The Structure of the Lipid A from the Halophilic Bacterium Spiribacter salinus M19-40T
Abstract
:1. Introduction
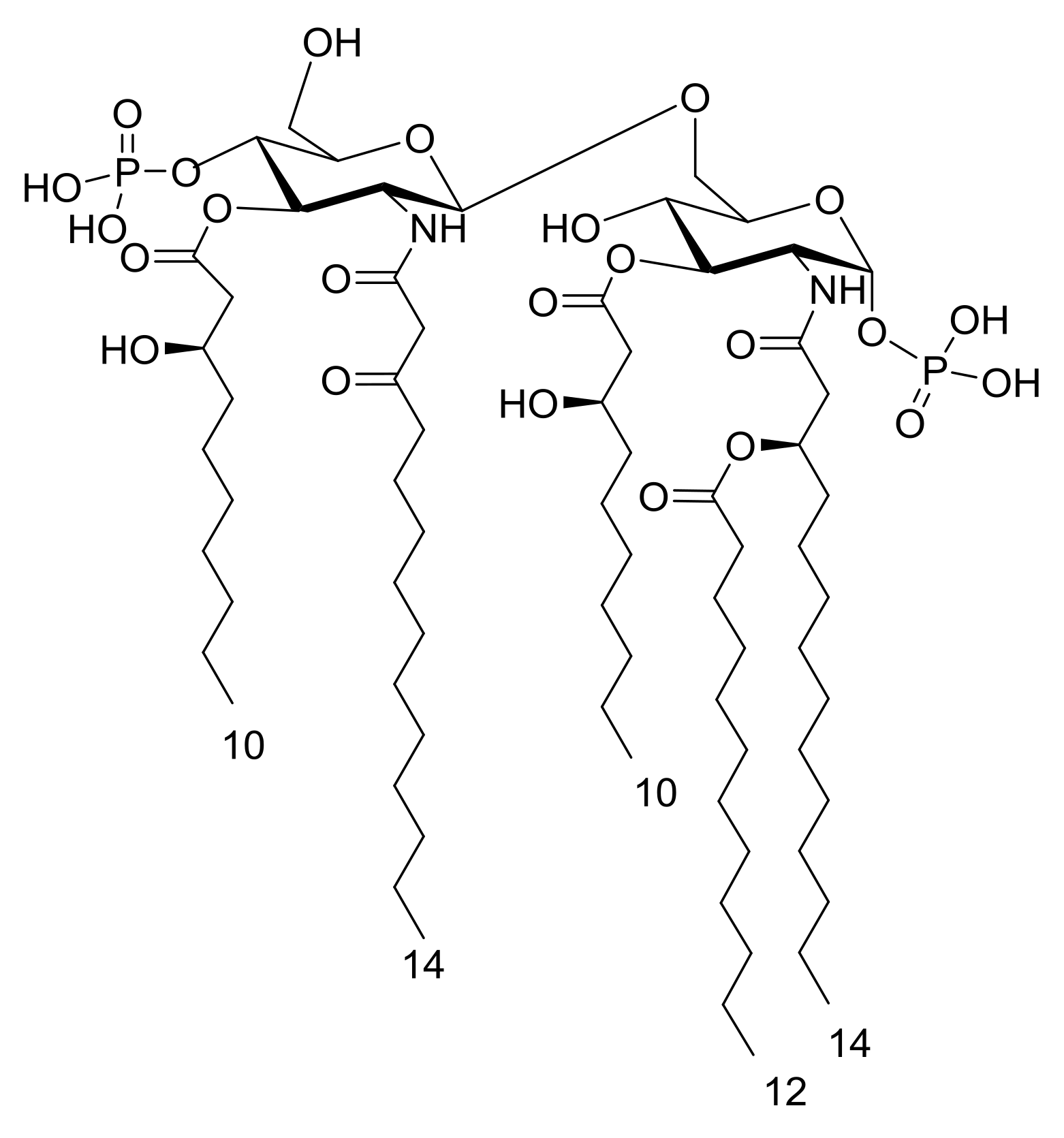
2. Results
2.1. Isolation and Compositional Analysis of the Lipid A from S. salinus M19-40T LPS
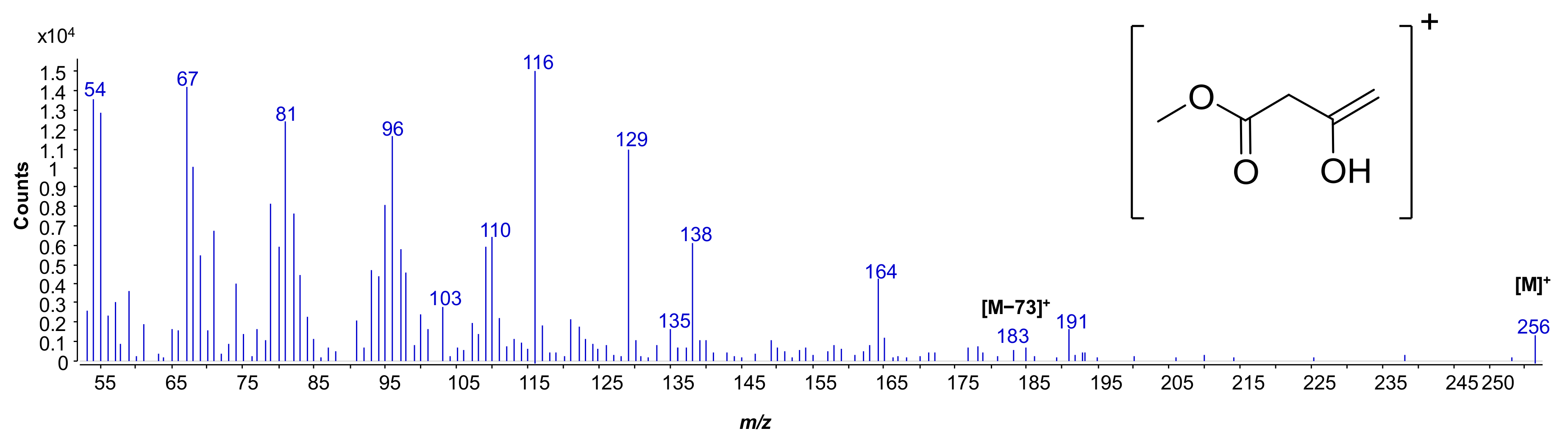
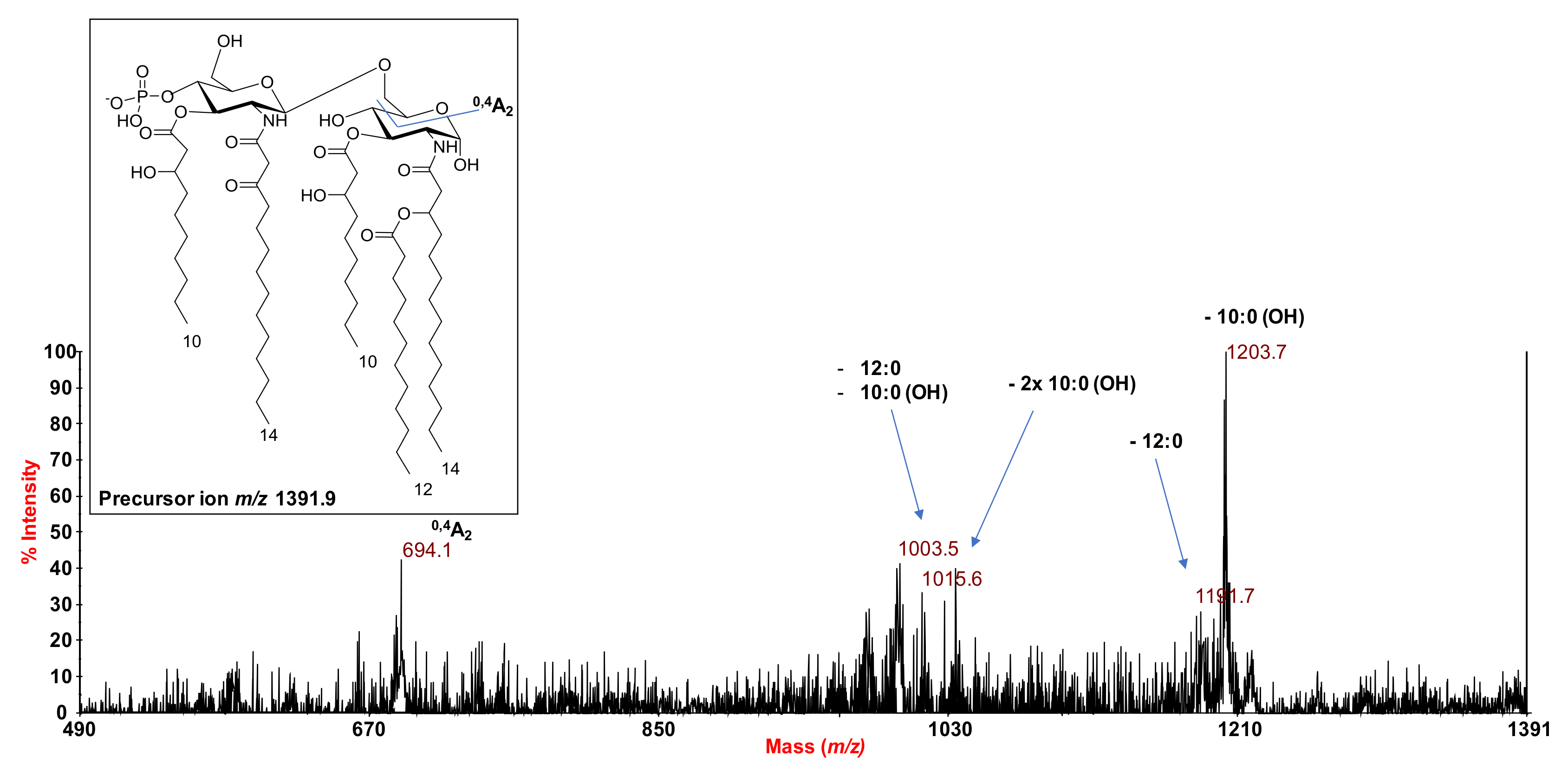
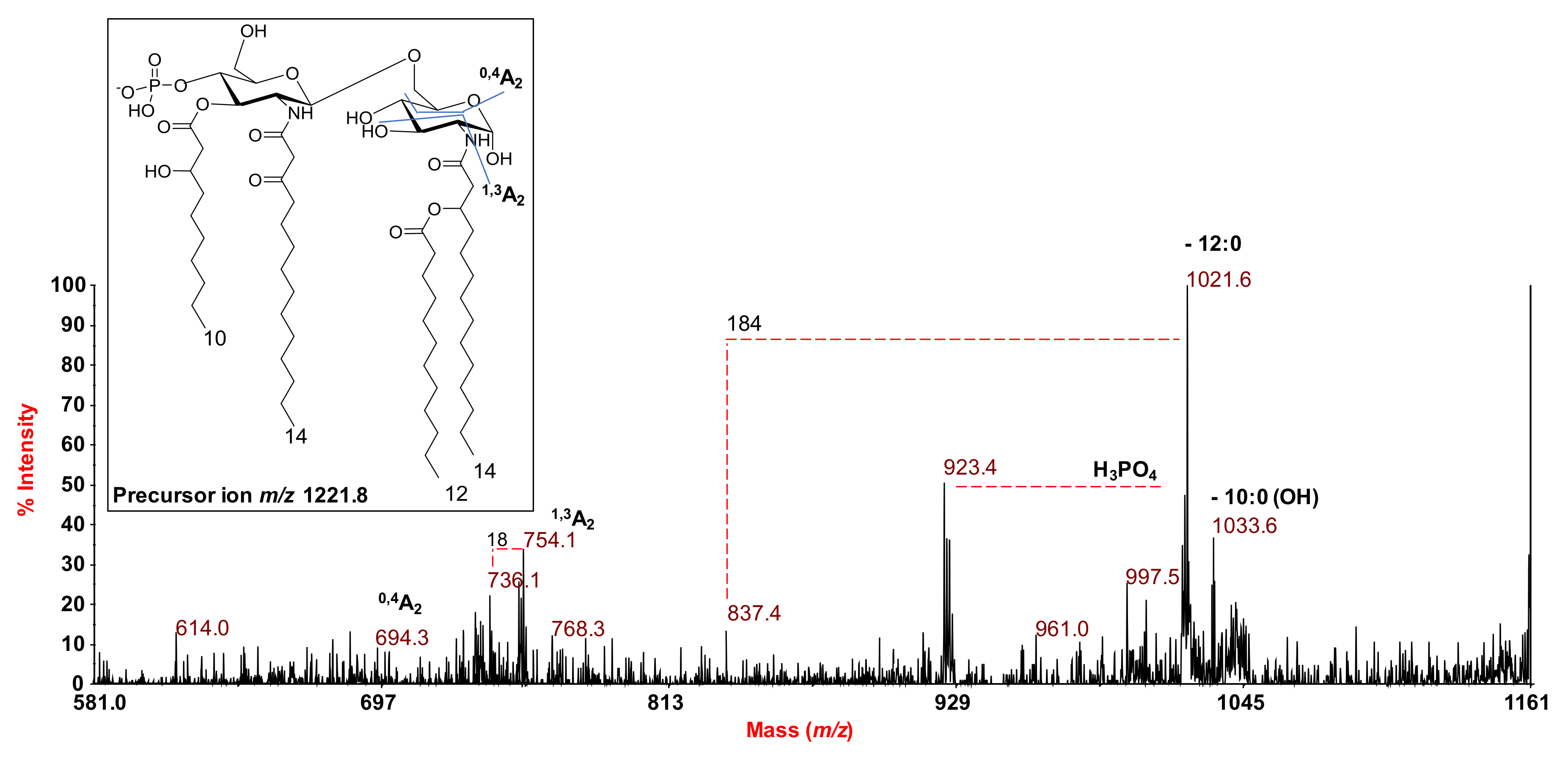
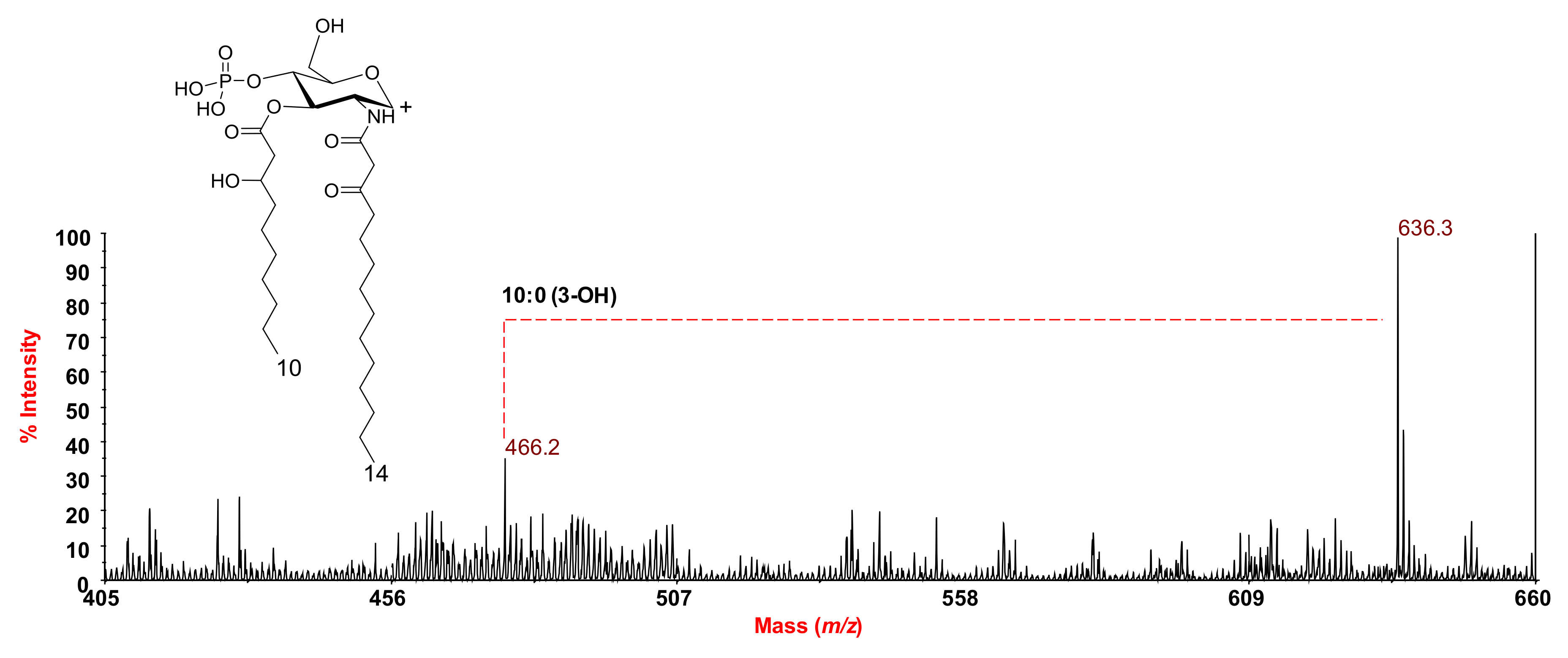
2.2. MALDI MS and MS2 Analysis on the Isolated Lipid A from S. salinus M19-40T LPS
3. Discussion
4. Materials and Methods
4.1. Bacterial Growth
4.2. Extraction and Purification of S. salinus M19-40T LPS
4.3. Chemical Analysis
4.4. Ammonium Hydroxide Hydrolysis of S. salinus M19-40T Lipid A
4.5. MALDI MS Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Raddadi, N.; Cherif, A.; Daffonchio, D.; Neifar, M.; Fava, F. Biotechnological applications of extremophiles, extremozymes and extremolytes. Appl. Microbiol. Biotechnol. 2015, 99, 7907–7913. [Google Scholar] [CrossRef] [PubMed]
- Krüger, A.; Schäfers, C.; Schröder, C.; Antranikian, G. Towards a sustainable biobased industry—Highlighting the impact of extremophiles. New Biotechnol. 2018, 40, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Babu, P.; Chandel, A.K.; Singh, O.V. Survival Mechanisms of Extremophiles In Extremophiles and Their Applications in Medical Processes, 1st ed.; Babu, P., Chandel, A.K., Singh, O.V., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 9–23. [Google Scholar]
- Head, I.M.; Jones, D.M.; Röling, W.F. Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 2006, 4, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.A.; Choi, S. Gram-Negative Marine Bacteria: Structural Features of Lipopolysaccharides and Their Relevance for Economically Important Diseases. Mar. Drugs 2014, 12, 2485–2514. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. Halophilic microbial communities and their Environments. Curr. Opin. Biotechnol. 2015, 33, 119–124. [Google Scholar] [CrossRef] [PubMed]
- León, M.J.; Fernández, A.B.; Ghai, R.; Sánchez-Porro, C.; Rodriguez-Valera, F.; Ventosa, A. From Metagenomics to Pure Culture: Isolation and Characterization of the Moderately Halophilic Bacterium Spiribacter salinus gen. nov., sp. nov. A. Appl. Environ. Microbiol. 2014, 80, 3850–3857. [Google Scholar]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, F.; De Castro, C.; Lanzetta, R.; Parrilli, M.; Silipo, A.; Molinaro, A. Carbohydrates in Drug Design and Discovery; Jiménez-Barbero, J., Javier Canada, F., Martín-Santamaría, S., Eds.; RSC Drug Discovery Series; RSC Publishing: Cambridge, UK, 2015; Volume 43, pp. 38–63. [Google Scholar]
- De Castro, C.; Lanzetta, R.; Leone, S.; Parrilli, M.; Molinaro, A. The structural elucidation of the Salmonella enterica subsp. enterica, reveals that it contains both O-factors 4 and 5 on the LPS antigen. Carbohydr. Res. 2013, 370, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.; Holst, O.; Di Lorenzo, F.; Callaghan, M.; Nurisso, A.; D’Errico, G.; Zamyatina, A.; Peri, F.; Berisio, R.; Jerala, R.; et al. Chemistry of Lipid A: At the Heart of Innate Immunity. Chem. Eur. J. 2015, 21, 500–519. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, F.; Billod, J.M.; Martín-Santamaría, S.; Silipo, A.; Molinaro, A. Gram-Negative Extremophile Lipopolysaccharides: Promising Source of Inspiration for a New Generation of Endotoxin Antagonists. Eur. J. Org. Chem. 2017, 28, 4055–4073. [Google Scholar] [CrossRef]
- Kieser, K.J.; Kagan, J.C. Multi-receptor detection of individual bacterial products by the innate immune system. Nat. Rev. Immunol. 2017, 6, 376–390. [Google Scholar] [CrossRef] [PubMed]
- Mata-Haro, V.; Cekic, C.; Martin, M.; Chilton, P.M.; Casella, C.R.; Mitchell, T.C. The Vaccine Adjuvant Monophosphoryl Lipid A as a TRIF-Biased Agonist of TLR4. Science 2007, 316, 1628–1632. [Google Scholar] [CrossRef] [PubMed]
- Cochet, F.; Peri, F. The Role of Carbohydrates in the Lipopolysaccharide (LPS)/Toll-Like Receptor 4 (TLR4) Signalling. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, F.; Palmigiano, A.; Albitar-Nehme, S.; Pallach, M.; Kokoulin, M.; Komandrova, N.; Romanenko, L.; Bernardini, M.L.; Garozzo, D.; Molinaro, A.; et al. Lipid A structure and immunoinhibitory effect of the marine bacterium Cobetia pacifica KMM 3879T. Eur. J. Org. Chem. 2018. [Google Scholar] [CrossRef]
- Westphal, O.; Jann, K. Bacterial lipopolysaccharides: Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 1965, 1, 83–91. [Google Scholar]
- Kittelberger, R.; Hilbink, F. Sensitive silver-staining detection of bacterial lipopolysaccharides in polyacrylamide gels. J. Biochem. Biophys. Methods 1993, 26, 81–86. [Google Scholar] [CrossRef]
- Ryhage, R.; Stenhagen, E. Mass Spectrometry in lipid research. J. Lipid Res. 1960, 1, 361–390. [Google Scholar] [PubMed]
- Strittmatter, W.; Weckesser, J.; Salimath, P.V.; Galanos, C. Nontoxic Lipopolysaccharide from Rhodopseudomonas sphaeroides ATCC 17023. J. Bacteriol. 1983, 155, 153–158. [Google Scholar] [PubMed]
- Rietschel, E.T. Absolute configuration of 3-hydroxy fatty acids present in lipopolysaccharides from various bacterial groups. Eur. J. Biochem. 1976, 64, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Domon, B.; Costello, C.E. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 1988, 5, 397–409. [Google Scholar] [CrossRef]
- Di Lorenzo, F.; Palmigiano, A.; Al Bitar-Nehme, S.; Sturiale, L.; Duda, K.A.; Gully, D.; Lanzetta, R.; Giraud, E.; Garozzo, D.; Bernadini, M.L.; et al. The Lipid A from Rhodopseudomonas palustris Strain BisA53 LPS possesses a unique structure and low immunostimulant properties. Chem. Eur. J. 2017, 23, 3637–3647. [Google Scholar] [CrossRef] [PubMed]
- Krasikova, I.N.; Kapustina, N.V.; Isakov, V.V.; Dmitrenok, A.S.; Dmitrenok, P.S.; Gorshkova, N.M.; Solovèva, T.F. Detailed structure of lipid A isolated from lipopolysaccharide from the marine proteobacterium Marinomonas vaga ATCC 27119T. Eur. J. Biochem. 2004, 271, 2895–2904. [Google Scholar] [CrossRef] [PubMed]
- Vorob’eva, E.V.; Dmitrenok, A.S.; Dmitrenok, P.S.; Isakov, V.V.; Krasikova, I.N.; Solov’eva, T.F. The Structure of Uncommon Lipid A from the Marine Bacterium Marinomonas communis ATCC 27118T. Russ. J. Bioorg. Chem. 2005, 31, 362–371. [Google Scholar] [CrossRef]
- Zahr, M.; Fobel, B.; Mayer, H.; Imhoff, J.F.; Campos, P.V.; Weckesser, J. Chemical composition of the lipopolysaccharides of Ectothiorhodospira shaposhnikovii, Ectothiorhodospira mobilis, and Ectothiorhodospira halophila. Arch. Microbiol. 1992, 157, 499–504. [Google Scholar]
- Menes, R.J.; Viera, C.E.; Farías, M.E.; Seufferheld, M.J. Halopeptonella vilamensis gen. nov, sp. nov., a halophilic strictly aerobic bacterium of the family Ectothiorhodospiraceae. Extremophiles 2016, 20, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Silipo, A.; Lanzetta, R.; Amoresano, A.; Parrilli, M.; Molinaro, A. Ammonium hydroxide hydrolysis: A valuable support in the MALDI-TOF mass spectrometry analysis of Lipid A fatty acid distribution. J. Lipid Res. 2002, 43, 2188–2195. [Google Scholar] [CrossRef] [PubMed]
- Silipo, A.; Sturiale, L.; Garozzo, D.; De Castro, C.; Lanzetta, R.; Parrilli, M.; Grant, W.D.; Molinaro, A. Structure elucidation of the highly heterogeneous lipid a from the lipopolysaccharide of the gram-negative extremophile bacterium Halomonas Magadiensis Strain 21 M1. Eur. J. Org. Chem. 2004, 10, 2263–2271. [Google Scholar] [CrossRef]
- Di Lorenzo, F. The lipopolysaccharide lipid A structure from the marine sponge-associated bacterium Pseudoalteromonas sp. 2A. J. Microbiol. 2017, 110, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Sturiale, L.; Palmigiano, A.; Silipo, A.; Knirel, Y.A.; Anisimov, A.P.; Lanzetta, R.; Parrilli, M.; Molinaro, A.; Garozzo, D. Reflectron MALDI TOF and MALDI TOF/TOF mass spectrometry reveal novel structural details of native lipooligosaccharides. J. Mass. Spectrom. 2011, 46, 1135–1142. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrau, C.; Di Lorenzo, F.; Menes, R.J.; Lanzetta, R.; Molinaro, A.; Silipo, A. The Structure of the Lipid A from the Halophilic Bacterium Spiribacter salinus M19-40T. Mar. Drugs 2018, 16, 124. https://doi.org/10.3390/md16040124
Barrau C, Di Lorenzo F, Menes RJ, Lanzetta R, Molinaro A, Silipo A. The Structure of the Lipid A from the Halophilic Bacterium Spiribacter salinus M19-40T. Marine Drugs. 2018; 16(4):124. https://doi.org/10.3390/md16040124
Chicago/Turabian StyleBarrau, Clara, Flaviana Di Lorenzo, Rodolfo Javier Menes, Rosa Lanzetta, Antonio Molinaro, and Alba Silipo. 2018. "The Structure of the Lipid A from the Halophilic Bacterium Spiribacter salinus M19-40T" Marine Drugs 16, no. 4: 124. https://doi.org/10.3390/md16040124
APA StyleBarrau, C., Di Lorenzo, F., Menes, R. J., Lanzetta, R., Molinaro, A., & Silipo, A. (2018). The Structure of the Lipid A from the Halophilic Bacterium Spiribacter salinus M19-40T. Marine Drugs, 16(4), 124. https://doi.org/10.3390/md16040124






