Unique Polyhalogenated Peptides from the Marine Sponge Ircinia sp.
Abstract
1. Introduction
2. Results and Discussion
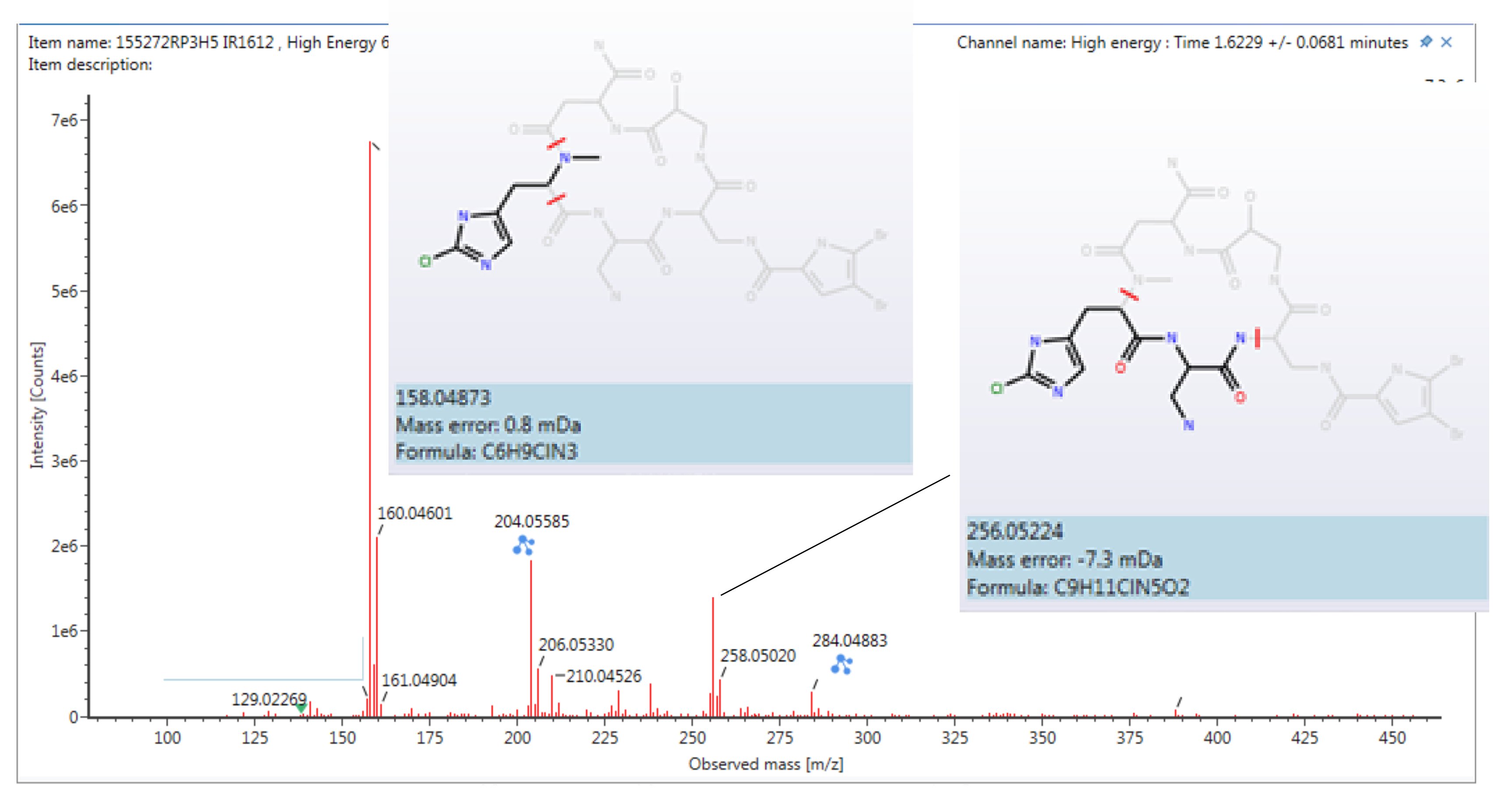
Isolation and Structure Elucidation
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Biological Material
3.3. Extraction and Isolation
3.4. Absolute Configuration
3.5. Biological Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanif, N.; Murni, A.; Tanaka, C.; Tanaka, J. Marine Natural Products from Indonesian Waters. Mar. Drugs 2019, 17, 364. [Google Scholar] [CrossRef] [PubMed]
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine Sponge Derived Natural Products between 2001 and 2010: Trends and Opportunities for Discovery of Bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, A.; Miyamoto, T.; Higuchi, R.; Uchiumi, T.; Kuwano, M.; Van Soet, R.W.M. Structure of a novel multidrug resistance modulator, irciniasulfonic acid, isolated from a marine sponge Ircinia sp. Tetrahedron Lett. 2001, 42, 3335–3337. [Google Scholar] [CrossRef]
- Sica, D.; Piccialli, V.; Pronzato, R. Sterols from the sponges Ircinia pipetta and Dysidea avara identification of cholestatrienol. Comp. Biochem. Physiol. 1987, 88, 293–296. [Google Scholar]
- Venkateswarlu, Y.; Reddy, M.V.R.; Rao, M.N. A new epoxy sterol from the sponge Ircinia fasciculata. J. Nat. Prod. 1996, 59, 876–877. [Google Scholar] [CrossRef]
- Issa, H.H.; Tanaka, J.; Higa, T. New cytotoxic furanosesterterpenes from an Okinawan marine sponge Ircinia sp. J. Nat. Prod. 2003, 66, 252–254. [Google Scholar] [CrossRef]
- Lai, Y.Y.; Lu, M.C.; Wang, L.H.; Chen, J.J.; Fang, L.S.; Wu, Y.C.; Sung, P.J. New scalarane sesterterpenoids from the Formosan sponge Ircinia felix. Mar. Drugs 2015, 13, 4296–4309. [Google Scholar] [CrossRef]
- Rashid, M.A.; Gustafson, K.R.; Boyd, M.R. New chondropsin macrolide lactams from marine sponges in the genus Ircinia. Tetrahedron Lett. 2001, 42, 1623–1626. [Google Scholar] [CrossRef]
- Chevallier, C.; Bugni, T.S.; Feng, X.; Harper, M.K.; Orendt, A.M.; Ireland, C.M. Tedanolide C: A potent new 18-membered-ring cytotoxic macrolide isolated from the Papua New Guinea marine sponge Ircinia sp. J. Org. Chem. 2006, 71, 2510–2513. [Google Scholar] [CrossRef]
- Feng, Y.; Caroll, A.R.; Pass, D.M.; Archbold, J.K.; Avery, V.M.; Quin, R.J. Polydiscamides B−D from a marine sponge Ircinia sp. as potent human sensory neuron-specific G protein coupled receptor agonists. J. Nat. Prod. 2008, 71, 8–11. [Google Scholar] [CrossRef]
- Mohamed, A.M.; Rao, V.; Hamann, M.T.; Kelly, M.; Hill, R.T. Monitoring Bacterial Diversity of the Marine Sponge Ircinia strobilina upon Transfer into Aquaculture. Appl. Environ. Microbiol. 2008, 74, 4133–4143. [Google Scholar] [CrossRef]
- Mau, C.; Nakao, Y.; Yoshida, W.Y.; Scheuer, P.J.; Kelly-Borges, M. Waiakeamide, a Cyclic Hexapeptide from the Sponge Ircinia dendroides. J. Org. Chem. 1996, 61, 6302–6304. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Takei, M.; Shindo, K.; Kitazume, C.; Tanaka, J.; Higa, T.; Fukamachi, H. Cyclotheonamide E4 and E5, New Potent Tryptase Inhibitors from an Ircinia Species of Sponge. J. Nat. Prod. 2002, 65, 259–261. [Google Scholar] [CrossRef]
- Hooper, G.J.; Orjala, J.; Schatzman, R.C.; Gerwick, W.H. Carmabins A and B, New Lipopeptides from the Caribbean Cyanobacterium Lyngbya majuscula. J. Nat. Prod. 1998, 614, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Ebada, S.S.; Wray, V.; De Voogd, N.J.; Deng, Z.; Lin, W.; Proksch, P. Two New Jaspamide Derivatives from the Marine Sponge Jaspis splendens. Mar. Drugs 2009, 7, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Do, T.; Schmitz, F.J.; Andrusevich, V.; Engel, M.H. New Cyclic Peptides from the Ascidian Lissoclinum patella. J. Nat. Prod. 1998, 61, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, M.; Cruz, L.J.; Hunkapiller, M.W.; Gray, W.R.; Olivera, B.M. Isolation and structure of a peptide toxin from the marine snail Conus magus. Arch. Biochem. Biophys. 1982, 218, 329–334. [Google Scholar] [CrossRef]
- Pettit, G.R.; Kamano, Y.; Herald, C.L.; Tuinman, A.A.; Boettner, F.E.; Kizu, H.; Schmidt, J.M.; Baczynskyj, L.; Tomer, K.B.; Bontems, R.J. The isolation and structure of a remarkable marine animal antineoplastic constituent: Dolastatin 10. J. Am. Chem. Soc. 1987, 109, 6883–6885. [Google Scholar] [CrossRef]
- Laird, D.W.; LaBarbera, D.V.; Feng, X.D.; Bugni, T.S.; Harper, M.K.; Ireland, C.M. Halogenanted Cyclic Peptides Isolated from the Sponge Corticium sp. J. Nat. Prod. 2007, 70, 741–746. [Google Scholar] [CrossRef]
- Tarazona, G.; Fernández, R.; Cruz, P.G.; Pérez, M.; Rodríguez, J.; Jiménez, C.; Cuevas, C. Combining JBCA and Marfey’s methodology to determine the absolute configuration of threonines: The case of gunungamide A, a new cyclic depsideptide containing chloropyrrole from the sponge Discodermia sp. Org. Chem. Front. 2019, 6, 15–21. [Google Scholar] [CrossRef]
- Yasuda, T.; Araki, A.; Kubota, T.; Ito, J.; Mikami, Y.; Fromont, J.; Kobayashi, J. Bromopyrrole Alkaloids from Marine Sponges of the Genus Agelas. J. Nat. Prod. 2009, 72, 488–491. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Kitamura, K.; Nagai, K.; Nakao, Y.; Fusetani, N.; van Soest, R.W.M.; Matsunaga, S. Carteramine A, an inhibitor of neutrophil chemotaxis, from the marine sponge Stylissa carteri. Tetrahedron Lett. 2007, 48, 2127–2129. [Google Scholar] [CrossRef]
- Emrich, R.; Weyland, H.; Weber, K. 2,3,4-Tribromopyrrole from the marine Polychaete Polyphysia crassa. J. Nat. Prod. 1990, 53, 703–705. [Google Scholar] [CrossRef]
- Tebben, J.; Motti, C.; Tapiolas, D.; Thomas-Hall, P.; Harder, T. A Coralline Algal-Associated Bacterium, Pseudoalteromonas Strain J010, Yields Five New Korormicins and a Bromopyrrole. Mar. Drugs 2014, 12, 2802–2815. [Google Scholar] [CrossRef]
- Marfey, P. Determination of D-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitro-benzene. Carlsberg Res. Commun. 1984, 49, 591–596. [Google Scholar] [CrossRef]


| Pos | δC, Mult a | δH, Mult (J in Hz) a | δC, Mult b | δH, Mult (J in Hz) b | |
|---|---|---|---|---|---|
| Dap1 | 1 | 169.5, C | 172.3, C | ||
| 2 | 50.3, CH | 4.23 ddd (6.2, 6.2, 2.8) | 52.2, CH | 4.54 dd (5.6, 2.9) | |
| 3 | 49.2, CH2 | 2.87 d (13.9, 6.2) | 50.5, CH2 | 3.17 dd (14.6, 5.6) | |
| 3.13 d (13.9, 2.8) | 3.54 d (14.2) | ||||
| NH | 9.11 d (6.2) | 8.97 d (6.6)* | |||
| NH2 | |||||
| NMeClHis | 1 | 169.0, CO | 170.9, CO | ||
| 2 | 65.6, CH | 3.99 dd (10.6, 3.9) | 67.0, CH | 4.06 dd (10.1, 4.5) | |
| 3 | 24.9, CH2 | 3.05 m | 25.9, CH2 | 3.19 dd (15.2, 10.1) | |
| 3.30 dd (15.2, 4.5) | |||||
| 4 | 110.5, C | 135.6, C | |||
| 5 | 109.5, CH | 6.76 s | 120.8, CH | 6.96 s | |
| 6 | 128.2, C | 131.0, C | |||
| NMe | 39.6, CH3 | 2.84 s | 40.4, CH3 | 3.02 s | |
| iAsn | 1 | 173.5, C | 175.8, C | ||
| 2 | 35.0, CH2 | 2.80 dd (16.6, 2.7) | 36.2, CH2 | 3.02 m | |
| 3.09 dd (16.6, 5.8) | 3.27 m | ||||
| 3 | 48.4, CH | 4.58 ddd (8.0, 5.8, 2.7) | 50.3, CH | 4.84 m | |
| 4 | 172.2, CO | 173.8, CO | |||
| NH | 7.13 d (8.0) | 7.61 d (6.3) * | |||
| NH2 | |||||
| iSer | 1 | 171.9, C | 173.3, C | ||
| 2 | 67.8, CH | 4.13 dd (9.0, 4.2) | 69.5, CH | 4.40 dd (9.5, 4.0) | |
| 3 | 42.8, CH2 | 2.75 ddd (9.0, 9.4, 5.4) | 44.2, CH2 | 3.00 m | |
| 3.47 m | 3.75 dd (13.2, 4.0) | ||||
| NH | 8.23 t (5.4) | 8.25 s * | |||
| Dap2 | 1 | 170.5, C | 172.8, C | ||
| 2 | 51.5, CH | 4.49 ddd (9.2, 9.2, 6.3) | 53.4, CH | 4.84 m | |
| 3 | 40.1, CH2 | 3.22 m | 41.6, CH2 | 3.57 dd (13.8, 8.6) | |
| 4.04 ddd (12.9, 6.3, 6.3) | 4.22 dd (13.8, 5.4) | ||||
| NH-1 | 7.72 d (9.2) | 7.98 d (9.5) * | |||
| NH-2 | 7.20 t (6.3, 6.3) | 7.37 t (6.0) * | |||
| Br2Py | 1 | 158.6, CO | 161.4, CO | ||
| 2 | 118.0, C | 120.2, C | |||
| 3 | 110.4, CH | 6.30 d (2.7) | 112.0, CH | 6.15 s | |
| 4 | 96.9, C | 99.4, C | |||
| 5 | 123.2, C | 124.2, C | |||
| NH | 12.68 d (2.7) | 12.03 s |
| Pos | δH, Mult (J in Hz) a | δC, Mult a | δH, Mult (J in Hz) b | δC, Mult b | |
|---|---|---|---|---|---|
| Br3Py | 1 | - | 161.8, CO | - | 159.1, CO |
| 2 | - | 128.6, C | - | 128.5, C | |
| 3 | - | 102.7, C | - | 100.7, C | |
| 4 | - | 101.3, C | - | 93.4, C | |
| 5 | - | 110.8, C | - | 107.9, C | |
| NMe | 3.76, s | 36.7, CH3 | 3.61, s | 35.7, CH3 | |
| Ile | 1 | - | 174.3, CO | - | 171.4, CO |
| 2 | 4.84, m | 55.7, CH | 4.65, dd, 8.35, 8.5 | 53.7, CH | |
| 3 | 1.96, m | 38.0, CH | 1.87, m | 35.8, CH | |
| 4 | 1.73, m, 1.22, m | 25.9, CH2 | 1.57, m; 1.21, m | 24.2, CH2 | |
| 5 | 0.95, t, 7.4 | 11.2, CH3 | 0.83, t, 7.4 | 10.7, CH3 | |
| 6 | 0.99, d, 6.8 | 15.7, CH3 | 0.85, d, 6.9 | 15.1, CH3 | |
| NH | 8.24, d, 8.1 | - | 8.56, d, 8.2 | - | |
| NMeLeu | 1 | - | 171.8, CO | - | 168.8, CO |
| 2 | 5.51, dd, 10.3, 4.7 | 54.3, CH | 5.35, dd, 10.1, 4.3 | 51.7, CH | |
| 3 | 1.77, m; 1.61, m | 38.0, CH2 | 1.59, m; 1.42, m | 36.7, CH2 | |
| 4 | 1.56, m | 25.7, CH | 1.43, m | 23.9, CH | |
| 5 | 0.97, d, 6.2 | 23.6, CH3 | 0.87, d, 6.2 | 23.1, CH3 | |
| 6 | 0.93, d, 6.1 | 22.3, CH3 | 0.83, d, 6.2 | 21.8, CH3 | |
| NMe | 3.21, s | 31.8, CH3 | 3.04, s | 30.5, CH3 | |
| Pro | 1 | - | 174.5, CO | - | 171.4, CO |
| 2 | 4.41, m | 61.6, CH | 4.31, dd, 8.3, 4.1 | 59.1, CH | |
| 3 | 2.23, m; 2.00, m | 30.5, CH2 | 2.02, m; 1.80, m | 28.9, CH2 | |
| 4 | 2.08, m; 1.92, m | 26.0, CH2 | 1.90, m; 1.77, m | 24.4, CH2 | |
| 5 | 3.75, m; 3.69, m | 48.8, CH2 | 3.53, m; 3.50; m | 46.7, CH2 | |
| Gln | 1 | - | 174.7, CO2H | 12.53, brs | 173.3, CO2H |
| 2 | 4.41, m | 52.8, CH | 4.10, ddd, 8.6, 8.5, 5.2 | 51.5, CH | |
| 3 | 2.27, m; 1.92, m | 30.5, CH2 | 1.93, m; 1.74, m | 27.0, CH2 | |
| 4 | 2.41, m; 2.32, m | 32.6, CH2 | 2.15, m; 2.11, m | 31.3, CH2 | |
| 5 | - | 177.9, CONH2 | 7.19, s; 6.77, s | 173.5, CONH2 | |
| NH | - | - | 8.08, brs | - |
| Compound | Target | %Inhibition at 1 × 10−5 M |
|---|---|---|
| 1 | Top-I | 3 |
| 1 | PD-1 | 0.3 |
| 2 | PD-1 | −3.5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández, R.; Bayu, A.; Aryono Hadi, T.; Bueno, S.; Pérez, M.; Cuevas, C.; Yunovilsa Putra, M. Unique Polyhalogenated Peptides from the Marine Sponge Ircinia sp. Mar. Drugs 2020, 18, 396. https://doi.org/10.3390/md18080396
Fernández R, Bayu A, Aryono Hadi T, Bueno S, Pérez M, Cuevas C, Yunovilsa Putra M. Unique Polyhalogenated Peptides from the Marine Sponge Ircinia sp. Marine Drugs. 2020; 18(8):396. https://doi.org/10.3390/md18080396
Chicago/Turabian StyleFernández, Rogelio, Asep Bayu, Tri Aryono Hadi, Santiago Bueno, Marta Pérez, Carmen Cuevas, and Masteria Yunovilsa Putra. 2020. "Unique Polyhalogenated Peptides from the Marine Sponge Ircinia sp." Marine Drugs 18, no. 8: 396. https://doi.org/10.3390/md18080396
APA StyleFernández, R., Bayu, A., Aryono Hadi, T., Bueno, S., Pérez, M., Cuevas, C., & Yunovilsa Putra, M. (2020). Unique Polyhalogenated Peptides from the Marine Sponge Ircinia sp. Marine Drugs, 18(8), 396. https://doi.org/10.3390/md18080396






