Biotechnological and Ecological Potential of Micromonospora provocatoris sp. nov., a Gifted Strain Isolated from the Challenger Deep of the Mariana Trench
Abstract
1. Introduction
2. Results and Discussion
2.1. Isolation, Maintenance and Characterization of Strain MT25T
2.2. Compound Identification
2.3. Genome Sequencing and Annotation
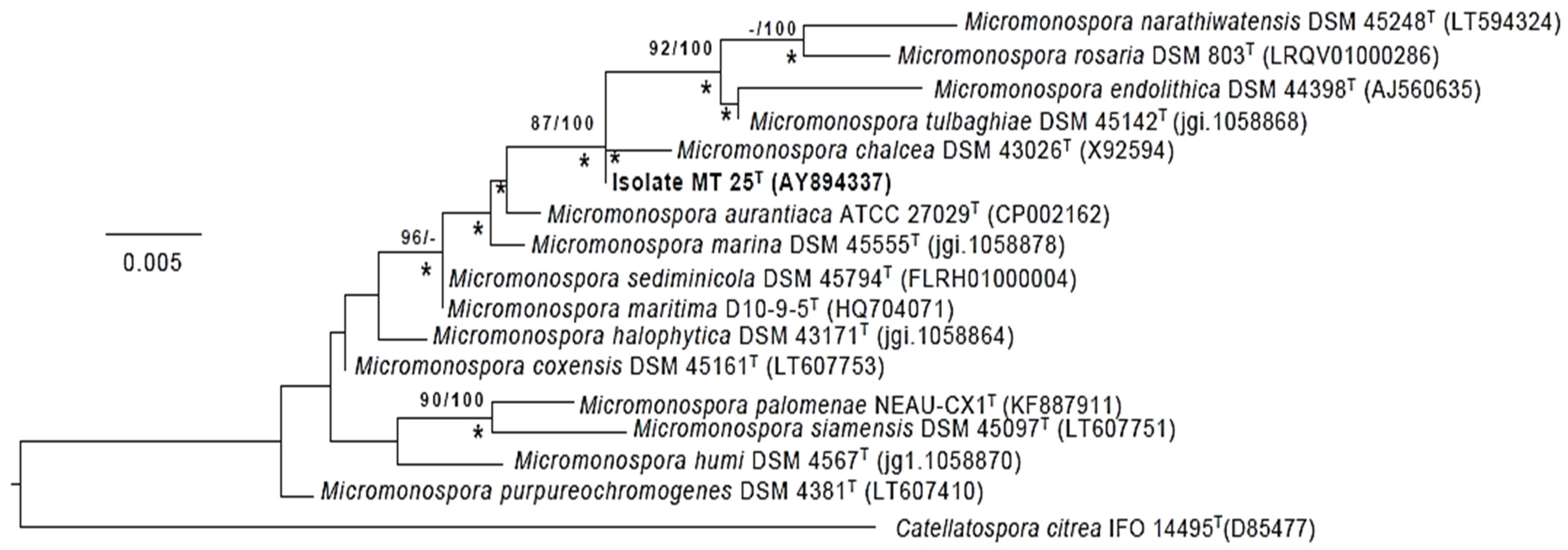
2.4. Phylogeny
2.5. Species Assignment
2.6. Description of Micromonospora provocatoris sp. nov.
2.7. Specialised Metabolite-Biosynthetic Gene Clusters
2.8. Genes Potentially Associated with Enviromental Stress
3. Materials and Methods
3.1. Microorganism
3.2. General Experimental Procedures
3.3. Fermentation Conditions
3.4. Isolation and Purification of Secondary Metabolites
3.5. Phylogeny
3.6. Phenotypic Characterisation
3.7. Whole-Genome Sequencing
3.7.1. DNA Extraction and Genome Sequencing
3.7.2. Annotation of Genome and Bioinformatics
3.7.3. Detection of the Gene Clusters
3.7.4. GenBank Accession Number
3.8. Comparison of Genomes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kamjam, M.; Sivalingam, P.; Deng, Z.; Hong, K. Deep sea actinomycetes and their secondary metabolites. Front. Microbiol. 2017, 8, 760. [Google Scholar] [CrossRef] [PubMed]
- Bull, A.T.; Goodfellow, M. Dark, rare and inspirational microbial matter in the extremobiosphere: 16,000 m of bioprospecting campaigns. Microbiology 2019, 165, 1252–1264. [Google Scholar] [CrossRef] [PubMed]
- Subramani, R.; Sipkema, D. Marine rare actinomycetes: A promising source of structurally diverse and unique novel natural products. Mar. Drugs 2019, 17, 249. [Google Scholar] [CrossRef]
- Riedlinger, J.; Reicke, A.; Zähner, H.; Krismer, B.; Bull, A.T.; Maldonado, L.A.; Ward, A.C.; Goodfellow, M.; Bister, B.; Bischoff, D.; et al. Abyssomicins, inhibitors of the para-aminobenzoic acid pathway produced by the marine Verrucosispora strain AB-18-032. J. Antibiot. 2004, 57, 271–279. [Google Scholar] [CrossRef]
- Nouioui, I.; Carro, L.; García-López, M.; Meier-Kolthoff, J.P.; Woyke, T.; Kyrpides, N.C.; Pukall, R.; Klenk, H.P.; Goodfellow, M.; Göker, M. Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 2018, 9, 2007. [Google Scholar] [CrossRef]
- Fiedler, H.P.; Bruntner, C.; Riedlinger, J.; Bull, A.T.; Knutsen, G.; Goodfellow, M.; Jones, A.; Maldonado, L.; Pathom-aree, W.; Beil, W.; et al. Proximicin A, B and C, novel aminofuran antibiotic and anticancer compounds isolated from marine strains of the actinomycete Verrucosispora. J. Antibiot. 2008, 61, 158–163. [Google Scholar] [CrossRef]
- Goodfellow, M.; Brown, R.; Ahmed, L.; Pathom-aree, W.; Bull, A.T.; Jones, A.L.; Stach, J.E.; Zucchi, T.D.; Zhang, L.; Wang, J. Verrucosispora fiedleri sp. nov., an actinomycete isolated from a fjord sediment which synthesizes proximicins. Antonie van Leeuwenhoek 2013, 103, 493–502. [Google Scholar] [CrossRef]
- Carro, L.; Nouioui, I.; Sangal, V.; Meier-Kolthoff, J.P.; Trujillo, M.E.; Montero-Calasanz, M.D.C.; Sahin, N.; Smith, D.L.; Kim, K.E.; Peluso, P.; et al. Genome-based classification of micromonosporae with a focus on their biotechnological and ecological potential. Sci. Rep. 2018, 8, 525. [Google Scholar] [CrossRef]
- Carro, L.; Castro, J.F.; Razmilic, V.; Nouioui, I.; Pan, C.; Igual, J.M.; Jaspars, M.; Goodfellow, M.; Bull, A.T.; Asenjo, J.A.; et al. Uncovering the potential of novel micromonosporae isolated from an extreme hyper-arid Atacama Desert soil. Sci. Rep. 2019, 9, 4678. [Google Scholar] [CrossRef]
- Carro, L.; Golinska, P.; Nouioui, I.; Bull, A.T.; Igual, J.M.; Andrews, B.A.; Klenk, H.P.; Goodfellow, M. Micromonospora acroterricola sp. nov., a novel actinobacterium isolated from a high altitude Atacama Desert soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 3426–3436. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Carro, L.; Razmilic, V.; Nouioui, I.; Richardson, L.; Pan, C.; Golinska, P.; Asenjo, J.A.; Bull, A.T.; Klenk, H.P.; Goodfellow, M. Hunting for cultivable Micromonospora strains in soils of the Atacama Desert. Antonie van Leeuwenhoek 2018, 111, 1375–1387. [Google Scholar] [CrossRef]
- Ørskov, J. Investigations into the Morphology of the Ray Fungi; Levin and Munksgaard: Copenhagen, Denmark, 1923. [Google Scholar]
- Krassilnikov, N.A. Ray Fungi and Related Organisms, Actinomycetales; Akademii Nauk S. S. S. R.: Moscow, Russia, 1938. [Google Scholar]
- Genilloud, O. Order XI. Micromonosporales ord. nov. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Goodfellow, M., Kämpfer, P., Busse, H.-J., Trujillo, M.E., Suzuki, K.-i., Ludwig, W., Whitman, W.B., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 5, p. 1035. [Google Scholar]
- Salam, N.; Jiao, J.Y.; Zhang, X.T.; Li, W.J. Update on the classification of higher ranks in the phylum Actinobacteria. Int. J. Syst. Evol. Microbiol. 2020, 70, 1331–1355. [Google Scholar] [CrossRef]
- Foulerton, A.G.R. New species of Streptothrix isolated from the air. Lancet 1905, 1, 1199–1200. [Google Scholar]
- Genilloud, O.; Ørskov, G.I.M. 156AL. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Goodfellow, M., Kämpfer, P., Busse, H.-J., Trujillo, M.E., Suzuki, K.-I., Ludwig, W., Whitman, W.B., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 5, pp. 1039–1057. [Google Scholar]
- Pathom-Aree, W.; Stach, J.E.; Ward, A.C.; Horikoshi, K.; Bull, A.T.; Goodfellow, M. Divesity of actinomycetes isolated from Challenger Deep sediment (10,898 m) from the Mariana Trench. Extremophiles 2006, 10, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Vickers, J.C.; Williams, S.T.; Ross, G.W. A taxonomic approach to selective isolation of streptomycetes from soil. In Biological, Biochemical and Biomedical Aspects of Actinomycetes; Ortiz-Ortiz, L., Bojalil, L.F., Yakoleff, V., Eds.; Academic Press: Orlando, FL, USA, 1984; pp. 553–561. [Google Scholar]
- Kato, C.; Li, L.; Tamaoka, J.; Horikoshi, K. Molecular analyses of the sediment of the 11,000-m deep Mariana Trench. Extremophiles 1997, 1, 117–123. [Google Scholar] [CrossRef]
- Smith, L.T.; Smith, G.M. An osmoregulated dipeptide in stressed Rhizobium meliloti. J. Bacteriol. 1989, 171, 4714–4717. [Google Scholar] [CrossRef] [PubMed]
- Masatomi, I.; Tetsuya, S.; Masahide, A.; Ryuichi, S.; Hiroshi, N.; Masaaki, I.; Tomio, T. IC202A, a new siderophore with immunosuppressive activity produced by Streptoalloteichus sp. 1454-19. II. Physico-chemical properties and structure elucidation. J. Antibiot. 1999, 52, 25–28. [Google Scholar]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Angiuoli, S.V.; Gussman, A.; Klimke, W.; Cochrane, G.; Field, D.; Garrity, G.; Kodira, C.D.; Kyrpides, N.; Madupu, R.; Markowitz, V.; et al. Toward an online repository of Standard Operating Procedures (SOPs) for (meta) genomic annotation. OMICS 2008, 12, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; Ciufo, S.; Fedorov, B.; O’Neill, K.; Tolstoy, I. RefSeq microbial genomes database: New representation and annotation strategy. Nucleic Acids Res. 2014, 42, D553–D559. [Google Scholar] [CrossRef] [PubMed]
- Sveshnikova, M.A.; Maksimova, T.S.; Kudrina, E.S. Species of the genus Micromonospora Oerskov, 1923 and their taxonomy. Mikrobiologiia 1969, 38, 883–893. [Google Scholar]
- Tanasupawat, S.; Jongrungruangchok, S.; Kudo, T. Micromonospora marina sp. nov., isolated from sea sand. Int. J. Syst. Evol. Microbiol. 2010, 60, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Songsumanus, A.; Tanasupawat, S.; Igarashi, Y.; Kudo, T. Micromonospora maritima sp. nov., isolated from mangrove soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 554–559. [Google Scholar] [CrossRef]
- Supong, K.; Suriyachadkun, C.; Tanasupawat, S.; Suwanborirux, K.; Pittayakhajonwut, P.; Kudo, T.; Thawai, C. Micromonospora sediminicola sp. nov., isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2013, 63, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Kirby, B.M.; Meyers, P.R. Micromonospora tulbaghiae sp. nov., isolated from the leaves of wild garlic, Tulbaghia violacea. Int. J. Syst. Evol. Microbiol. 2010, 60, 1328–1333. [Google Scholar] [CrossRef]
- Contreras-Castro, L.; Maldonado, L.A.; Quintana, E.T.; Carro, L.; Klenk, H.-P. Genomic insight into three marine Micromonospora sp. strains from the Gulf of California. Microbiol. Resour. Announc. 2019, 8, e01673-18. [Google Scholar] [CrossRef]
- Riesco, R.; Carro, L.; Román-Ponce, B.; Prieto, C.; Blom, J.; Klenk, H.P.; Normand, P.; Trujillo, M.E. Defining the species Micromonospora saelicesensis and Micromonospora noduli Under the framework of genomics. Front. Microbiol. 2018, 9, 1360. [Google Scholar] [CrossRef]
- Songsumanus, A.; Tanasupawat, S.; Thawai, C.; Suwanborirux, K.; Kudo, T. Micromonospora humi sp. nov., isolated from peat swamp forest soil. Int. J. Syst. Evol. Microbiol. 2011, 61, 1176–1181. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.; Cognat, V.; Goodfellow, M.; Koechler, S.; Heintz, D.; Carapito, C.; Van Dorsselaer, A.; Mahmoud, H.; Sangal, V.; Ismail, W. Phylogenomic classification and biosynthetic potential of the fossil fuel-biodesulfurizing Rhodococcus strain IGTS8. Front. Microbiol. 2020, 11, 1417. [Google Scholar] [CrossRef]
- Li, X.; Huang, Y.; Whitman, W.B. The relationship of the whole genome sequence identity to DNA hybridization varies between genera of prokaryotes. Antonie van Leeuwenhoek 2015, 107, 241–249. [Google Scholar] [CrossRef]
- Palmer, M.; Steenkamp, E.T.; Blom, J.; Hedlund, B.P.; Venter, S.N. All ANIs are not created equal: Implications for prokaryotic species boundaries and integration of ANIs into polyphasic taxonomy. Int. J. Syst. Evol. Microbiol. 2020, 70, 2937–2948. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Klenk, H.P.; Göker, M. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 2014, 64, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar]
- Sosio, M.; Gaspari, E.; Iorio, M.; Pessina, S.; Medema, M.H.; Bernasconi, A.; Simone, M.; Maffioli, S.I.; Ebright, R.H.; Donadio, S. Analysis of the pseudouridimycin biosynthetic pathway provides insights into the formation of c-nucleoside antibiotics. Cell Chem. Biol. 2018, 25, 540–549.e4. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, H.; Chen, H.; Jing, X.; Zheng, W.; Li, R.; Sun, T.; Liu, J.; Fu, J.; Huo, L.; et al. Discovery of recombinases enables genome mining of cryptic biosynthetic gene clusters in Burkholderiales species. Proc. Natl. Acad. Sci. USA 2018, 115, E4255–E4263. [Google Scholar]
- Awakawa, T.; Fujita, N.; Hayakawa, M.; Ohnishi, Y.; Horinouchi, S. Characterization of the biosynthesis gene cluster for alkyl-O-dihydrogeranyl-methoxyhydroquinones in Actinoplanes missouriensis. ChemBioChem 2011, 12, 439–448. [Google Scholar] [CrossRef]
- Yanai, K.; Murakami, T.; Bibb, M. Amplification of the entire kanamycin biosynthetic gene cluster during empirical strain improvement of Streptomyces kanamyceticus. Proc. Natl. Acad. Sci. USA 2006, 103, 9661–9666. [Google Scholar] [CrossRef]
- Shigemori, H.; Komaki, H.; Yazawa, K.; Mikami, Y.; Nemoto, A.; Tanaka, Y.; Sasaki, T.; In, Y.; Ishida, T.; Kobayashi, J.; et al. Brasilicardin A. A novel tricyclic metabolite with potent immunosuppressive activity from actinomycete Nocardia brasiliensis. J. Org. Chem. 1998, 63, 6900–6904. [Google Scholar] [CrossRef]
- Ogasawara, Y.; Yackley, B.J.; Greenberg, J.A.; Rogelj, S.; Melançon, C.E., 3rd. Expanding our understanding of sequence-function relationships of type II polyketide biosynthetic gene clusters: Bioinformatics-guided identification of Frankiamicin A from Frankia sp. EAN1pec. PLoS ONE 2015, 10, e0121505. [Google Scholar] [CrossRef]
- Abdel-Mageed, W.M.; Juhasz, B.; Lehri, B.; Alqahtani, A.S.; Nouioui, I.; Pech-Puch, D.; Tabudravu, J.N.; Goodfellow, M.; Rodríguez, J.; Jaspars, M.; et al. Whole genome sequence of Dermacoccus abyssi MT1.1 isolated from the Challenger Deep of the Mariana Trench reveals phenazine biosynthesis locus and environmental adaptation factors. Mar. Drugs 2020, 18, 131. [Google Scholar] [CrossRef] [PubMed]
- Gumley, A.W.; Inniss, W.E. Cold shock proteins and cold acclimation proteins in the psychrotrophic bacterium Pseudomonas putida Q5 and its transconjugant. Can. J. Microbiol. 1996, 42, 798–803. [Google Scholar] [CrossRef]
- Fujii, S.; Nakasone, K.; Horikoshi, K. Cloning of two cold shock genes, cspA and cspG, from the deep-sea psychrophilic bacterium Shewanella violacea strain DSS12. FEMS Microbiol. Lett. 1999, 178, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mageed, W.M.; Lehri, B.; Jarmusch, S.A.; Miranda, K.; Al-Wahaibi, L.H.; Stewart, H.A.; Jamieson, A.J.; Jaspars, M.; Karlyshev, A.V. Whole genome sequencing of four bacterial strains from South Shetland Trench revealing biosynthetic and environmental adaptation gene clusters. Mar. Genom. 2020, 54, 100782. [Google Scholar] [CrossRef]
- Lelivelt, M.J.; Kawula, T.H. Hsc66, an Hsp70 homolog in Escherichia coli, is induced by cold shock but not by heat shock. J. Bacteriol. 1995, 177, 4900–4907. [Google Scholar] [CrossRef]
- Lee, S.; Sowa, M.E.; Choi, J.M.; Tsai, F.T. The ClpB/Hsp104 molecular chaperone—A protein disaggregating machine. J. Struct. Biol. 2004, 146, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Redder, P.; Hausmann, S.; Khemici, V.; Yasrebi, H.; Linder, P. Bacterial versatility requires DEAD-box RNA helicases. FEMS Microbiol. Rev. 2015, 39, 392–412. [Google Scholar] [CrossRef]
- Chattopadhyay, M.; Jagannadham, M. Maintenance of membrane fluidity in Antarctic bacteria. Polar Biol. 2001, 24, 386–388. [Google Scholar]
- Chattopadhyay, M.K.; Jagannadham, M.V. A branched chain fatty acid promotes cold adaptation in bacteria. J. Biosci. 2003, 28, 363–364. [Google Scholar] [CrossRef]
- Goude, R.; Renaud, S.; Bonnassie, S.; Bernard, T.; Blanco, C. Glutamine, glutamate, and alpha-glucosylglycerate are the major osmotic solutes accumulated by Erwinia chrysanthemi strain 3937. Appl. Environ. Microbiol. 2004, 70, 6535–6541. [Google Scholar] [CrossRef]
- Kuhlmann, A.U.; Hoffmann, T.; Bursy, J.; Jebbar, M.; Bremer, E. Ectoine and hydroxyectoine as protectants against osmotic and cold stress: Uptake through the SigB-controlled betaine-choline- carnitine transporter-type carrier EctT from Virgibacillus pantothenticus. J. Bacteriol. 2011, 193, 4699–4708. [Google Scholar] [CrossRef]
- Gouffi, K.; Blanco, C. Is the accumulation of osmoprotectant the unique mechanism involved in bacterial osmoprotection? Int. J. Food Microbiol. 2000, 55, 171–174. [Google Scholar] [CrossRef]
- Nau-Wagner, G.; Opper, D.; Rolbetzki, A.; Boch, J.; Kempf, B.; Hoffmann, T.; Bremer, E. Genetic control of osmoadaptive glycine betaine synthesis in Bacillus subtilis through the choline-sensing and glycine betaine-responsive GbsR repressor. J. Bacteriol. 2012, 194, 2703–2714. [Google Scholar] [CrossRef] [PubMed]
- Boncompagni, E.; Osteras, M.; Poggi, M.C.; le Rudulier, D. Occurrence of choline and glycine betaine uptake and metabolism in the family Rhizobiaceae and their roles in osmoprotection. Appl. Environ. Microbiol. 1999, 65, 2072–2077. [Google Scholar] [CrossRef] [PubMed]
- Sagot, B.; Gaysinski, M.; Mehiri, M.; Guigonis, J.M.; Le Rudulier, D.; Alloing, G. Osmotically induced synthesis of the dipeptide N-acetylglutaminylglutamine amide is mediated by a new pathway conserved among bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 12652–12657. [Google Scholar] [CrossRef] [PubMed]
- Campanaro, S.; Treu, L.; Valle, G. Protein evolution in deep sea bacteria: An analysis of amino acids substitution rates. BMC Evol. Biol. 2008, 8, 313. [Google Scholar] [CrossRef]
- Goordial, J.; Raymond-Bouchard, I.; Zolotarov, Y.; de Bethencourt, L.; Ronholm, J.; Shapiro, N.; Woyke, T.; Stromvik, M.; Greer, C.; Bakermans, C.; et al. Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol. Ecol. 2016, 92, fiv154. [Google Scholar]
- Bartlett, D.H. Microbial adaptations to the psychrosphere/piezosphere. J. Mol. Microbiol. Biotechnol. 1999, 1, 93–100. [Google Scholar]
- Sekar, K.; Linker, S.M.; Nguyen, J.; Grünhagen, A.; Stocker, R.; Sauer, U. Bacterial glycogen provides short-term benefits in changing environments. Appl. Environ. Microbiol. 2020, 86, e00049-20. [Google Scholar] [CrossRef]
- Cannon, G.C.; Heinhorst, S.; Kerfeld, C.A. Carboxysomal carbonic anhydrases: Structure and role in microbial CO2 fixation. Biochim. Biophys. Acta 2010, 1804, 382–392. [Google Scholar] [CrossRef]
- Busarakam, K.; Bull, A.T.; Trujillo, M.E.; Riesco, R.; Sangal, V.; van Wezel, G.P.; Goodfellow, M. Modestobacter caceresii sp. nov., novel actinobacteria with an insight into their adaptive mechanisms for survival in extreme hyper-arid Atacama Desert soils. Syst. Appl. Microbiol. 2016, 39, 243–251. [Google Scholar] [CrossRef]
- Castro, J.F.; Nouioui, I.; Sangal, V.; Choi, S.; Yang, S.J.; Kim, B.Y.; Trujillo, M.E.; Riesco, R.; Montero-Calasanz, M.D.C.; Rahmani, T.P.D.; et al. Blastococcus atacamensis sp. nov., a novel strain adapted to life in the Yungay core region of the Atacama Desert. Int. J. Syst. Evol. Microbiol. 2018, 68, 2712–2721. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.F.; Nouioui, I.; Sangal, V.; Trujillo, M.E.; Montero-Calasanz, M.D.C.; Rahmani, T.; Bull, A.T.; Asenjo, J.A.; Andrews, B.A.; Goodfellow, M. Geodermatophilus chilensis sp. nov., from soil of the Yungay core-region of the Atacama Desert, Chile. Syst. Appl. Microbiol. 2018, 41, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Golińska, P.; Świecimska, M.; Montero-Calasanz, M.D.C.; Yaramis, A.; Igual, J.M.; Bull, A.T.; Goodfellow, M. Modestobacter altitudinis sp. nov., a novel actinobacterium isolated from Atacama Desert soil. Int. J. Syst. Evol. Microbiol. 2020, 70, 3513–3527. [Google Scholar] [CrossRef]
- Golinska, P.; Montero-Calasanz, M.D.C.; Świecimska, M.; Yaramis, A.; Igual, J.M.; Bull, A.T.; Goodfellow, M. Modestobacter excelsi sp. nov., a novel actinobacterium isolated from a high altitude Atacama Desert soil. Syst. Appl. Microbiol. 2020, 43, 126051. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, M.; Williams, S.T. Ecology of actinomycetes. Annu. Rev. Microbiol. 1983, 37, 189–216. [Google Scholar] [CrossRef] [PubMed]
- Zenova, G.M.; Zviagintsev, D.G. Aktinomitsety roda Micromonospora v lugovykh ékosistemakh [Actinomycetes of the genus Micromonospora in meadow ecosystems]. Mikrobiologiia 2002, 71, 662–666. [Google Scholar]
- Hong, K.; Gao, A.H.; Xie, Q.Y.; Gao, H.; Zhuang, L.; Lin, H.P.; Yu, H.P.; Li, J.; Yao, X.S.; Goodfellow, M.; et al. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 2009, 7, 24–44. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Chalita, M.; Ha, S.M.; Na, S.I.; Yoon, S.H.; Chun, J. ContEst16S: An algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2053–2057. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M.; Spröer, C.; Klenk, H.P. When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 2013, 195, 413–418. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Fitch, W. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Zool. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Pattengale, N.D.; Alipour, M.; Bininda-Emonds, O.R.; Moret, B.M.; Stamatakis, A. How many bootstrap replicates are necessary? J. Comput. Biol. 2010, 17, 337–354. [Google Scholar] [CrossRef]
- Goloboff, P.A.; Farris, J.S.; Nixon, K.C. TNT, a free program for phylogenetic analysis. Cladistics 2008, 24, 774–786. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*. Phylogenetic analysis using parsimony (*and Other Methods); Vers. 4; Sinauer: Sunderland, MA, USA, 1998. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Jukes, T.; Cantor, C. Evolution of protein molecules. In Mammalian Protein Metabolism; Volume III. Chapter 24; Munro, H., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Staneck, J.L.; Roberts, G.D. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 1974, 28, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Lechevalier, M.P.; Lechevalier, H. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Bacteriol. 1970, 20, 435–443. [Google Scholar] [CrossRef]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Kroppenstedt, B.M.; Goodfellow, M. The family Thermomonosporaceae, Actinocoralliria, Spirillospora and Thermomonospora. In The Prokaryotes, 2nd ed.; Volume 3: Archaea. Bacteria: Firmicutes, Actinomycetes; Dworkin, M., Falkow, S., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 682–724. [Google Scholar]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; MIDI Technical Note; MIDI: Newark, DE, USA, 1990; Volume 101, p. 1. [Google Scholar]
- O’Donnell, A.G.; Falconer, C.; Goodfellow, M.; Ward, A.C.; Williams, E. Biosystematics and diversity amongst novel carboxydotrophic actinomycetes. Antonie van Leeuwenhoek 1993, 64, 325–340. [Google Scholar] [CrossRef]
- Williams, S.T.; Goodfellow, M.; Alderson, G.; Wellington, E.M.H.; Sneath, P.H.A.; Sackin, M.J. Numerical classification of Streptomyces and related genera. J. Gen. Microbiol. 1983, 129, 1743–1813. [Google Scholar] [CrossRef]
- Lehmann, P.F.; Murray, P.R.; Baron, E.J.; Pfaller, M.A.; Tenover, F.C.; Yolken, R.H. (Eds.) Manual of Clinical Microbiology, 7th ed.; Sigma-Aldrich: St. Louis, MO, USA, 2015; Volume 146, pp. 107–108. [Google Scholar] [CrossRef]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Current Protocols in Molecular Biology; John Wiley and Sons, Inc.: New York, NY, USA, 1994. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]




| No. | 1 | 2 | ||||
|---|---|---|---|---|---|---|
| δC, Mult. | δN, Mult. | δH, Mult (J in Hz) | δC, Mult. | δN, Mult. | δH, Mult (J in Hz) | |
| 1 | - | 108.5, NH2 | a. 7.28, brs
b. 6.75, brs | 31.4 t | ||
| 2 | 173.8, C | - | - | 41.4 t | 2.76 (t,8.0) | |
| 3 | 31.4, CH2 | - | 2.06, m | 29.2 t | 1.51 m | |
| 4 | 27.2, CH2 | - | a. 1.89, m b. 1.74, m | 25.4, t | 1.38 m | |
| 5 | 52.7, CH | - | 4.15, m | 28.3 t | 1.51 m | |
| 6 | 171.4, C | - | - | 49.3 t | 3.46 m | |
| 7 | - | 117.3, NH | 7.97, d (7.8) | - | 174.4, s | 9.68 brs |
| 8 | 52.1, CH | - | 4.12, m | 173.9 s | - | |
| 9 | 27.8, CH2 | - | a. 1.87, m b. 1.67, m | 30.0 t | 2.58 m | |
| 10 | 31.6, CH2 | - | 2.08, m | 31.3 t | 2.27 m | |
| 11 | 173.8, C | - | - | 173.9 s | - | |
| 12 | - | 108.6, NH2 | a. 7.28, brs b. 6.75, brs | - | 116.2 d | 7.79 brs |
| 13 | - | 123.2, NH | 8.11, d (7.8) | 41.0 t | 3.00 (q, 8.0) | |
| 14 | 169.7, C | - | - | 31.4 t | 1.38 m | |
| 15 | 22.6, CH3 | - | 1.86, s | 26.0 t | 1.26 m | |
| 16 | 173.3, C | - | - | 28.6 t | 1.51 m | |
| 17 | - | 104.8, NH2 | a. 7.27, brs b. 7.05, brs | 49.4 t | 3.46 m | |
| 18 | 174.4 s | 9.67 brs | ||||
| 19 | 173.9 s | - | ||||
| 20 | 30.1 t | 2.58 m | ||||
| 21 | 32.4 t | 2.27 m | ||||
| 22 | 173.9 s | - | ||||
| 23 | 116.2 d | 7.79 brs | ||||
| 24 | 41.0 t | 3.00 (q, 8.0) | ||||
| 25 | 31.4 t | 1.38 m | ||||
| 26 | 26.0 t | 1.26 m | ||||
| 27 | 28.6 t | 1.51 m | ||||
| 28 | 49.6 t | 3.46 m | ||||
| 29 | - | 175.6 s | 9.63 brs | |||
| 30 | 173.5 s | - | ||||
| 31 | 22.9 q | 1.96 s | ||||
| Features | Strain MT25T |
|---|---|
| Assembly size, bp | 6,053,796 |
| No. of contigs | 1170 |
| G + C (%) | 71.6 |
| Fold coverage | 39.94× |
| N50 | 8214 |
| L50 | 203 |
| Genes | 6643 |
| CDs | 6573 |
| Pseudo genes | 2188 |
| Protein encoding genes | 4385 |
| rRNA | 8 |
| tRNA | 59 |
| ncRNAs | 3 |
| Accession No. | NZ_QNTW00000000 |
| Assembly method | SPAdes v. 5.0.0.0 |
| Phylogenomic Neighbors | Similarity ANI | Values (%) dDDH |
|---|---|---|
| M. aurantiaca ATCC 27029T | 95.2 | 62.7 |
| M. chalcea DSM 43026T | 93.5 | 53.0 |
| M. marina DSM 45555T | 94.6 | 58.6 |
| M. tulbaghiae DSM 45142T | 96.0 | 68.1 |
| Characteristics | Strain MT25T | M. tulbaghiae DSM 45142T |
|---|---|---|
| Morphology: | ||
| Spores borne on sporophores | - | + |
| Spore ornamentation | Rugose | Smooth |
| Substrate mycelial color on yeast extract-malt extract agar | Orange | Brown |
| AP1-ZYM tests: | ||
| Acid and alkaline phosphatases, β-glucosidase, lipase (C14), naphthol-AS-BI-phosphohydrolase | + | - |
| α-galacosidase, β-glucoronidase, N-acetyl-β-glucosaminidase | - | + |
| Biochemical tests: | ||
| H2S production | - | + |
| Nitrate reduction | + | - |
| Degradation tests: | ||
| L-tyrosine | + | - |
| Casein | - | + |
| Gelatin | - | + |
| Starch | - | + |
| Tween-80 | - | + |
| Tolerance tests: | ||
| Growth at 4 °C | - | + |
| Growth at pH 6.0 and pH 10 | + | - |
| Growth in presence of 5% w/v NaCl | - | + |
| Chemotaxonomy: | ||
| Major whole-organism sugars | Glucose, mannose, ribose and xylose | Glucose, ribose and xylose |
| BGCs | No. | Nucleotide (nt) bp | Type | Accession Number | Homologue | Accession Number | Identity |
|---|---|---|---|---|---|---|---|
| Siderophore | 1 | 6963 | Desferrioxamine E | QNTW01000257 | Desferrioxamine EBGC from Streptomyces sp. ID38640 | MG459167.1 | 100% |
| T2PKS * | 1 | 3695 | Frankiamicin | QNTW01000523 | Frankiamicin BGC from Frankia sp. EAN1pec | CP000820.1 | 28% |
| Terpene | 1 | 20066 | Isorenieratene | QNTW01000028 | Isorenieratene BGC from Streptomyces griseus subsp. griseus NBRC 13350 | AP009493.1 | 28% |
| Terpene | 1 | 11057 | Phosphonoglycans | QNTW01000118 | Phosphonoglycans BGC from Glycomyces sp. NRRL B-16210 | KJ125437.1 | 3% |
| Oligosaccharides | 1 | Brasilicardin A | Brasilicardin A BGC from Nocardia terpenica IFM 0406 | KV411304.1 | 23% | ||
| NRPS *** | 1 | 10526 | Rhizomide (A-C) | QNTW01000131 | Rhizomide A BGC from Paraburkholderia rhizoxinica HKI 454 | NC_014718.1 | 100% |
| T3PKS ** | 1 | 12,601 | Alkyl-O-dihydrogeranyl-methoxyhydroquinones | QNTW01000093 | alkyl-O-dihydrogeranyl-methoxyhydroquinones biosynthetic gene cluster from Actinoplanes missouriensis 431 | AP012319.1 | 28% |
| Lanthipeptide-class-i | 1 | 26,371 | Kanamycin | QNTW01000003 | kanamycin biosynthetic gene cluster from Streptomyces kanamyceticus | AB254080.1 | 1% |
| Lanthipeptide-class-i | 1 | 18,770 | No match found | QNTW01000004 | - | - | - |
| Lanthipeptide-class-iii | 1 | 7750 | No match found | QNTW01000229 | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdel-Mageed, W.M.; Al-Wahaibi, L.H.; Lehri, B.; Al-Saleem, M.S.M.; Goodfellow, M.; Kusuma, A.B.; Nouioui, I.; Soleh, H.; Pathom-Aree, W.; Jaspars, M.; et al. Biotechnological and Ecological Potential of Micromonospora provocatoris sp. nov., a Gifted Strain Isolated from the Challenger Deep of the Mariana Trench. Mar. Drugs 2021, 19, 243. https://doi.org/10.3390/md19050243
Abdel-Mageed WM, Al-Wahaibi LH, Lehri B, Al-Saleem MSM, Goodfellow M, Kusuma AB, Nouioui I, Soleh H, Pathom-Aree W, Jaspars M, et al. Biotechnological and Ecological Potential of Micromonospora provocatoris sp. nov., a Gifted Strain Isolated from the Challenger Deep of the Mariana Trench. Marine Drugs. 2021; 19(5):243. https://doi.org/10.3390/md19050243
Chicago/Turabian StyleAbdel-Mageed, Wael M., Lamya H. Al-Wahaibi, Burhan Lehri, Muneera S. M. Al-Saleem, Michael Goodfellow, Ali B. Kusuma, Imen Nouioui, Hariadi Soleh, Wasu Pathom-Aree, Marcel Jaspars, and et al. 2021. "Biotechnological and Ecological Potential of Micromonospora provocatoris sp. nov., a Gifted Strain Isolated from the Challenger Deep of the Mariana Trench" Marine Drugs 19, no. 5: 243. https://doi.org/10.3390/md19050243
APA StyleAbdel-Mageed, W. M., Al-Wahaibi, L. H., Lehri, B., Al-Saleem, M. S. M., Goodfellow, M., Kusuma, A. B., Nouioui, I., Soleh, H., Pathom-Aree, W., Jaspars, M., & Karlyshev, A. V. (2021). Biotechnological and Ecological Potential of Micromonospora provocatoris sp. nov., a Gifted Strain Isolated from the Challenger Deep of the Mariana Trench. Marine Drugs, 19(5), 243. https://doi.org/10.3390/md19050243










