1H, d,
J = 8.3 Hz, H-2), a methoxy group at
δH 3.50 (3H, s, H-15), a methyl group at
δH 1.06 (3H, d,
J = 6.4 Hz, H-16), a methylene at
δH 2.81 (1H, dd,
J = 14.4, 12.9 Hz, H-9a) and 2.25 (1H, dd,
J = 14.5, 5.3 Hz, H-9b), a oxygenated methine at
δH 4.20 (1H, dd,
J = 10.5, 6.0 Hz, H-7), a methine at
δH 1.97–1.86 (1H, m, H-8), three hydroxyl groups at
δH 11.35 (1H s, OH-1), 8.09 (1H, s, OH-11), and 5.91 (1H, d,
J = 6.0 Hz, OH-7). The
13C NMR (
Table 3) revealed sixteen carbon signals: three carbonyl group C-10 (
δC 198.6), C-12 (
δC 191.8) and C-14 (
δC 168.5), three aromatic carbon C-3 (
δC 138.7), C-2 (
δC 109.5), and C-4 (
δC 107.4), a nonoxygenated quaternary aromatic carbons at C-13 (
δC 106.5), two oxygenated quaternary aromatic carbons at C-1 (
δC 161.9) and C-5 (
δC 158.9), two sp
3-quaternary carbon C-11 (
δC 90.0) and C-6 (
δC 73.0), a methoxy group C-15 (
δC 52.7), a methyl group C-16 (
δC 18.6), an oxygenated methine C-7 (
δC 71.8), a methine C-8 (
δC 31.1) and a methylene C-9 group (
δC 43.1). Analysis of the
1H and
13C NMR data of
6 revealed the presence of the same 5-hydroxy-4H-chromen-4-one moiety as found in xanthones [
18,
19]. In the
1H-
1H COSY spectrum, the correlations from H-7 to H-8 and OH-7, from H-8 to H-9 and H-16. Further confirmation was found for HMBC correlations of H-7 to C-16, C-6, C-11 and C-9; H
3-16 to C-7, C-8 and C-9, indicated that C-16 was attached to C-8, and OH-7 was located at C-7. HMBC correlations from the O-methyl proton signal H
3-15 to the carboxylic carbon C-14 confirmed that the O-methyl group was located at C-14. HMBC correlations from OH-11 to C-11, C-6 and C-10, OH-1 to C-2, C-13 and C-1 indicated that OH-1 and OH-11 were attached to C-1 and C-11, respectively [
22,
23]. Selected key correlations in the observed NMR spectrum are shown in
Figure 3. Thus, the planar structure of
6 was established. Moreover, the relative configuration of
6 was established to be 6
R*, 7
S*, 8
R* and 11
R* by X-ray crystallography using Mo K
a radiation (
Figure 4).
Further analysis of the structures allowed us to raise a plausible biosynthetic pathway of compounds
1–
6. As outlined in the
Scheme 1, compounds
1–
5 were structurally related to the known metabolite 6-methoxymethyleugenin, which was derived from the widely existing 5,7-dihydroxy-2-methylchromone via the hydroxymethylation with formaldehyde and the methylation with SAM (
S-adenosyl methionine). The compound
1 was the dimerization of 6-methoxymethyleugenin, and the sequential methylation with SAM could afford the related compound
2. For compound
3–
5, we proposed that the sulfur in these structures was from
l-cysteine. Thus, the Michael addition of
l-cysteine to the
ortho-quinone methide intermediate from 6-methoxymethyleugenin gave the compound
I. Then, transamination, decarboxylation and reduction sequence of
I furnished the 2-sulfo-ethanol
II occurred. An oxidation of sulfur in
II gave the compound
III. Finally, compound
3 was obtained through the double oxidation of sulfur in
II. The oxidation of the hydroxyl group in
III to the corresponding carboxylic acid occurred and followed with a decarboxylation afforded for compound
5. Furthermore, compound
4 was the oxidation product of
5 [24,25]. In addition, compound
6 was the oxidation product of the known natural product blennolide B, which was proposed by Franck to be a derivative of emodin (
Scheme 2) [
26].
The structures of five known compounds were identified as 6-ethoxymethyleugenin (7), 6-methoxymethyleugenin (8), 6-hydroxymethyleugenin (9), emodin (10) and the ascomycete metabolite chaetoquadrin D (11) by comparison of spectroscopic data with reported values and are described here for the first time as produced by Amycolatopsis sp.
The AlkB family of DNA repair enzymes utilize an α-ketoglutarate/Fe(II)-dependent mechanism to oxidize the aberrant alkyl groups, finally repairing alkyl DNA bases [
27,
28]. Compounds
1–
11 were evaluated for their in vitro ABH2 inhibitory activities. Compounds
1–
11 exhibited weak inhibitory activity against the ABH2 enzyme. However, in 2019, a paper was published that tested emodin (
10). It exhibited strong inhibitory activity for the ALKH3 enzyme with IC
50 of 8.8 μM [
29]. This hinted that these compounds might inhibit other members of the AlkB family of enzymes.
In conclusion, the chemical investigation of a streptomycin-resistant strain of the deep-sea marine actinomycete, Amycolatopsis sp. WP1, led to the isolation and identification of six novel compounds, amycolachromones A–F (1–6) and five known analogues (7–11). Among them, amycolachromones A–B (1–2) represents an unusual fused skeleton between two 6-hydroxymethyleugenin, and the relative configuration of amycolachromones F (6) was determined by the signal-crystal X-ray diffraction. The discovery of amycolachromones A–F not only expanded the chemical diversity of natural products and inspire further synthetic studies, but also provided a template for the exploration of inhibitors of other members of the AlkB family of enzymes.
3. Materials and Methods
General experimental procedures. All chemical reagents and solvents were purchased from Sigma–Aldrich (Shanghai, China). UV spectra were acquired with a DU 800 UV/vis spectrophotometer (Beckman Coulter, Brea, CA, USA). IR spectra were acquired with a Nicolet 380 FT-IR (Thermo Electron Corporation, Beverly, MA, USA). NMR experiments were conducted using an Agilent NMR 500 MHz spectrometer (Santa Clara, CA, USA) and BRUKER NMR 600 MHz spectrometer (San Jose, CA, USA) with (CD3)2SO as the solvent (referenced to residual DMSO at δH 2.54 and δC 39.5) at 25 °C. Electrospray ionization mass spectra (ESIMS) were acquired using an AB Sciex TripleTOF 4600 spectrometer (Boston, MA, USA) in the positive and negative ion mode. HPLC experiments were performed on a Hitachi Elite LaChrom system (Tokyo, Japan) equipped with a diode array detector model L-2450, pump L-2130 and autosampler L-2200. Semipreparative HPLC experiments were completed with a Waters XBridge Prep C18 (Miflord, CO, USA) 5 μm, 10 mm × 250 mm column and Phenomenex Luna C18 5 μm, 250 mm × 21.2 mm column.
Bacterial Strain and Culture Conditions. The WP1 strain (CGMCC No. 10738) was isolated from deep-sea sediments of the Southwest Indian Ocean and identified as Amycolatopsis sp. by 16S rRNA sequence comparison. WP1 was grown in ISP2 medium consisting of 1.0% (w/v) malt extract, 0.4% (w/v) yeast extract, 0.4% (w/v) glucose and 3% (w/v) sea salt, the pH of medium was adjusted to 7.4 using 2 M HCl and 2 M NaOH.
Mutants of strain WP1. The WP1 strain suspensions were spread onto ISP2 plates containing different concentrations (0, 10, 20, 30, 40, 50 and 60 mg/mL) of streptomycin. The plates were incubated at 37 °C for 7 days. Mutant colonies producing the white pigment different than the WP1 strain were selected, generating mutant strain L-30-6, which was obtained on the IPS2 plate containing 30 mg/mL streptomycin.
Extraction and isolation. The mutant L-30-6 strain was inoculated into ISP2 broth with 3% sea salt in 250 mL Erlenmeyer flasks, at 30 °C on a rotary shaker at 180 rpm for 2 days as seed culture. Each of the seed cultures (32 mL) was transferred into 1 L Erlenmeyer flasks containing 400 mL of ISP2 supplemented with 3% sea salt. These flasks were incubated at 30 °C on a rotary shaker at 180 rpm for 6 days. The resulting cultures (60 L) were centrifuged to yield the supernatant and a mycelial pellet. The supernatant was adsorbed onto macroporous resin XAD16N (DOW, St. Louis, Missouri, CA, USA) and eluted with linear gradient of 0–100% EtOH in H2O to afford six fractions (A–F).
Fraction C (3.8 g) was subjected to semipreparative HPLC (Phenomenex Luna C18, 250 mm × 21.2 mm, 5 µm, 10 mL/min) using a gradient solvent from 40–90% MeOH in H2O over 30 min to give five fractions (C1–C5). Fraction C2 was further purified by semipreparative HPLC (Waters XBridge Prep C18 5 μm, 10 mm× 250 mm, 4 mL/min) using an isocratic solvent system of CH3CN:H2O (15:85) over 30 min to afford compound 6 (10.2 mg) and C2A. Subfraction C2A was further purified by preparative HPLC with MeOH-H2O (45:55) to provide compound 7 (2.6 mg). Fraction C3 was further purified by semipreparative HPLC with MeOH-H2O (45:55) to yield compound 12 (6.5 mg), 3 (3.1 mg) and 4 (2.2 mg). Fraction C4 was further purified by semipreparative HPLC with MeOH:H2O (45:55) to afford compound 5 (2.2 mg).
Fraction D (2.1 g) was subjected to semipreparative HPLC (Phenomenex Luna C18, 250 mm × 21.2 mm, 5 µm,10 mL/min) using a gradient solvent from 50–80% MeOH in H2O over 30 min to generate five fractions (D1–D5). Fraction D3 was further purified by semipreparative HPLC using an isocratic solvent system of MeCN:H2O (50:50) to afford compound 1 (1.7 mg) and compound 2 (1.8 mg). D4 was subjected to preparative HPLC with MeCN:H2O (30:70) to provide compounds 9 (35.3 mg) and 10 (13.5 mg). D5 was further purified by preparative HPLC with MeOH-H2O (45:55) to yield compounds 8 (1.9 mg) and 11 (6.8 mg). The following are details of the extraction and isolation of the compounds.
Amycolachromone A (
1): White, amorphous powder; UV (MeOH) λmax (log ε) 253 (3.28) nm; IR (ZnSe)
νmax 3426, 3195, 2844, 1656, 1445, 1008 cm
−1;
1H and
13C NMR data,
Table 1; HRESIMS
m/
z 477.1172 [M + Na]
+ (calcd for C
24H
22O
9, 477.1162).
Amycolachromone B (
2): White, amorphous powder; UV (MeOH) λmax (log ε) 254 (3.46) nm; IR (ZnSe)
νmax 3460, 3190, 2894, 1658, 1445, 1008 cm
−1;
1H and
13C NMR data,
Table 1; HRESIMS
m/
z 469.1502 [M + H]
+ (calcd for C
25H
24O
9, 469.1499),
m/
z 491.1333 [M + Na]
+ (calcd 469.1318).
Amycolachromone C (
3): White, amorphous powder; UV (MeOH) λmax (log ε) 250 (3.43), 233 (3.46) nm; IR (ZnSe)
νmax 3420, 3199, 2993, 1650, 1310, 1089 cm
−1;
1H and
13C NMR data,
Table 2; HRESIMS
m/
z 351.0515 [M + Na]
+ (calcd for C
14H
16O
7S, 351.0514).
Amycolachromone D (
4): White, amorphous powder; UV (MeOH) λmax (log ε) 250 (3.56), 233 (3.58) nm; IR (ZnSe)
νmax 3520, 32,469, 2990, 1750, 1281, 1008 cm
−1;
1H and
13C NMR data,
Table 2; HRESIMS
m/
z 321.0406 [M + Na]
+ (calcd for C
13H
14O
6S, 321.0409).
Amycolachromone E (
5): White, amorphous powder; UV (MeOH) λmax (log ε) 250 (3.61), 240 (3.61) nm; IR (ZnSe)
νmax 3470, 3122, 2880, 1603, 1210, 1089 cm
−1;
1H and
13C NMR data,
Table 2; HRESIMS
m/
z 305.0462 [M + Na]
+ (calcd for C
13H
14O
5S, 305.0460).
Amycolachromone F (
6): White, crystal; UV (MeOH) λmax (log ε) 356 (3.20), 277 (3.68) nm; IR (ZnSe)
νmax 3477, 2956, 2916, 1748, 1622, 1475, 1349, 1083 cm
−1;
1H and
13C NMR data,
Table 3; HRESIMS
m/
z 337.0915 [M + H]
+ (calcd for C
16H
16O
8, 337.0923).
6-Ethoxymethyleugenin (7): White, amorphous powder; HR-ESIMS m/z 287.0891 [M + Na]+ (calcd for C14H16O5, 287.0985). 1H-NMR (600 MHz, DMSO-d6): δH 13.19 (s, OH-5), 6.70 (s, 1H, H-8), 6.28 (s, 1H, H-3), 4.41 (s, 2H, CH2OCH3), 3.89 (s, 3H, OCH3), 3.44 (q, J = 7.0 Hz, 2H, 6-CH2OCH2), 2.40 (s, 3H, 2-CH3), 1.08 (t, J = 7.0 Hz, 3H, OCH2CH3). 13C-NMR (150 MHz, DMSO-d6): δC182.6 (C-4), 168.5 (C-2), 164.3 (C-7), 159.9 (C-5), 158.0 (C-8a), 109.1 (C-3), 108.9 (C-6), 104.4 (C-4a), 89.6 (C-8), 65.1 (CH2OCH3), 59.3 (6-CH2OCH2), 56.9 (7-OCH3), 20.4 (2-CH3) and 15.3 (CH2CH3).
6-Methoxymethyleugenin (8): White, amorphous powder; HR-ESIMS m/z 273.0738 [M + Na]+ (calcd for C13H14O, 273.0739). 1H-NMR (600 MHz, CDCl3): δH 13.04 (s, OH-5), 6.36 (s, 1H, H-8), 6.04 (s, 1H, H-3), 4.55 (s, 2H, CH2OCH3), 3.90 (s, 3H, OCH3), 3.40 (s, 3H, CH2OCH3), 2.35 (s, 3H, CH3). 13C-NMR (150 MHz, CDCl3): δC182.4 (C-4), 166.6 (C-2), 164.2 (C-7), 160.6 (C-5), 158.2 (C-8a), 109.1 (C-3), 108.9 (C-6), 105.1 (C-4a), 89.6 (C-8), 61.6 (CH2OCH3), 58.2 (CH2OCH3), 56.2 (OCH3) and 20.4 (2-CH3).
6-Hydroxymethyleugenin (9): White, amorphous powder; HRESIMS m/z 259.0579 [M + Na]+ (calcd for C12H12O5, 259.0582). 1H-NMR (600 MHz, DMSO-d6): δH 13.09 (s, OH-5), 6.65 (s, 1H, H-8), 6.24 (s, 1H, H-3), 4.55 (t, J = 5.3 Hz, CH2OH), 4.43 (d, J = 5.2 Hz, 2H, H-9), 3.87 (s, 3H, OCH3), 2.38 (s, 3H, CH3). 13C-NMR (150 MHz, DMSO-d6): δC182.5 (C-4), 168.4 (C-2), 164.1 (C-7), 159.1 (C-5), 157.7 (C-8a), 112.5 (C-6), 08.8 (C-3), 104.6 (C-4a), 90.7 (C-8), 56.7 (OCH3), 50.9 (CH2OH) and 20.4(CH3).
Emodin (10): White, amorphous powder; HRESIMS m/z 269.0448 [M − H]− (calcd for C15H12O5, 269.0450). 1H-NMR (500 MHz, DMSO-d6): δH 12.07 (d, J = 19.5 Hz, 2H, OH), 7.49 (d, J = 1.1 Hz, 1H, H-5), 7.16 (s, 1H, H-7), 7.10 (d, J = 2.4 Hz, 1H, H-4), 6.57 (d, J = 2.4 Hz, 1H, H-2), 2.41 (s, 3H, CH3). 13C-NMR (150 MHz, DMSO-d6): δC190.0 (C-9), 182.0 (C-10), 166.6 (C-6), 165.0 (C-8), 161.9 (C-1), 148.6 (C-3), 135.6 (C-10a), 133.3 (C-4a), 24.6 (C-2), 120.9 (C-4), 113.9 (C-9a), 109.6 (C-5), 109.2 (C-7), 108.4 (C-8a) and 22.0 (CH3).
Chaetoquadrin D (11): White, amorphous powder; HRESIMS m/z HR-ESIMS m/z 370.0969 [M + H]+ (calcd for C16H19NO7S, 370.0960). 1H-NMR (500 MHz, DMSO-d6): δH 13.43 (s, 5-OH), 8.08 (t, NH), 6.79 (s, H-8), 6.32 (s, H-3), 4.36 (s, H-1′), 3.92 (s, 7-OCH3), 3.45 (dd, J = 13.6, 6.1 Hz, H-4′), 3.20 (t, J = 7.0 Hz, H-3′), 2.41 (s, 2-CH3), 1.81 (s, H-7′). 13C-NMR (125 MHz, DMSO-d6): δC 182.4 (C-4), 70.1 (C-6′), 169.0 (C-2), 163.7 (C-7), 159.9 (C-5), 158.2 (C-8a), 108.9 (C-3), 104.5 (C-4a), 100.8 (C-6), 91.4 (C-8), 57.3 (7-OCH3), 52.9 (C-3′), 48.5 (C-1′), 32.9 (C-4′), 22.9 (C-7′) and 20.42 (2-CH3).
X-ray Crystallographic Analysis of Compound 6. Crystals of 6 were obtained in the mixed solvent comprising MeOH and H2O, and crystallographic data were deposited at the Cambridge Crystallographic Data Centre (CCDC) under the reference number CCDC 1873441. The X-ray diffraction data were collected with Mo Kα radiation (λ = 0.71073 Å). The structure was solved by direct methods using the SHELXS-97 program. Orthorhombic C16H16O8, CH4O, H2O, a = 7.7760(3) Å, b = 8.6993(4) Å, c = 26.8196(11) Å, α = 90°, β = 90°, γ = 90°, V = 1814.23(13) Å3, Z = 4, ρCalcd = 1.414 g/cm3, μ = 0.118 mm−1, and F (0 0 0) = 816. Measurements were in the range 3.038° ≤ θ ≤ 26.368°, with 3697 independent reflections, of which 3133 unique reflections with I > 2σ (I) were collected for the analysis, Rint = 0.0332. The final R indices: R1 = 0.0455, wR2 = 0.1177 [I > 2σ(I)], R indices (all data): R1 = 0.0566, wR2 = 0.1256, and largest difference peak and hole: 0.560 and-0.231 e Å-3.
The ABH2 family DNA repair enzyme assay. Effects of compounds 1–11 on the ABH2 family demethylase activity reactions on m3c-ss-DNA were evaluated. All reactions were performed at 37 °C in reaction buffer [5 μM Fe(NH4)2(SO4)2, 0.93 mMα-ketoglutarate, 1.86 mM ascorbic acid, and 46.56 mM HEPES (pH 8.0)] for 1 h. Varying concentrations of compounds 1–11 (0, 5.0, 7.5, 20, 30, 40, 50, 75 and 100 μM) were used for tests. The m3c-ss-DNA was pre-mixed with reaction buffer in a concentration of 5.0 μM. The reactions were initiated by adding 2.0 μM ABH2. The reactions were stopped by adding 10.0 mM EDTA followed by heating to 95 °C for 5 min. All the results of reaction were analyzed by HPLC. All reaction samples were quantified by DNApac PA-100 column (4 mm × 250 mm, Thermo Scientific, (Waltham, MA, USA) with isocratic 60 % mobile B, 1.5 M ammonium acetate, under a constant flow rate of 1.0 mL/min. Mobile A was water. The UV detection wavelength was 260 nm.