Abstract
Chemical investigation on EtOAc extract of the deep-sea-derived fungus Trichobotrys effuse FS524 resulted in the isolation of six new highly substituted phenol derivatives, trieffusols A–F (1–6), along with ten known relative analogs (7–16). Their structures with absolute configurations were extensively characterized on the basis of spectroscopic data analyses, single-crystal X-ray diffraction experiments, and electronic circular dichroism (ECD) calculations. Structurally, trieffusols A and B shared an unprecedented ploy-substituted 9-phenyl-hexahydroxanthone skeleton with an intriguing 6-6/6/6 tetracyclic fused ring system, which is often encountered as significant moieties in the pharmaceutical drugs but rarely discovered in natural products. In the screening towards their anti-inflammatory activities of 1–6, trieffusols C and D exhibited moderate inhibitory activities against nitric oxide (NO) production in LPS-induced RAW 264.7 macrophages with IC50 values ranging from 51.9 to 55.9 μM.
1. Introduction
Marine-derived fungi have emerged as one of the most promising strategic resources to search pharmacologically significant leads for drug discovery and have aroused widespread attention from natural product chemists, pharmacologists, as well as biosynthetic chemists, due to their structurally abundant and diverse secondary metabolites in recent years [1,2]. Over the past decades, the research articles on marine natural products (MNPs) have surged dramatically, bringing about a lot of conspicuous natural products with novel chemical scaffolds and unique biological functional arrays [3,4,5,6,7]. In terms of pharmacological research, secondary metabolites derived from marine fungi are increasingly recognized as important sources of biologically meaningful natural products [8,9]. These MNPs have exhibited a wide range of biological activities such as anti-cancer [10], fungicidal [11], pro-angiogenic [12], anti-lymphangiogenic [13], and osteoclast differentiation inhibitory activities [14]. Therefore, in-depth chemical research on the MNPs would pave the way to providing potential model structures and precursor drugs for new drug developments.
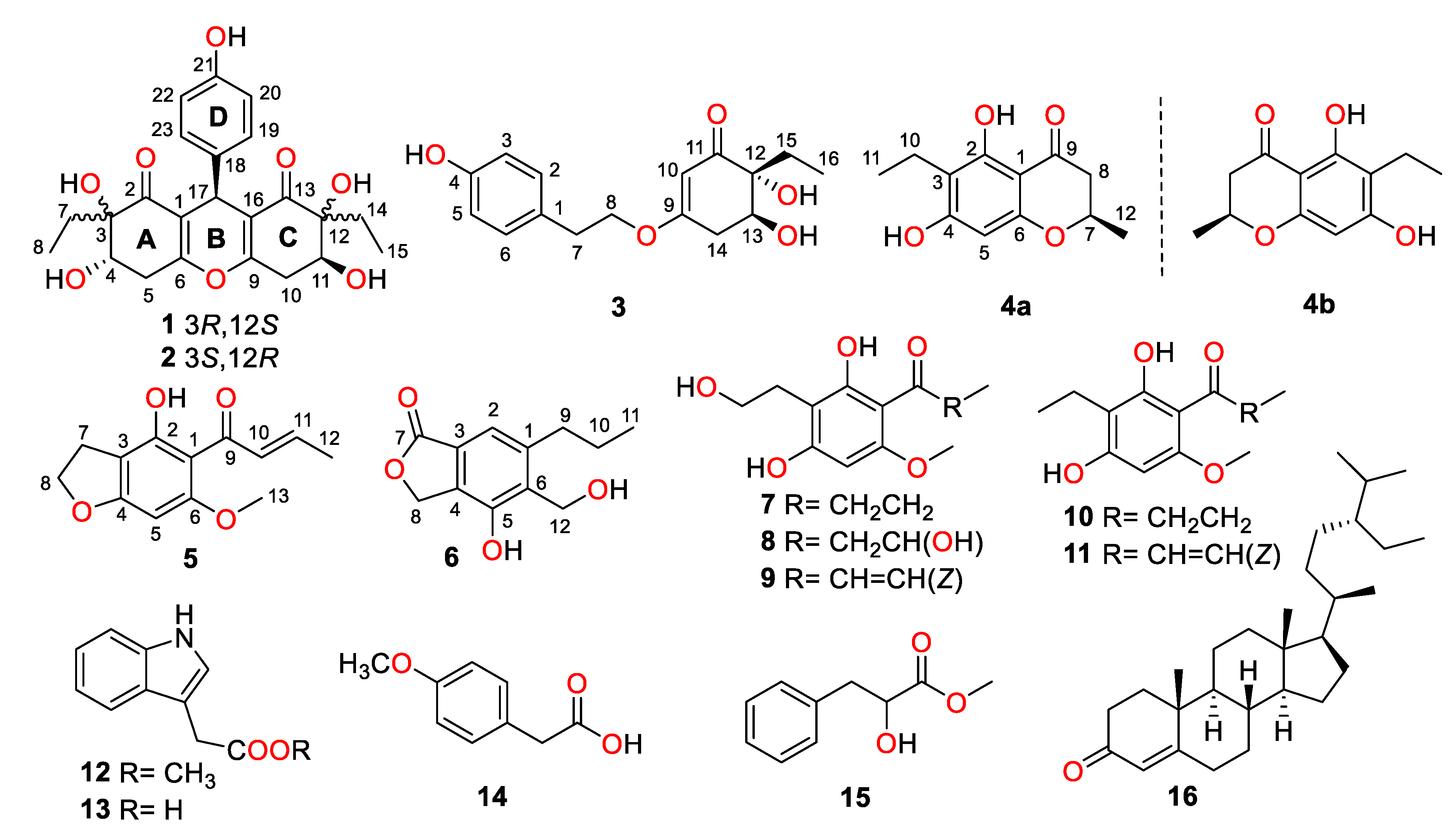
During our continuing research for structurally unique and biologically significant NPs from marine fungi [15,16,17], the fungus strain Trichobotrys effuse FS524, isolated from a sediment sample collecting at the South China Sea, attracted our attention, and chemical investigation of the strain resulted in the isolation of six new highly substituted phenol derivatives, trieffusols A–F (1–6), along with eleven known analogues (Figure 1), including phomalone (7) [18], 2,4-dihydroxy-3-(2-hydroxyethyl)-6-methoxyphenyl)-3-hydroxybutan-1-one (8) [18], (E)-1-(2,4-dihydroxy-3-(2-hydroxyethyl)-6-methoxyphenyl)but-2-en-1-one (9) [18], deoxyphomalone (10) [18], phomalichenone A (11) [18], methylindole-3-acetate (12) [19], 3-indole acetic acid (13) [20], 4-methoxyphenylacetic acid (14) [21], papuline (15) [22], and stigmast-4-en-3-one (16) [23]. Furthermore, their structures with absolute configurations were successfully established with the aid of spectroscopic data analyses, single-crystal X-ray diffraction experiments, and ECD calculations. Among them, trieffusols A and B share an intriguing 6-6/6/6 tetracyclic ring system with the formation of an unprecedented ploy-substituted 9-phenyl-hexahydroxanthone skeleton, which is often encountered as one of the most ubiquitous and intriguing functional moieties in the pharmaceutical drugs but rarely discovered in natural products. Herein, we present the isolation, structure elucidation, and biological evaluation of them in this study.
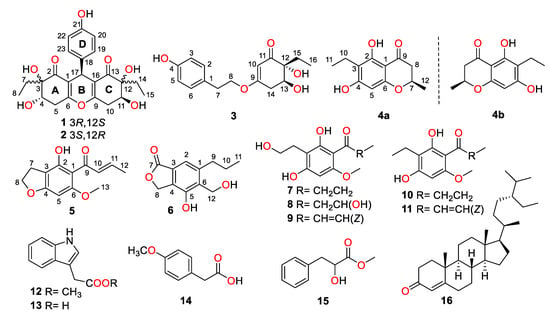
Figure 1.
Structures of compounds 1–16.
2. Results and Discussion
2.1. Structure Elucidation
Compound 1, a colorless crystal, was given the molecular formula as C23H26O8 determined by the HRESIMS cationic peak at m/z 431.1696 [M + H]+ (calcd 431.1700), which corresponded to eleven degrees of unsaturation. The IR spectrum of 1 logically revealed the presence of hydroxyl and carbonyl functional groups through the characteristic resonance absorptions at 3443 cm−1 and 1668 cm−1, respectively. The further inspection of its 1H NMR spectrum (Table 1) clarified the existence of a para-substituted benzene ring resonating at [δH 7.11 (2H, d, J = 8.6 Hz, H-19, 23), 6.63 (2H, d, J = 8.6 Hz, H-20, 22)], three methine moieties at [δH 4.03 (1H, m, H-4), 4.16 (1H, m, H-11), and 4.42 (1H, s, H-17)], along with two methyl groups at [δH 0.77 (3H, t, J = 7.6 Hz, H3-8) and 0.83 (3H, t, J = 7.4 Hz, H3-15)]. The 13C NMR spectrum combined with HSQC data of 1 resolved 23 carbon resonances attributable to two methyls, four methylenes, seven methines, and ten quaternary carbons containing two keto-carbonyl ones. The aforementioned aromatic ring and functionalities logically accounted for eight degrees of unsaturation, and the remaining three degrees of unsaturation necessitated that 1 should possess an additional tricyclic ring system.

Table 1.
1H (600 MHz) and 13C (150 MHz) NMR data for compounds 1 and 2 in CD3COCD3.
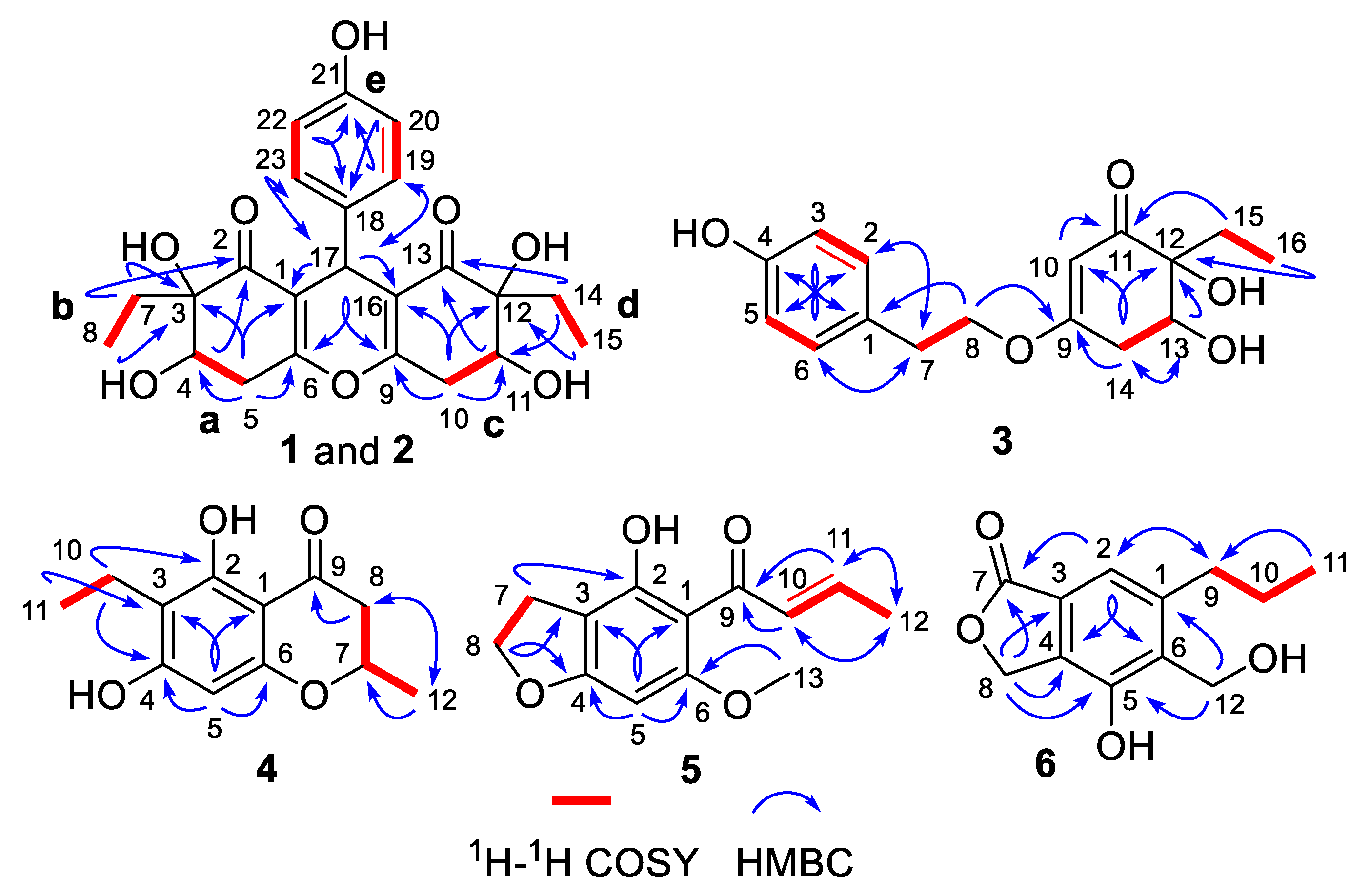
The chemo-logical construction of the planar structure for compound 1 featuring a tetracyclic 6-6/6/6 ring system was elucidated by the analysis of 2D NMR spectra (Figure 2). In the 1H–1H COSY spectrum, the cross-peaks of H-4/H2-5, H2-7/H3-8, H2-10/H-11, H2-14/H3-15, and H-19/23/H-20/22 suggested the presence of five independent fragments, a (C-4/C-5), b (C-7/C-8), c (C-10/C-11), d (C-14/C-15), and e (C-19/23/C-20/22). The HMBC correlations from H-19/23 to C-17 and C-21, H-20/22 to C-18 and C-21, coupled with the fragment e, could readily confirmed the existence of the para-substituted benzene ring (ring D). In addition, the obvious HMBC correlations from H-4 to C-2 and C-3, H2-5 to C-1, C-3, C-4, and C-6, H2-7 to C-2 and C-3, H3-8 to C-3 in conjunction with fragments a and b unambiguously concluded the presence of the cyclohexenone moiety (ring A).
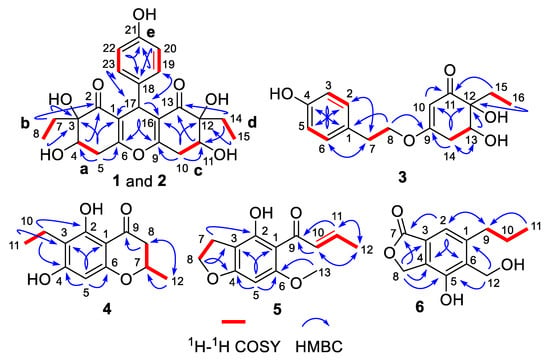
Figure 2.
1H–1H COSY and key HMBC correlations of 1–6.
Similarly, the establishment of the other cyclohexenone moiety (ring C) was confirmed by the key HMBC correlations from H2-10 to C-9, C-11, C-12, and C-16, H-11 to C-13, H2-14 to C-11 and C-13, H3-15 to C-12 as well as fragments c and d. Moreover, considering the remaining one degree of unsaturation and chemical shift of C-6 (δC 162.8) and C-9 (δC 161.8), we suspected that an oxygen atom should be connected between C-6 and C-9 with the formation of an oxygen bridge, which finally constructed the core pentasubstituted-4H-pyran skeleton (ring B). The aforementioned deduction was successfully reconfirmed by the informative HMBC correlations from H-17 to C-1, C-2, C-6, C-9, C-13, C-16, C-18, C-19, and C-23. Therefore, the planar structure of 1 was elucidated as a phenol–polyketone derivative consisting of an intriguing natural rarely-encountered 6-6/6/6 fused-ring system and given the trivial name “trieffusol A”.
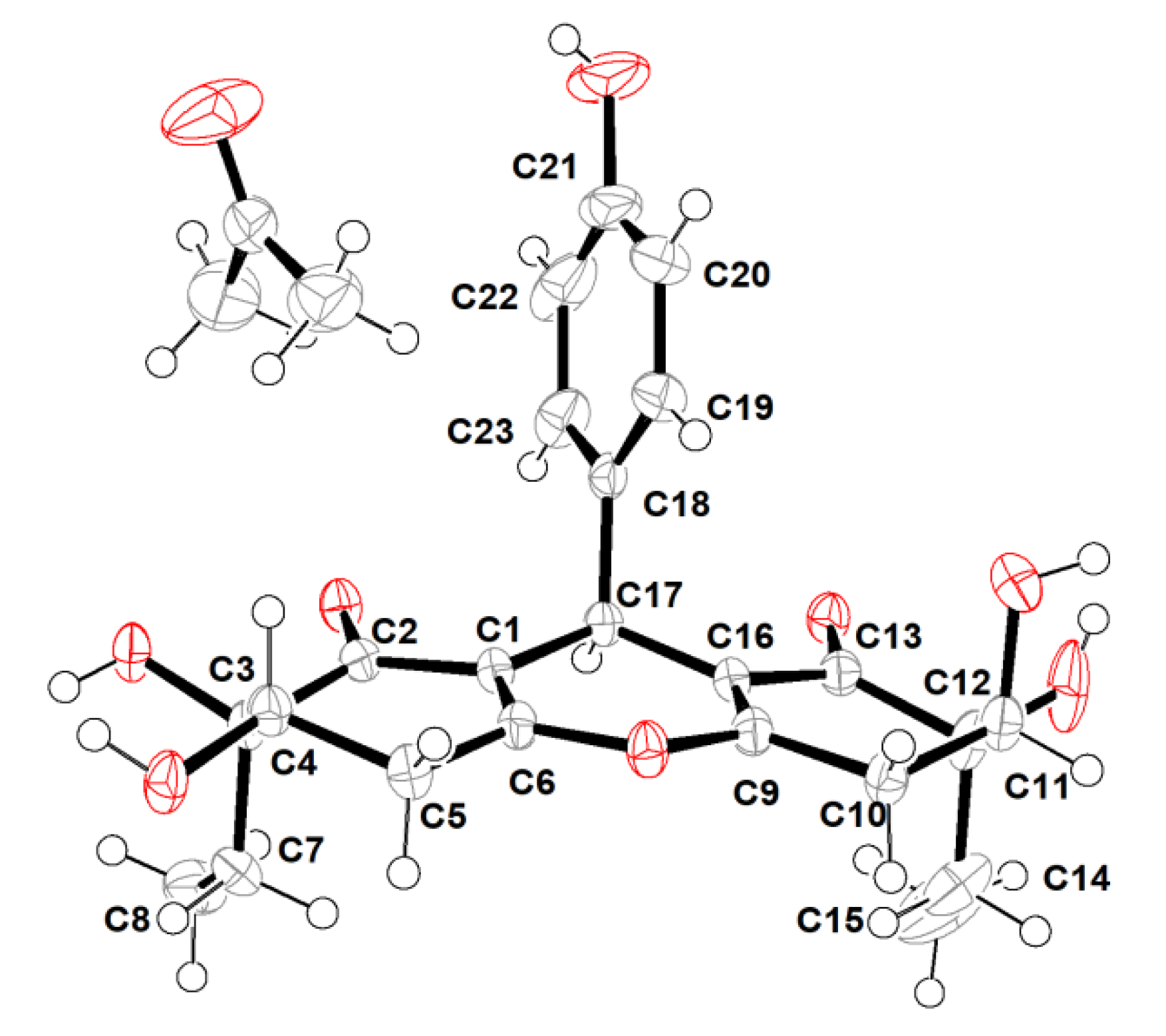
However, the high overlap of critical proton signals for H-4 and H-11, H2-7 and H2-14, H2-5, and H2-10, in conjunction with H3-8 and H3-15, made further construction of the relative configurations for the chiral genetic centers C-3 and C-4, as well as C-11 and C-12 in the cyclohexenone rings A and C, become a challenging issue. Moreover, all of these aforementioned carbons were far away from the central C-17 chiral genetic center, which would further give rise to two pairs of alternative diastereomeric configurations. Therefore, the assignment of the relative and absolute configurations of compound 1 through NMR and CD spectra seemed to be bleak. In order to corroborate the above structural deduction and establish absolute stereochemistry of 1, we attempted to get X-ray crystals in the methanol/water (30:1) system. Fortunately, the single crystals with good quality were obtained and subjected to an X-ray diffraction experiment with Cu Kα radiation. The crystal data (Figure 3) not only confirmed our deduction about the planar structure of 1 but also unambiguously established its absolute configuration as 3R,4S,11S,12S,17R. Therefore, the complete structure with the absolute configuration of compound 1 was finally established and given the trivial name “trieffusol A”, which possesses an unprecedented ploy-substituted 9-phenyl-hexahydroxanthone skeleton with an intriguing 6-6/6/6 tetracyclic fused ring system.
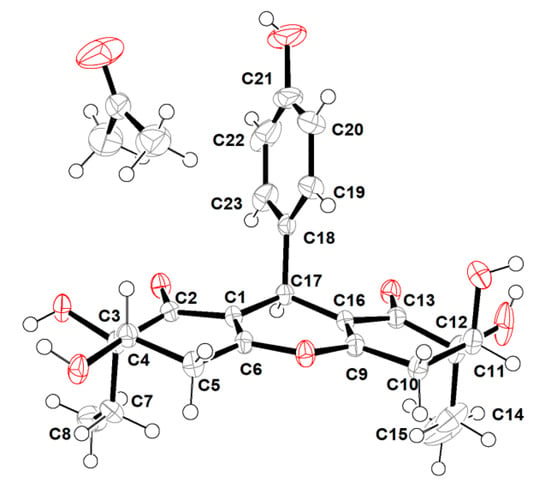
Figure 3.
X-ray crystallographic analysis of 1.
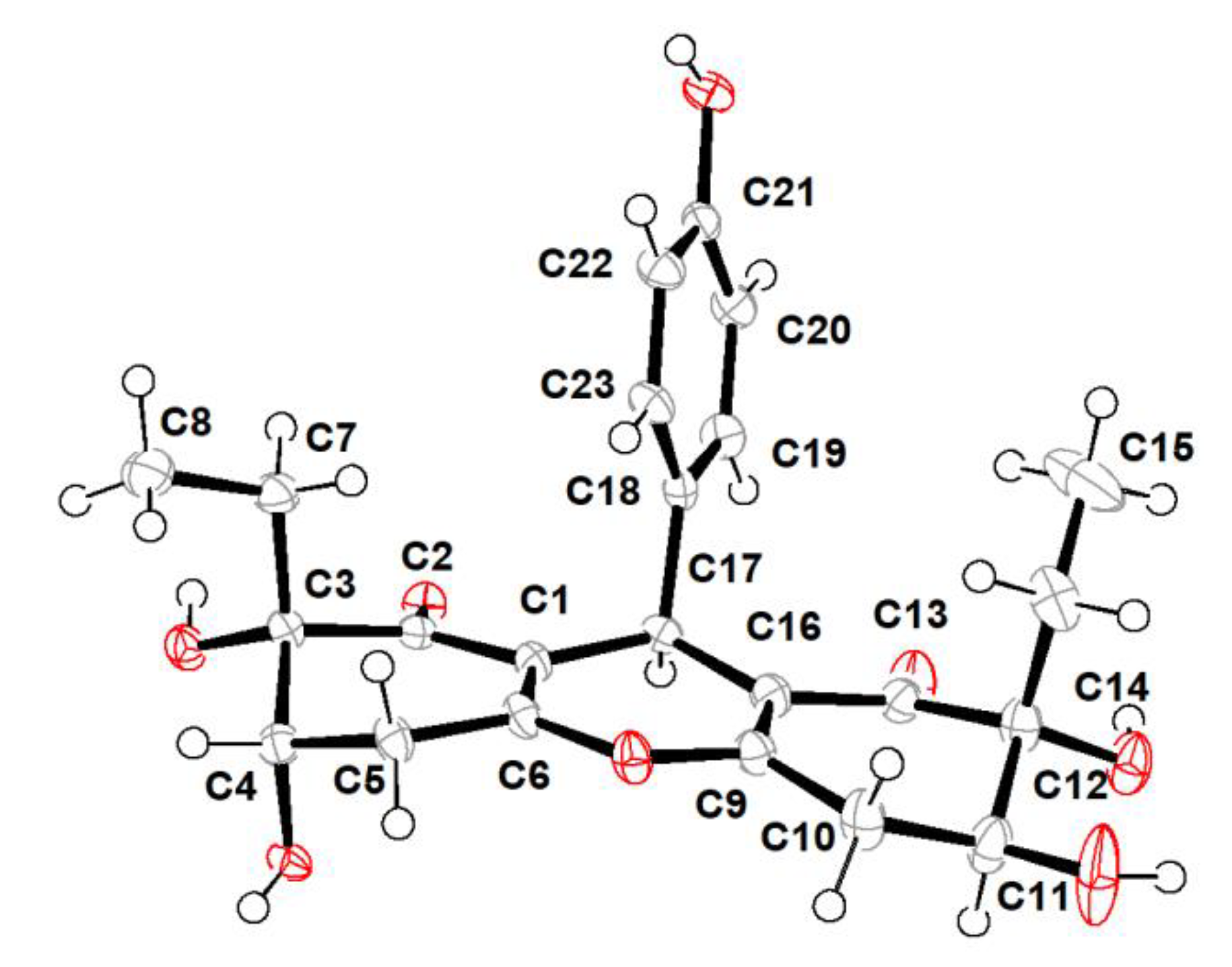
Trieffusol B was also obtained as a colorless crystal and had the same molecular formula C23H26O8 as that of 1 based on the negative mode HERSIMS (m/z 429.1554 [M − H]–, calcd 429.1555), indicating the presence of eleven degrees of hydrogen deficiency. Its 1H and 13C NMR data closely resembled those of 1, except for chemical shift changes at C-4, C-7, C-8, C-11, C-14, and C-15. A comprehensive analysis of the 1D and 2D NMR data deduced that compounds 1 and 2 share the same planar structures, indicating that these two compounds should be a pair of diastereoisomers sharing the same ploy-substituted 9-phenyl-hexahydroxanthone skeleton with an intriguing 6-6/6/6 tetracyclic fused ring system. The aforementioned deduction was further substantiated by the X-ray single-crystallographic analysis (Figure 4), which finally clarified the absolute configuration of compound 2 as 3S,4S,11S,12R,17S.
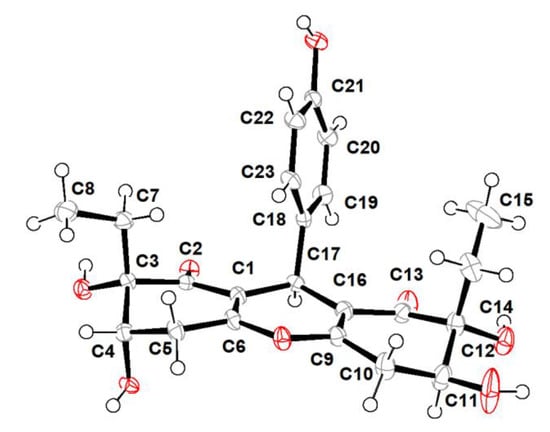
Figure 4.
X-ray crystallographic analysis of 2.
Trieffusol C was purified as a brown oil. Its molecular formula was determined as C16H20O5 based on the protonated molecule peak at m/z 293.1385 [M + H]+ (calcd 293.1384) by HRESIMS, which requires seven degrees of unsaturation. The 1H NMR data of 3 (Table 2) showed various characteristic resonances responsive for a para-substituted aromatic ring at [δH 7.07 (2H, d, J = 8.5 Hz, H-2, 6), 6.72 (2H, d, J = 8.5 Hz, H-3, 5)], one trisubstituted olefinic bond at δH 5.34 (1H, s, H-10), one oxygenated methine moiety at δH 3.98 (1H, dd, J = 9.9, 5.9 Hz, H-13), one oxygenated methylene at δH 4.07 (2H, td, J = 6.8, 1.6 Hz, H2-8), together with a methyl functionality at δH 0.81 (3H, t, J = 7.5 Hz, H3-16). Analysis of the 13C NMR and HSQC data suggested the presences of 16 carbons, comprising one carbonyl carbon (δC 202.1), four nonprotonated carbons (δC 80.0, 129.7, 157.2, 176.5), five olefinic carbons (δC 100.8, 116.3, 116.3, 131.0, 131.0), one methine carbon (δC 72.6), four methylene carbons (δC 24.1, 35.1, 36.8, 71.5), together with one methyl carbon (δC 7.2).

Table 2.
1H (600 MHz) and 13C (150 MHz) NMR data for compounds 3 and 4 in CD3OD.
Construction of the planar structure for 3 was accomplished by analysis of its 2D NMR data. Firstly, the presence of a para-substituted phenyl moiety was confirmed by the HMBC correlations from H-3/5 to C-1 and C-4, H-2/6 to C-1 and C-4, together with 1H-1H COSY correlations of H-2/6/H-3/5. Secondly, the HMBC correlations from H-10 to C-9, C-11, C-12, and C-14, H-13 to C-12 and C-15, H2-14 to C-9, C-10, C-12, and C-13, H2-15 to C-11 and C-12, H3-16 to C-12 and C-15, as well as 1H-1H COSY correlations of H-13/H2-14 and H2-15/H3-16 strongly indicated the existence of the trisubstituted cyclohex-2-en-1-one moiety. Finally, the connections of the two independent phenyl and cyclohex-2-en-1-one fragments through the linkage of C-1/C-7/C-8/O/C-9 were supported by the HMBC correlations from H-2/6 to C-7, H2-7 to C-1, C-2, and C-6, H2-8 to C-1 and C-9 as well as 1H-1H COSY fragment H2-7/H2-8. Hence, the planar structure of 3 was successfully constructed, as shown in Figure 1.
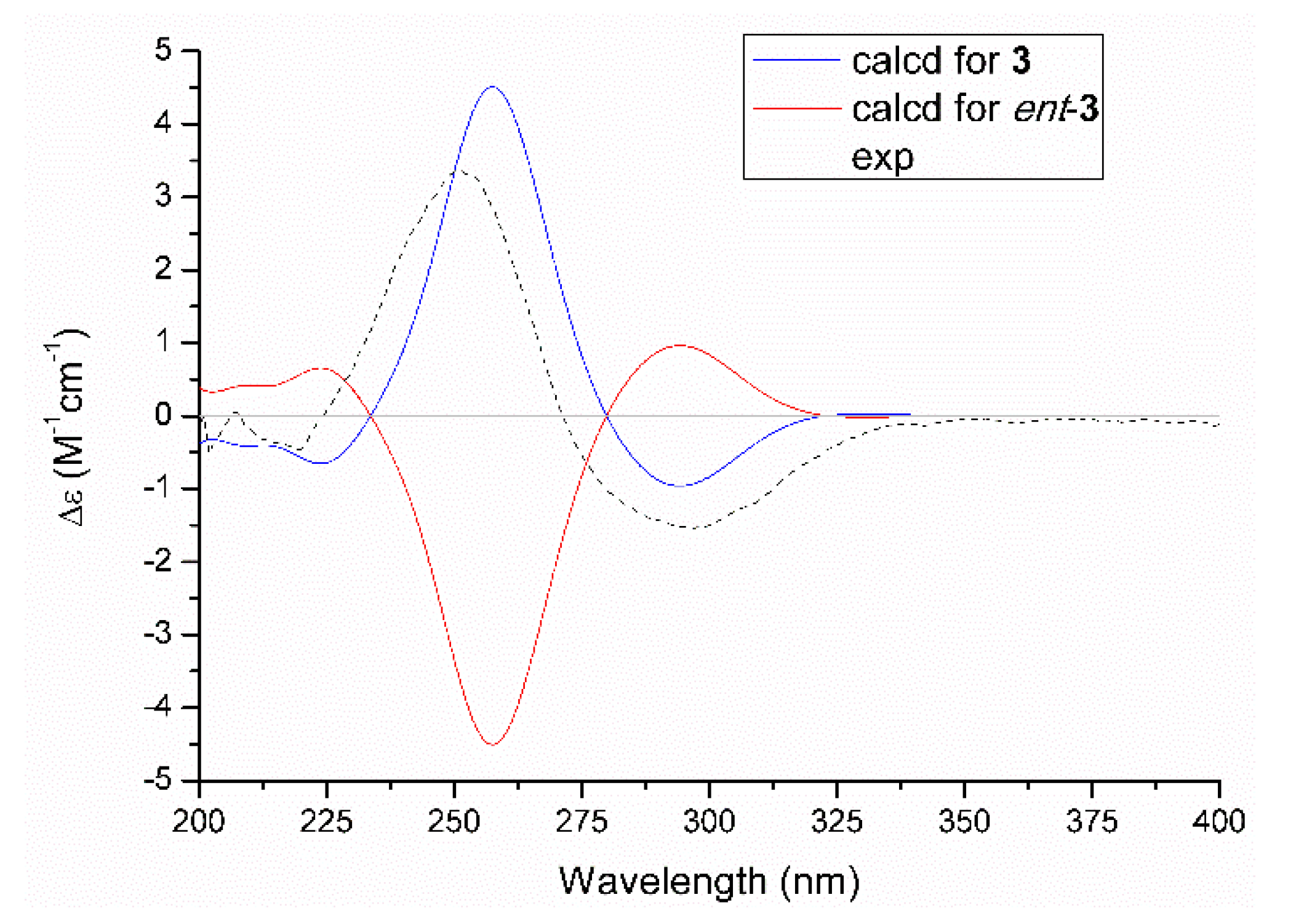
The relative configuration of 3 was assessed by the NOESY correlation, and the absence of the critical NOE correlation of H-13/H2-15 tentatively suggested that these two protons should orientate oppositely. The absolute stereochemistry for chiral genetic centers of C-12 and C-13 in compound 3 was determined on the basis of the comparison of experimental and the quantum mechanically calculated electric circular dichroism (ECD) data by using the time-dependent density functional theory (TDDFT) at the B3LYP/6-31+G (d,p) level in MeOH. Satisfactorily, the calculated ECD spectrum of 12R,13S-3 (Figure 5) matched well with that of the experimental one, with a positive Cotton effect at 255 nm and a negative one at 300 nm, respectively, which unambiguously clarified the absolute configuration of 3 to be 12R,13S.
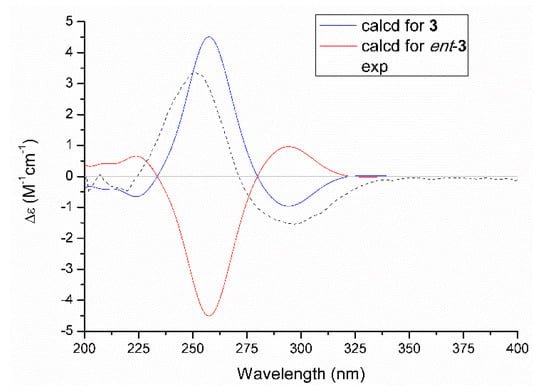
Figure 5.
Experimental and calculated ECD spectra of 3.
Trieffusol D was obtained as a white powder with the molecular formula of C12H14O4, as deduced by HRESIMS data (m/z 223.0966 [M + H]+, calcd 223.0965). The 1H NMR data of 4 (Table 2) displayed one aromatic proton [δH 5.89 (1H, s, H-5)], one methine [δH 4.47 (1H, dqd, J = 12.3, 6.3, 3.2 Hz, H-7)], one methylene [δH 2.53 (2H, q, J = 7.4 Hz, H2-10)], and two methyls [δH 1.04 (3H, t, J = 7.4 Hz, H3-11), 1.43 (3H, d, J = 6.3 Hz, H3-12)]. Analysis of the 13C NMR spectrum of 4 in association with the aid of HSQC spectrum, helped to unlock 12 carbon resonances attributable to one conjugated carbonyl functional group (δC 198.0), six aromatic or olefinic carbons (δC 95.2, 103.1, 111.6, 162.4, 162.5, 165.8), one oxygenated methine (δC 75.2), two methylenes (δC 16.0, 44.2), and two methyls (δC 13.8, 21.0).
The 1H-1H COSY correlations of H2-10/H3-11 and the HMBC correlations from H-5 to C-1, C-3, C-4, and C-6, H2-10 to C-2, C-3, and C-4, H3-11 to C-3 and C-10 led to the establishment of the pentasubstituted benzene ring. Besides, the sequential HMBC correlations from H2-8 to C-7, C-9, and C-12, H3-12 to C-7 and C-8 along with the 1H-1H COSY correlations of H2-8/H-7/H3-12 suggested the existence of a tetrasubstituted tetrahydro-4H-pyran-4-one scaffold. Therefore, the gross structure of trieffusol D was established undoubtedly.
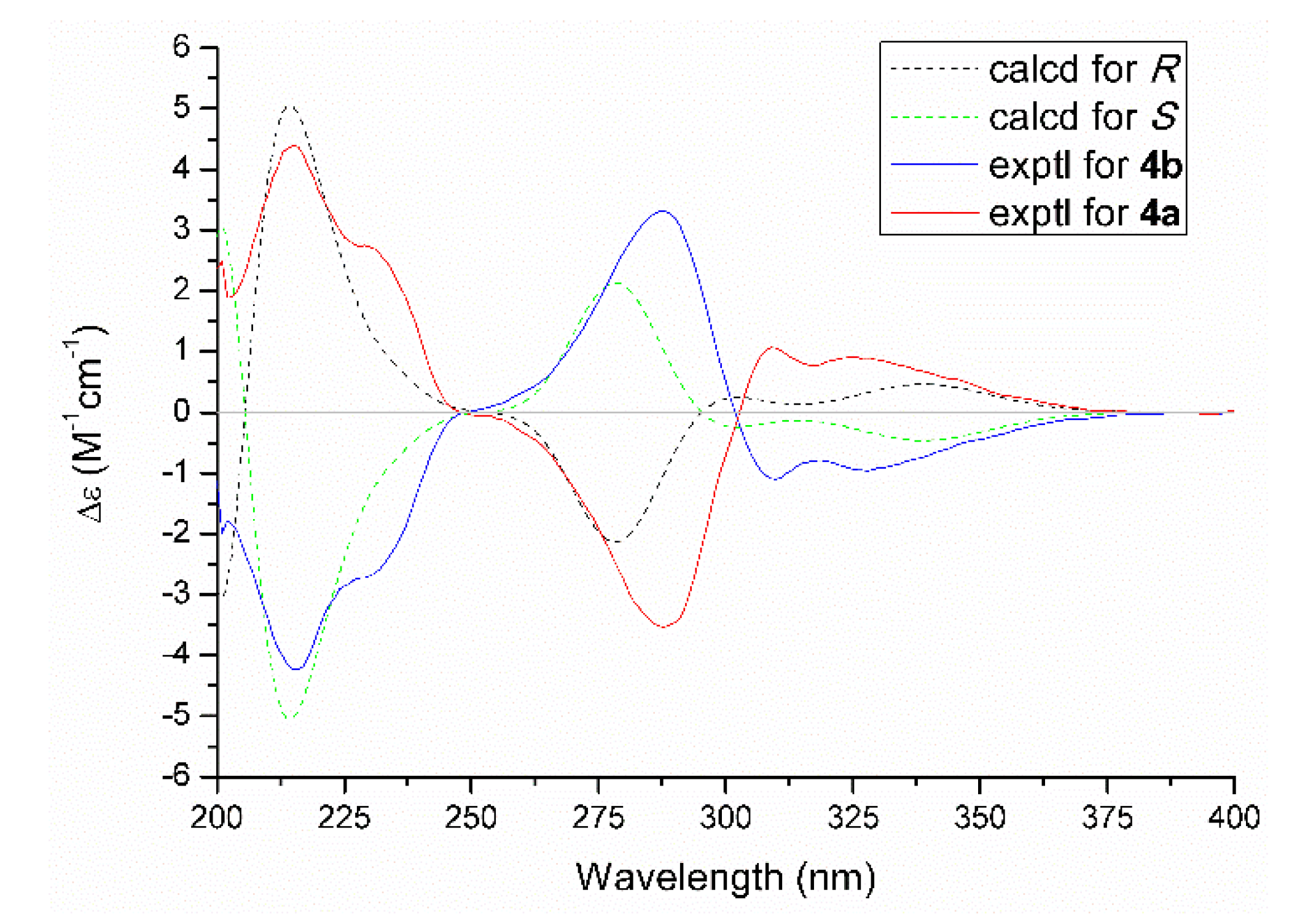
As for the absolute configuration, the specific optical rotation of 4 was close to zero, which logically suggested that it might exist as a pair of enantiomers. The further chiral-phase separation via chiral HPLC yielded two optically pure enantiomers 4a and 4b, respectively. Subsequently, the theoretical ECD spectra for 4a and 4b (Figure 6) were calculated by using the time-dependent density functional theory (TDDFT) at the B3LYP/6-31+G (d,p) level in MeOH. As a result, the calculated ECD spectra matched well with those of the experimental ones. Therefore, the absolute configurations of 4a and 4b were finally assigned as R and S, respectively.
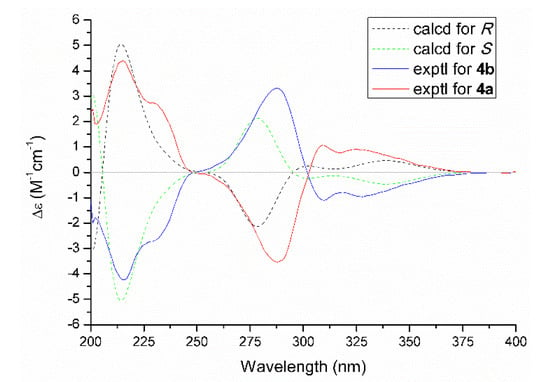
Figure 6.
Experimental and calculated ECD spectra of 4a and 4b.
Trieffusol E was afforded as a colorless powder and possessed a molecular formula of C13H14O4 based on the HRESIMS ion peak at m/z 235.0971 [M + H]+ (calcd 235.0965). Analysis of the 1D (Table 3) and 2D NMR data of 5 showed great similarity to these of 4-hydroxy-6-methoxy-5-(1′-oxobutyl)benzo[b]dihydrofuran [18]. The main difference was that the two methylenes at C-10 and C-11 positions in the known compound were oxidized to be a disubstituted double bond in 5. This conclusion was further verified by the key HMBC correlations from H-10 to C-9 and C-12, H-11 to C-9 and C-11, H3-12 to C-10 and C-11, together with the 1H–1H COSY correlations of H-10/H-11/H3-12. Besides, the other structural identification details, as shown in Figure 2, could also support this conclusion. Moreover, the coupling constant (J = 15.1 Hz) between H-10 and H-11 obviously illustrated the configuration of the disubstituted double bond as an E-configuration. Therefore, the structure of 5 was elucidated unambiguously, as depicted in Figure 1.

Table 3.
1H (600 MHz) and 13C (150 MHz) NMR data for compounds 5 and 6 in CD3OD.
Trieffusol F was isolated as a brownish solid and determined to have a molecular formula as C12H14O4 from the HRESIMS data (m/z 223.0971 [M + H]+, calcd 223.0965). The 1H NMR data (Table 3) showed characteristic resonances for one aromatic proton [δH 7.21 (1H, s, H-2)], two oxygenated methylenes [δH 4.89 (2H, s, H2-12), 5.27 (2H, s, H2-8)], and one methyl [δH 1.00 (3H, t, J = 7.3 Hz, H3-11)]. The 13C NMR data and HSQC spectrum of 6 showed 12 carbons, which were assigned to one methyl, four methylenes (two oxygenated ones), one aromatic carbon, five quaternary carbons (one oxygenated one), and one ester carbonyl functionality. The key HMBC correlations from H-2 to C-4, C-6, and C-7, H2-8 to C-3, C-4, C-5, and C-7 indicated the establishment of the isobenzofuran-1(3H)-one moiety. Furthermore, the attachments of the C-9 at C-1 and C-12 at C-6 were confirmed by the HMBC correlations from H2-9 to C-1, C-2, and C-6, H-2 to C-9, H2-12 to C-1, C-5, and C-6, along with the 1H-1H COSY correlations of H2-9/H2-10/H3-11. Thus, the structure of 6 was defined, as shown in Figure 1.
2.2. Biological Activity
Compounds 1–6 were evaluated for their inhibition effect of NO production in the lipopolysaccharide (LPS)-induced mouse macrophages. As shown in Table 4, compounds 3 and 4 exhibited the inhibitory activities with IC50 values ranging from 51.9 to 55.9 μM, comparable to that of the positive control aminoguanidine (IC50: 24.8 μM). At the same time, both of them showed no cytotoxicities against macrophages, of which IC50 values were all greater than 200 μM.

Table 4.
Inhibitory effects of 1-6 on the NO production and the cytotoxicity.
3. Materials and Methods
3.1. General Experimental Procedures
HRESIMS data were collected on an MAT95XP machine (Thermo Fisher Scientific, Bremen, Germany). NMR spectra were acquired by an Avance-600 spectrometer (Bruker, Fällanden, Switzerland). Circular dichroism (CD) spectra were afforded by a Jasco 820 spectropolarimeter (Jasco Corporation, Kyoto, Japan). Optical rotations were obtained by an MCP-500 spectropolarimeter (Anton Paar, Graz, Austria). UV spectra were acquired using a UV-2600 spectrophotometer (Shimadzu, Kyoto, Japan). IR data were done with an Affinity-1 spectrometer (Shimadzu, Kyoto, Japan). Preparative HPLC was performed using an ODS-A column (250 × 20 mm, 5 μm, 12 nm, YMC Co., Ltd, Kyoto, Japan). An ODS-A/AQ column (250 × 10 mm, 5 μm, 12 nm, YMC CO., Ltd, Kyoto, Japan) was used for semipreparative HPLC separation and the CHIRALPAK IC column (250 × 10 mm, 5 μm) for chiral semipreparative HPLC separation. Silica gel (100–200 and 200-300 mesh, Qingdao Marine Chemical Inc., Qingdao, China), C18 reversed-phase silica gel (40–63 μm, Merck, Darmstadt, Germany), and Sephadex LH-20 gel (Pharmacia Fine Chemical Co. Ltd., Uppsala, Sweden) were used in the chromatography processes. Fractions were monitored by TLC, and spots were detected on heated TLC plates (silica gel GF254 plates, Qingdao Marine Chemical Inc., Qingdao, China) with 10% H2SO4 in EtOH under UV light.
3.2. Fungal Material
The strain FS524 used in this work was isolated from a sediment sample, which was collected at the depth of 1428 m in the South China Sea (110°59′04′′E, 18°00′47′′N) in June 2017. The sequence data for this strain have been submitted to the GenBank under accession no. MN545626. By using BLAST (nucleotide sequence comparison program) to search the GenBank database, FS524 has 99.8% similarity to Trichobotrys effuse DFFSCS021 (accession no. JX156367). The strain was preserved at the Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Institute of Microbiology.
3.3. Fermentation and Extraction
The marine fungus T. effuse FS524 was cultured on potato dextrose agar (PDA) at 28 °C for 7 days to prepare the seed culture, and then inoculated into flasks (3 L) each containing 9 g sea salts, 250 g of rice, and 300 mL of water. After that, all flasks were incubated at 28 °C for one month and extracted repeatedly with EtOAc. After evaporation of the solvent, a dark brown solid extract (67.3 g) was obtained. The crude extract was fractionated by silica gel column chromatography (100–200 mesh) with two gradient systems of increasing polarity (petroleum ether–EtOAc, 30:1→1:1; CH2Cl2/CH3OH, 10:1→1:1) to furnish nine fractions (A–I).
Fraction C (14.8 g) was subjected to silica gel CC (petroleum ether/EtOAc, 30:1→1:1) to afford seven subfractions (C1–C7). C2 was further divided into two parts (C2.1, C2.2) by Sephadex LH-20 CC (CH2Cl2–MeOH, 1:1). C2.2 was purified by semi-preparative HPLC equipped with a chiral column (isopropanol–hexane, 70:30, 2 mL/min) to yield 7 (4.1 mg, tR = 15.8 min). C3 was separated by Sephadex LH-20 CC (CH2Cl2–MeOH, 1:1) to afford three subfractions (C3.1–C3.3). Then, semi-preparative HPLC equipped with a chiral column (isopropanol-hexane, 80:20, 2 mL/min) analysis of C3.2 afforded 9 (7.8 mg, tR = 21.3 min) and 8 (3.4 mg, tR = 30.1 min), respectively. Additionally, C4 was purified by semi-preparative HPLC equipped with a chiral column (isopropanol–hexane, 50:50, 2 mL/min) to obtain 5 (1.3 mg, tR = 21.9 min) and 16 (5.4 mg, tR = 19.5 min), respectively.
Fraction E (3.9 g) was divided into five subfractions (E1–E5) by Sephadex LH-20 CC (CH2Cl2/MeOH, 1:1). E5 was separated by semi-preparative HPLC (MeCN–H2O, 80:20, 2 mL/min) to yield 4 (3.1 mg, tR = 9.2 min), 11 (4.7 mg, tR = 11.0 min), and 10 (138.2 mg, tR = 12.5 min). Furthermore, 4 was purified by semi-preparative HPLC equipped with a CHIRALPAK IC column (250 × 10 mm, 5 μm) (isopropanol–hexane, 20:80, 2 mL/min) to yield 4b (1.5 mg, tR = 12.0 min) and 4a (1.4 mg, tR = 14.5 min), respectively.
Fraction G (11.1 g) was subjected to C-18 reversed-phase silica gel CC (gradient elution with MeOH–H2O, 30:70→100:0) to afford seven subfractions (G1–G7). G1 was separated by silica gel CC (petroleum ether-EtOAc, 8:1→1:1) to afford five subfractions (G1.1–G1.5). Then semi-preparative HPLC (MeCN–H2O, 30:70, 2 mL/min) analysis of G1.4 afforded 3 (2.5 mg, tR = 19.0 min). G1.5 was separated by semi-preparative HPLC (MeCN–H2O, 25:75, 2 mL/min) to get G1.5.2 (99.8 mg, tR = 8.0 min) and G1.5.4 (20.5 mg, tR = 13.8 min). G1.5.4 was further purified by semi-preparative HPLC equipped with a chiral column (isopropanol–hexane, 60:40, 2 mL/min) to yield 1 (10.8 mg, tR = 14.4 min). Additionally, semi-preparative HPLC equipped with a chiral column (isopropanol–hexane, 45:55, 2 mL/min) analysis of G1.5.2 afforded 2 (2.3 mg, tR = 13.2 min). G3 was divided into two subfractions (G3.1, G3.2) by Sephadex LH-20 CC (CH2Cl2–MeOH, 1:1). Then G3.1 was separated into five subfractions (G3.1.1–G3.1.5) by semi-preparative HPLC (MeCN–H2O, 40:60, 2 mL/min). G3.1.5 was purified by semi-preparative HPLC (MeCN–H2O, 45:55, 2 mL/min) and semi-preparative HPLC equipped with a chiral column (isopropanol–hexane, 60:40, 2 mL/min) to obtain 12 (8.5 mg, tR = 10.0 min). Further semi-preparative HPLC equipped with a chiral column (isopropanol–hexane, 35:65, 2 mL/min) analysis of G3.1.2 and G3.1.4 afforded 13 (15.2 mg, tR = 8.9 min), 14 (5.6 mg, tR = 11.2 min) and 15 (19.2 mg, tR = 13.6 min), respectively. G4 was divided into eight subfractions (G4.1–G4.8) by preparative HPLC (MeOH–H2O, 65:35, 5 mL/min). Additionally, semi-preparative HPLC equipped with a chiral column (isopropanol–hexane, 30:70, 2 mL/min) and further purified by semi-preparative HPLC (MeCN–H2O, 55:45, 2 mL/min) analysis of G4.5 afforded 6 (4.3 mg, tR = 10.6 min).
Trieffusol A (1): mp 149–150 °C; colorless crystal; [α] +14.5 (c 0.12, MeOH). CD (0.35 mg/mL, MeOH): 226 (2.99), 281 (1.09), 325 (−1.38) nm. UV (MeOH) λmax (log ε): 225 (4.12), 288 (3.69) nm. IR νmax: 3443, 2359, 1668, 1362, 1157 cm−1. 1H (600 MHz) and 13C (150 MHz) NMR spectral data, see Table 1. HRESIMS: m/z 431.1696 [M + H]+ (calcd for C23H27O8, 431.1700).
Trieffusol B (2): mp 148–149 °C; colorless crystal; [α] +33.6 (c 0.09, MeOH). CD (0.37 mg/mL, MeOH): 224 (2.57), 293 (0.39), 328 (−0.34) nm. UV (MeOH) λmax (log ε): 224 (4.03), 293 (3.58) nm. IR data was the same as 1. 1H (600 MHz) and 13C (150 MHz) NMR spectral data, see Table 1. HRESIMS: m/z 429.1554 [M − H]− (calcd for C23H25O8, 429.1555).
Trieffusol C (3): brown oil; [α] +8.1 (c 0.12, MeOH). CD (0.39 mg/mL, MeOH): 251 (3.38), 297 (−1.53) nm. UV (MeOH) λmax (log ε): 225 (3.93), 250 (3.97) nm. IR νmax: 3389, 1595, 1516, 1221, 1153, 831 cm−1. 1H (600 MHz) and 13C (150 MHz) NMR spectral data, see Table 2. HRESIMS: m/z 293.1385 [M + H]+ (calcd for C16H21O5, 293.1384).
(+)-Trieffusol D (4a): white powder; [α] +20.9 (c 0.09, MeOH). CD (0.33 mg/mL, MeOH): 215 (4.39), 288 (−3.53), 309 (1.07) nm. UV (MeOH) λmax (log ε): 213 (4.03), 292 (3.90) nm. IR νmax: 2930, 1585, 1306, 1119, 716 cm−1. 1H (600 MHz) and 13C (150 MHz) NMR spectral data, see Table 2. HRESIMS: m/z 223.0966 [M + H]+ (calcd for C12H15O4, 223.0965).
(−)-Trieffusol D (4b): white powder; [α] −24.7 (c 0.07, MeOH). CD (0.34 mg/mL, MeOH): 216 (−4.24), 287 (3.31), 310 (−1.10) nm. UV (MeOH) λmax (log ε): 213 (3.98), 292 (3.88) nm. IR data was the same as 4a. 1H (600 MHz) and 13C (150 MHz) NMR spectral data, see Table 2. HRESIMS: m/z 221.0823 [M − H]− (calcd for C12H13O4, 221.0819).
Trieffusol E (5): colorless powder; UV (MeOH) λmax (log ε): 207 (3.88), 241 (3.59), 324 (3.62) nm. IR νmax: 3362, 1418, 1020, 642 cm−1. 1H (600 MHz) and 13C (150 MHz) NMR spectral data, see Table 3. HRESIMS: m/z 235.0971 [M + H]+ (calcd for C13H15O4, 235.0965).
Trieffusol F (6): brownish solid; UV (MeOH) λmax (log ε): 208 (3.74), 250 (3.19), 301 (2.81) nm. IR νmax: 3310, 1748, 1522, 1009, 773 cm−1. 1H (600 MHz) and 13C (150 MHz) NMR spectral data, see Table 3. HRESIMS: m/z 223.0971 [M + H]+ (calcd for C12H15O4, 223.0965).
3.4. X-ray Crytallographic Data of Compounds 1 and 2
The single-crystal X-ray diffraction data for compounds 1 and 2 were collected on an Agilent Xcalibur Nova single-crystal diffractometer using CuKα radiation at 293 and 100 K, respectively. The crystal structures were refined by full-matrix least-squares calculation (for details see X-ray crystallographic analysis, Tables S1 and S2 in the supporting information). Crystallographic data have been deposited at the Cambridge Crystallographic Data Center with the deposition number of CDCC 1974673 for 1 and CDCC 1974674 for 2, respectively. Copies of these data can be obtained free of charge via www.ccdc.cam.au.ck/conts/retrieving.html.
3.5. Quantum Chemical Calculations
Merck molecular force field (MMFF) and DFT/TD-DFT calculations were carried out with the Spartan’14 software (Wavefunction Inc., Irvine, CA, USA) and the Gaussian 09 program, respectively [24]. Conformers within the 10 kcal mol−1 energy window were generated and optimized using DFT calculations at the B3LYP/6-31+G (d,p) level. Frequency calculations were performed at the same level to confirm that each optimized conformer was true minimum and to estimate their relative thermal free energy (ΔG) at 298.15 K. Conformers with the Boltzmann distribution over 5% were chosen for ECD calculations in methanol at the B3LYP/6-311+G (d,p) level. Solvent effects were taken into consideration using the self-consistent reaction field (SCRF) method with the polarizable continuum model (PCM) [25]. Details of the individual conformers are provided in the supporting information. The ECD spectrum was generated by the SpecDis program [26] using a Gaussian band shape with 0.26 eV exponential half-width from dipole-length dipolar and rotational strengths.
3.6. Nitric Oxide Inhibitory Activities Assay
Compounds 1–6 were evaluated for the inhibitory activity of nitric oxide (NO) production in lipopolysaccharide (LPS)-induced RAW 246.7 mouse macrophages [27]. The cells (180 μL) with a density of 5 × 105 cells/mL of media on a 96-well plate were put under 37 °C at a 5% CO2 condition. After a 24 h preincubation, the seeded cells were treated with gradient dilutions of 1-6 with a maximum concentration of 100 μM, followed by stimulation with LPS (1 μg/mL) for 24 h. Then 50 μL cell culture supernatant solution was moved to a new plate that contained NO detection Griess A (50 μL) and Griess B (50 μL). Finally, the absorbance was measured at 540 nm. Aminoguanidine was used as a positive control and all data were obtained in triplicate. The viability of RAW264.7 cells was evaluated according to the SRB method simultaneously to exclude the interference of the cytotoxicity of 1–6. The RAW264.7 cells were purchased from the Cell Bank of the Chinese Academy of Sciences.
4. Conclusions
In summary, six new highly substituted phenol derivatives, trieffusols A–F (1–6), along with eleven known analogs (7–16), were identified from the deep-sea-derived fungus Trichobotrys effuse FS524. Interestingly, trieffusols A and B share an intriguing 6-6/6/6 tetracyclic ring system with the formation of an unprecedented ploy-substituted 9-phenyl-hexahydroxanthone skeleton, which is often encountered as one of the most ubiquitous and intriguing functional moieties in the pharmaceutical drugs but rarely discovered in natural products. All the compounds were screened for their nitric oxide (NO) inhibitory activities, compounds 3 and 4 exhibited moderate inhibitory activities against NO production in LPS-induced RAW 264.7 macrophages.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/18/3/134/s1. Figures S1–S2: B3LYP/6-31+G (d,p) optimized low-energy conformers of 3 and 4; Figures S3–S58: 1D, 2D NMR, HRESIMS, CD, UV and IR spectra of compounds 1–6.
Author Contributions
Formal analysis, Z.L. and H.T.; funding acquisition, W.Z.; investigation, S.C., Y.C. and S.Z.; project administration, W.Z.; resources, S.L.; supervision, H.L., W.Z. and S.Z.; validation, S.C. and H.L.; writing—original draft, S.C.; writing—review and editing, H.L., W.Z. and S.Z. All authors have read and agreed to the published version of the manuscript.
Funding
We thank the National Natural Science Foundation of China (41906106, 31272087), the Pearl River Science and Technology New Star Fund of Guangzhou (201806010080), the GDAS’ Project of Science and Technology Development (2019GDASYL-0103007), the Science and Technology Program of Guangzhou (201607020018), the innovation and university promotion project of Guangdong Pharmaceutical University (2017KCXTD020) and the Team Project of the Natural Science Foundation of Guangdong Province (2016A030312014).
Acknowledgments
We sincerely thank Can Li of Central Laboratory of Southern Medical University for NMR data measurements.
Conflicts of Interest
The authors declare no competing financial interest.
References
- Jimenez, C. Marine natural products in medicinal chemistry. ACS Med. Chem. Lett. 2018, 9, 959–961. [Google Scholar] [CrossRef] [PubMed]
- El-Kashef, D.H.; Daletos, G.; Plenker, M.; Hartmann, R.; Mandi, A.; Kurtan, T.; Weber, H.; Lin, W.; Ancheeva, E.; Proksch, P. Polyketides and a dihydroquinolone alkaloid from a marine-derived strain of the fungus Metarhizium marquandii. J. Nat. Prod. 2019, 82, 2460–2469. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.D.; Fan, P.; Zhou, L.M.; Ma, Q.Y.; Xie, Q.Y.; Zheng, H.Z.; Zheng, Z.H.; Zhang, R.S.; Yuan, J.Z.; Dai, H.F.; et al. Penerpenes A–D, four indole terpenoids with potent protein tyrosine phosphatase inhibitory activity from the marine-derived fungus Penicillium sp. KFD28. Org. Lett. 2019, 21, 4864–4867. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Wang, J.; Wei, X.; Chen, Y.; Fu, T.; Xiang, Y.; Huang, X.; Tian, X.; Xiao, Z.; Zhang, W.; et al. Variecolortins A−C, three pairs of spirocyclic diketopiperazine enantiomers from the marine-derived fungus Eurotium sp. SCSIO F452. Org. Lett. 2018, 20, 4593–4596. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, Y.; Chen, S.; Liu, Y.; Lu, Y.; Chen, D.; Lin, Y.; Huang, X.; She, Z. Aspterpenacids A and B, two sesterterpenoids from a mangrove endophytic fungus Aspergillus terreus H010. Org. Lett. 2016, 18, 1406–1409. [Google Scholar] [CrossRef]
- Zhao, D.L.; Cao, F.; Wang, C.Y.; Yang, L.J.; Shi, T.; Wang, K.L.; Shao, C.L.; Wang, C.Y. Alternatone A, an unusual perylenequinone-related compound from a soft-coral-derived strain of the fungus Alternaria alternate. J. Nat. Prod. 2019, 82, 3201–3204. [Google Scholar] [CrossRef]
- Limbadri, S.; Luo, X.; Lin, X.; Wang, J.; Yang, B.; Zhou, X.; Liu, Y. Versispiroketal A, an unusual tetracyclic bridged spiroketal from the sponge-associated fungus Aspergillus versicolor SCSIO 41013. Org. Biomol. Chem. 2019, 17, 2182–2186. [Google Scholar]
- Jiao, W.H.; Li, J.; Wang, D.; Zhang, M.M.; Liu, L.Y.; Sun, F.; Li, J.Y.; Capon, R.J.; Lin, H.W. Cinerols, nitrogenous meroterpenoids from the marine sponge Dysidea cinerea. J. Nat. Prod. 2019, 82, 2586–2593. [Google Scholar] [CrossRef]
- Luo, M.; Cui, Z.; Huang, H.; Song, X.; Sun, A.; Dang, Y.; Lu, L.; Ju, J. Amino acid conjugated anthraquinones from the marine-derived fungus Penicillium sp. SCSIO sof101. J. Nat. Prod. 2017, 80, 1668–1673. [Google Scholar] [CrossRef]
- Luo, X.; Lin, X.; Tao, H.; Wang, J.; Li, J.; Yang, B.; Zhou, X.; Liu, Y. Isochromophilones A−F, cytotoxic chloroazaphilones from the marine mangrove endophytic fungus Diaporthe sp. SCSIO 41011. J. Nat. Prod. 2018, 81, 934–941. [Google Scholar] [CrossRef]
- Wang, J.; Cong, Z.; Huang, X.; Hou, C.; Chen, W.; Tu, Z.; Huang, D.; Liu, Y. Soliseptide A, a cyclic hexapeptide possessing piperazic acid groups from Streptomyces solisilvae HNM30702. Org. Lett. 2018, 20, 1371–1374. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Tang, X.; Luo, X.; Wang, Q.; Liu, K.; Zhang, Y.; de Voogd, N.J.; Yang, J.; Li, P.; Li, G. Agelanemoechine, a dimeric bromopyrrole alkaloid with a pro-angiogenic effect from the south china sea sponge Agelas nemoechinata. Org. Lett. 2019, 21, 9483–9486. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.R.; Wang, S.W.; Chang, F.R.; Cheng, Y.B. Anti-lymphangiogenic alkaloids from the zoanthid Zoanthus vietnamensis collected in Taiwan. J. Nat. Prod. 2019, 82, 2790–2799. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lee, J.; Kim, K.J.; Sung, Y.; Park, K.H.; Oh, E.; Park, C.; Son, Y.J.; Kang, H. Austalides, osteoclast differentiation inhibitors from a marine-derived strain of the fungus Penicillium rudallense. J. Nat. Prod. 2019, 82, 3083–3088. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Q.; Li, S.; Cui, H.; Sun, Z.; Chen, D.; Lu, Y.; Liu, H.; Zhang, W. Polypropionate derivatives with mycobacterium tuberculosis protein tyrosine phosphatase B inhibitory activities from the deep-sea-derived fungus Aspergillus fischeri FS452. J. Nat. Prod. 2019, 82, 3440–3449. [Google Scholar] [CrossRef]
- Xu, J.; Tan, H.; Chen, Y.; Li, S.; Huang, Z.; Guo, H.; Li, H.; Gao, X.; Liu, H.; Zhang, W. Lithocarpins A–D: Four tenellone-macrolide conjugated [4 + 2] hetero-adducts from the deep-sea derived fungus Phomopsis lithocarpus FS508. Org. Chem. Front. 2018, 5, 1792–1797. [Google Scholar] [CrossRef]
- Chen, S.C.; Liu, Z.M.; Tan, H.B.; Chen, Y.C.; Li, S.N.; Li, H.H.; Guo, H.; Zhu, S.; Liu, H.X.; Zhang, W.M. Tersone A-G, new pyridone alkaloids from the deep-dea fungus Phomopsis tersa. Mar. Drugs 2019, 17, 394. [Google Scholar] [CrossRef]
- Kim, J.W.; Ko, W.; Kim, E.; Kim, G.S.; Hwang, G.J.; Son, S.; Jeong, M.H.; Hur, J.S.; Oh, H.; Ko, S.K.; et al. Anti-inflammatory phomalichenones from an endolichenic fungus Phoma sp. J. Antibiot. 2018, 71, 753–756. [Google Scholar] [CrossRef]
- Kang, U.; Ryu, S.M.; Lee, D.; Seo, E.K. Chemical constituents of the leaves of Brassica oleracea var. acephala. Chem. Nat. Compd. 2018, 54, 1023–1026. [Google Scholar] [CrossRef]
- Rathnayake, G.R.N.; Kumar, N.S.; Jayasinghe, L.; Araya, H.; Fujimoto, Y. Chemical investigation of metabolites produced by an endophytic fungi Phialemonium curvatum from the leaves of Passiflora edulis. Nat. Prod. Res. 2018, 32, 2483–2486. [Google Scholar] [CrossRef]
- Li, X.J.; Gao, J.M.; Chen, H.; Zhang, A.L.; Tang, M. Toxins from a symbiotic fungus, Leptographium qinlingensis associated with Dendroctonus armandi and their in vitro toxicities to Pinus armandi seedlings. Eur. J. Plant. Pathol. 2012, 134, 239–247. [Google Scholar] [CrossRef]
- Valerio, F.; Masi, M.; Cimmino, A.; Moeini, S.A.; Lavermicocca, P.; Evidente, A. Antimould microbial and plant metabolites with potential use in intelligent food packaging. Nat. Prod. Res. 2018, 2, 1605–1610. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Liu, Z.Z.; Kim, K.W.; Wang, X.; Li, Z.; Kim, Y.C.; Yook, C.S.; Liu, X.Q. Chemical constituents from leaves of Pileostegia viburnoides Hook.f.et Thoms. Nat. Prod. Sci. 2016, 22, 154–161. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Rev. D.01 ed; Gaussian, Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Wu, P.; Xue, J.; Yao, L.; Xu, L.; Li, H.; Wei, X. Bisacremines E-G, three polycyclic dimeric acremines produced by Acremonium persicinum SC0105. Org. Lett. 2015, 17, 4922–4925. [Google Scholar] [CrossRef]
- Bruhn, T.; Schaumloffel, A.; Hemberger, Y.; Bringmann, G. SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef]
- Miranda, K.M.; Espey, M.G.; Wink, D.A. A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 2001, 5, 62–71. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).