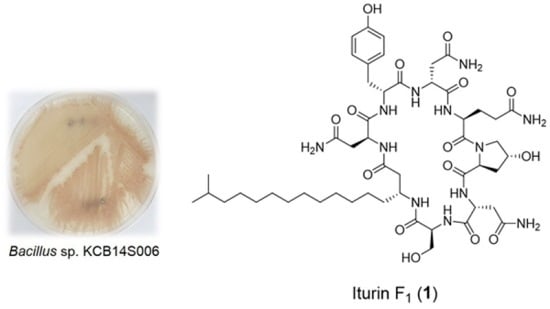
New Cyclic Lipopeptides of the Iturin Class Produced by Saltern-Derived Bacillus sp. KCB14S006
Abstract
:1. Introduction
2. Results
2.1. Isolation of Compounds
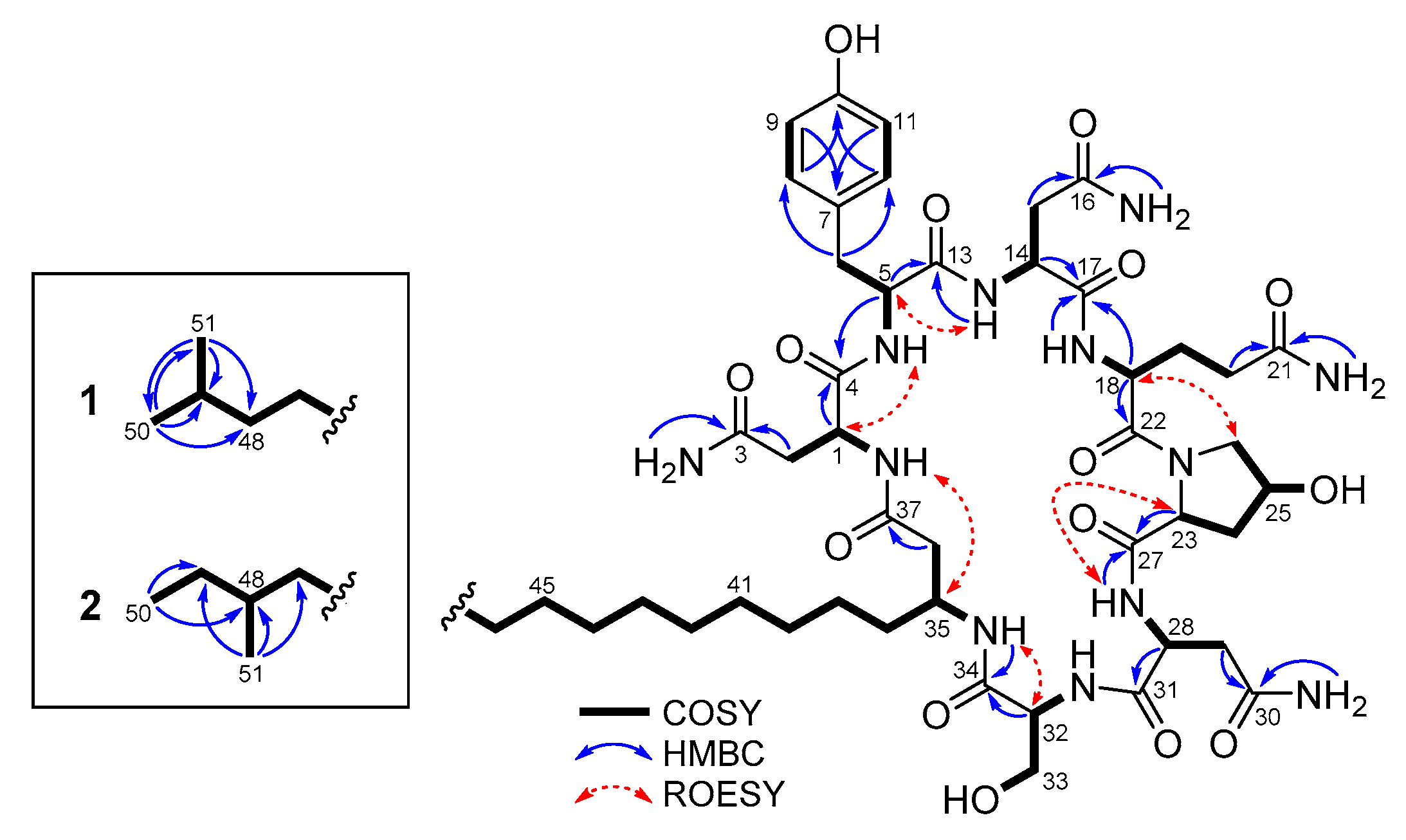
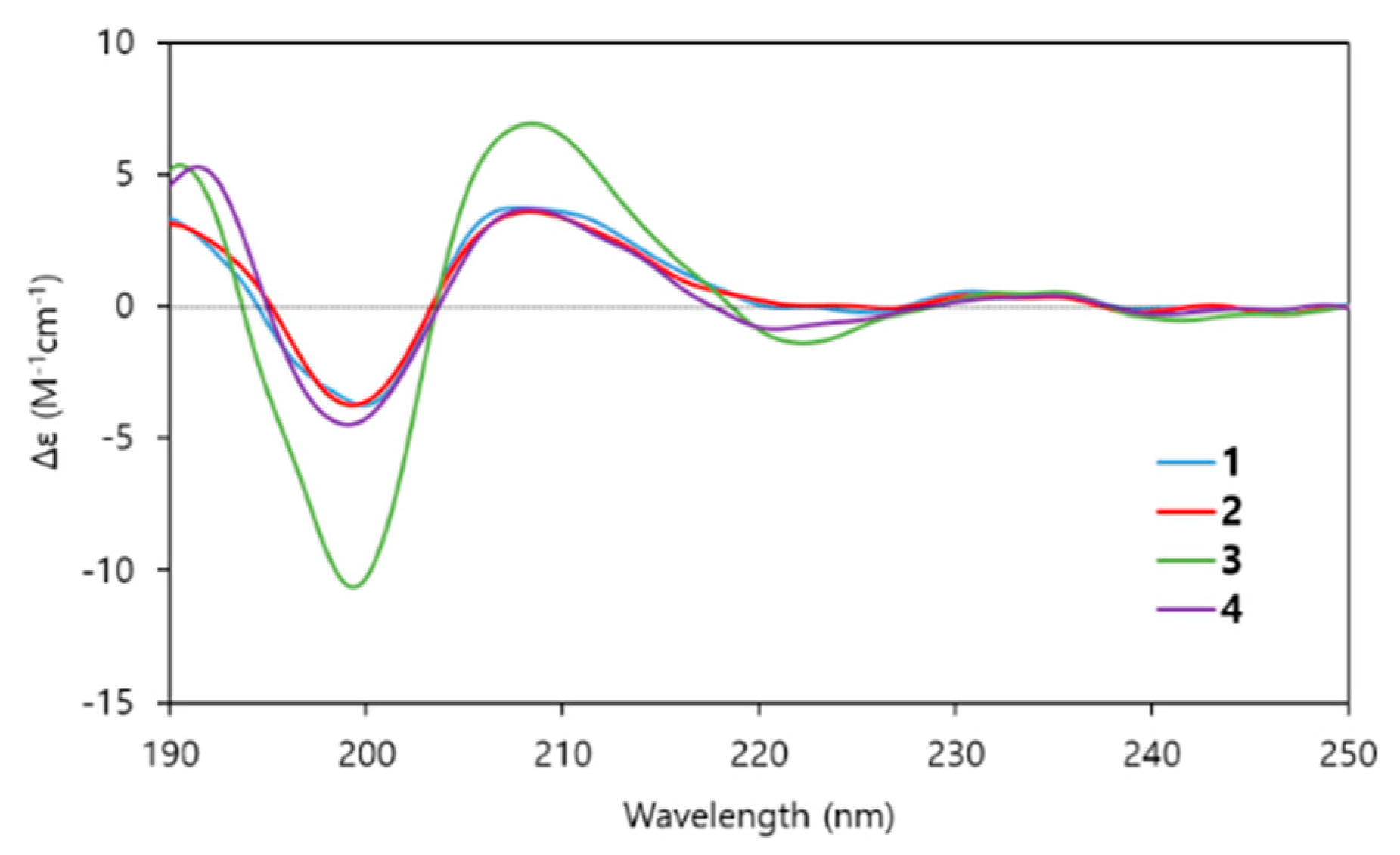
2.2. Structure Determination
2.3. The Bioactivities of Compounds 1–4
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Isolation of the Strain KCB14S006
3.3. Culture, Extraction and Isolation
3.4. Antibacterial Assay
3.5. Antifungal Assay
3.6. Cytotoxicity Assay
3.7. Indoleamine 2,3-Dioxygenase (IDO) Inhibition Assay
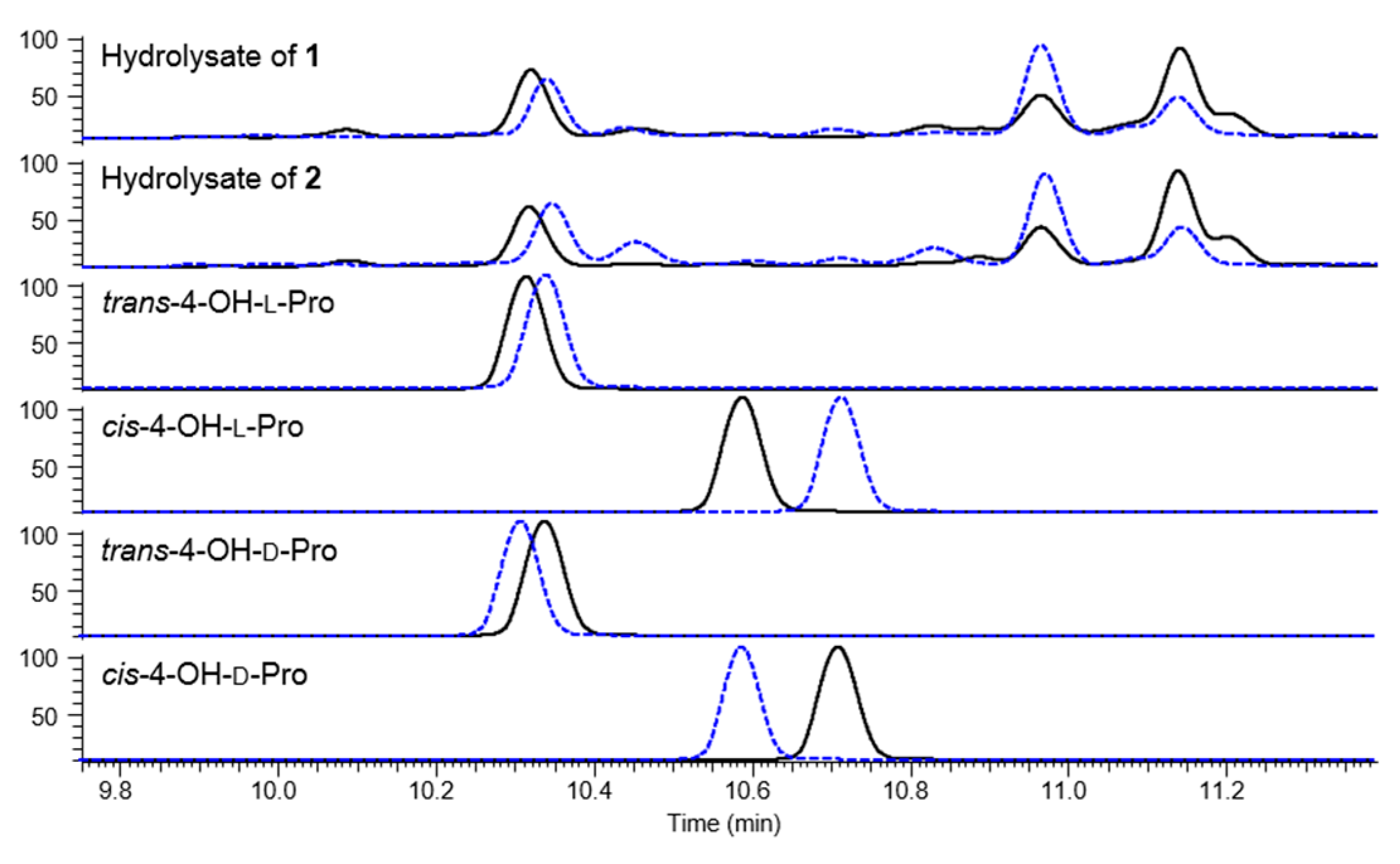
3.8. Advanced Marfey’s Analysis
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends. Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Aranda, F.J.; Teruel, J.A.; Ortiz, A. Further aspects on the hemolytic activity of the antibiotic lipopeptide iturin A. Biochim. Biophys. Acta 2005, 1713, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Bland, J.M. The first synthesis of a member of the iturin family, the antifungal cyclic lipopeptide, iturin-A2. J. Org. Chem. 1996, 61, 5663–5664. [Google Scholar] [CrossRef]
- Isogai, A.; Takayama, S.; Murakoshi, S.; Suzuki, A. Structure of β-amino acids in antibiotics iturin A. Tetrahedron Lett. 1982, 23, 3065–3068. [Google Scholar] [CrossRef]
- Monciardini, P.; Iorio, M.; Maffioli, S.; Sosio, M.; Donadio, S. Discovering new bioactive molecules from microbial sources. Microb. Biotechnol. 2014, 3, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Jebakumar, S.R.D. Unexplored hypersaline habitats are sources of novel actinomycetes. Front. Microbiol. 2014, 5, 242. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, J.; Liu, P.; Wang, W.; Zhu, W. Three new compounds from Aspergillus terreus PT06-2 grown in a high salt medium. Mar. Drugs 2011, 9, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xiao, L.; Wei, J.; Schmitz, J.C.; Liu, M.; Wang, C.; Cheng, L.; Wu, N.; Chen, L.; Zhang, Y.; et al. Identification of Streptomyces sp. nov. WH26 producing cytotoxic compounds isolated from marine solar saltern in China. World J. Microbiol. Biotechnol. 2013, 7, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Ha, T.-K.-Q.; Oh, W.K.; Shin, J.; Oh, D.-C. Antiviral indolosesquiterpenoid xiamycins C-E from a halophilic Actinomycete. J. Nat. Prod. 2016, 79, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Scifinder. Available online: http://scifinder.cas.org (accessed on 27 March 2015).
- Dictionary of Natural Products. Available online: http://dnp.chemnetbase.com (accessed on 27 March 2015).
- Besson, F.; Michel, G. Isolation and characterization of new iturins: Iturin D and iturin E. J. Antibiot. 1987, 40, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Fujii, K.; Hayashi, K.; Suzuki, M.; Ikai, Y.; Oka, H. Application of d,l-FDLA derivatization to determination of absolute configuration of constituent amino acids in peptide by advanced Marfey’s method. Tetrahedron Lett. 1996, 37, 3001–3004. [Google Scholar] [CrossRef]
- Etzbach, L.; Plaza, A.; Garcia, R.; Baumann, S.; Müller, R. Cystomanamides: Structure and biosynthetic pathway of a family of glycosylated lipopeptides from Myxobacteria. Org. Lett. 2014, 16, 2414–2417. [Google Scholar] [CrossRef] [PubMed]
- Gerwick, W.H.; Jiang, Z.D.; Agarwal, S.K.; Farmer, B.T. Total structure of hormothamnin A, a toxic cyclic undecapeptide from the tropical marine cyanobacterium Hormothamnion enteromorphoides. Tetrahedron 1992, 48, 2313–2324. [Google Scholar] [CrossRef]
- Nagai, U.; Besson, F.; Peypoux, F. Absolute configuration of an iturinic acid as determined by CD spectrum of its DNP-p-methoxyanilide. Tetrahedron Lett. 1979, 25, 2359–2360. [Google Scholar] [CrossRef]
- Marion, D.; Genest, M.; Caille, A.; Peypoux, F.; Michel, G.; Ptak, M. Conformational study of bacterial lipopeptides: Refinement of the structure of iturin A in solution by two-dimensional 1H-NMR and energy calculations. Biopolymers 1986, 25, 153–170. [Google Scholar] [CrossRef] [PubMed]
- Besson, F.; Raimbault, C.; Hourdou, M.L.; Buchet, R. Solvent-induced conformational modifications of iturin A: An infrared and circular dichroic study of a l,d-lipopeptide of Bacillus subtilis. Spectrochim. Acta A Mol. Biomol. Spectrosc. 1996, 52, 793–803. [Google Scholar] [CrossRef]
- Hiradate, S.; Yoshida, S.; Sugie, H.; Yada, H.; Fujii, Y. Mulberry anthracnose antagonists (iturins) produced by Bacillus amyloliquefaciens RC-2. Phytochemistry 2002, 61, 693–698. [Google Scholar] [CrossRef]
- Maget-Dana, R.; Peypoux, F. Iturins, a special class of pore-forming lipopeptides: Biological and physicochemical properties. Toxicology 1994, 87, 151–174. [Google Scholar] [CrossRef]
- Bonmatin, J.M.; Laprévote, O.; Peypoux, F. Diversity among microbial cyclic lipopeptides: Iturins and surfactins. Activity-structure relationships to design new bioactive agents. Comb. Chem. High Throughput Screen. 2003, 6, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Dey, G.; Bharti, R.; Dhanarajan, G.; Das, S.; Dey, K.K.; Kumar, B.N.P.; Sen, R.; Mandal, M. Marine lipopeptide Iturin A inhibits Akt mediated GSK3β and FoxO3a signaling and triggers apoptosis in breast cancer. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Platten, M.; von Knebel Doeberitz, N.; Oezen, I.; Wick, W.; Ochs, K. Cancer immunotherapy by targeting IDO1/TDO and their downstream effectors. Front. Immunol. 2015, 5, 673. [Google Scholar] [CrossRef] [PubMed]
- Uyttenhove, C.; Pilotte, L.; Théate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; Van den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Newton, R.C.; Friedman, S.M.; Scherle, P.A. Indoleamine 2, 3-dioxygenase, an emerging target for anti-cancer therapy. Curr. Cancer Drug Targets 2009, 9, 938–952. [Google Scholar] [CrossRef] [PubMed]
- Cady, S.G.; Sono, M. 1-Methyl-dl-tryptophan, β-(3-benzofuranyl)-dl-alanine (the oxygen analog of tryptophan), and β-[3-benzo(b)thienyl]-dl-alanine (the sulfur analog of tryptophan) are competitive inhibitors for indoleamine 2,3-dioxygenase. Arch. Biochem. Biophys. 1991, 291, 326–333. [Google Scholar] [CrossRef]
- Mondol, M.A.; Shin, H.J.; Islam, M.T. Diversity of secondary metabolites from marine Bacillus species: Chemistry and biological activity. Mar. Drugs 2013, 11, 2846–2872. [Google Scholar] [CrossRef] [PubMed]
- Mondol, M.A.; Kim, J.H.; Lee, M.A.; Tareq, F.S.; Lee, H.S.; Lee, Y.J.; Shin, H.J. Ieodomycins A–D, antimicrobial fatty acids from a marine Bacillus sp. J. Nat. Prod. 2011, 74, 1606–1612. [Google Scholar] [CrossRef] [PubMed]
- Barsby, T.; Kelly, M.T.; Andersen, R.J. Tupuseleiamides and basiliskamides, new acyldipeptides and antifungal polyketides produced in culture by a Bacillus laterosporus isolate obtained from a tropical marine habitat. J. Nat. Prod. 2002, 65, 1447–1451. [Google Scholar] [CrossRef] [PubMed]
- Pettit, G.R.; Knight, J.C.; Herald, D.L.; Pettit, R.K.; Hogan, F.; Mukku, V.J.; Hamblin, J.S.; Dodson, M.J.; Chapuis, J.C. Antineoplastic agents. 570. Isolation and structure elucidation of bacillistatins 1 and 2 from a marine Bacillus silvestris. J. Nat. Prod. 2009, 72, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Donio, M.B.; Ronica, S.F.; Viji, V.T.; Velmurugan, S.; Jenifer, J.A.; Michaelbabu, M.; Citarasu, T. Isolation and characterization of halophilic Bacillus sp. BS3 able to produce pharmacologically important biosurfactants. Asian Pac. J. Trop. Med. 2013, 6, 876–883. [Google Scholar] [CrossRef]
- Carballeira, N.M.; Miranda, C.; Lozano, C.M.; Nechev, J.T.; Ivanova, A.; Ilieva, M.; Tzvetkova, I.; Stefanov, K. Characterization of novel methyl-branched chain fatty acids from a halophilic Bacillus species. J. Nat. Prod. 2001, 64, 256–259. [Google Scholar] [CrossRef] [PubMed]


| Position | 1 1 | 2 2 | 3 3 | |||
|---|---|---|---|---|---|---|
| δC | δH, m (J in Hz) | δC | δH, m (J in Hz) | δC | δH, m (J in Hz) | |
| 1 | 50.9 | 4.39, ovl | 50.9 | 4.39, ovl | 50.8 | 4.43, ovl |
| 2 | 36.2 | 2.30, dd (15.9, 8.8) | 36.2 | 2.29, m | 36.3 | 2.29, dd (15.7, 8.4) |
| 2.13, dd (15.9, 4.6) | 2.13, dd (16.3, 5.0) | 2.16, dd (15.7, 5.4) | ||||
| 3 | 170.5 | 170.6 | 170.6 | |||
| 4 | 173.4 | 173.4 | 173.4 | |||
| 1-NH | 7.78, s | 7.78, d (4.2) | 7.72, d (4.1) | |||
| 3-NH2 | 7.34, s | 7.35, s | 7.33, ovl | |||
| 6.92, ovl | 6.91, ovl | 6.92, s | ||||
| 5 | 56.3 | 4.03, m | 56.3 | 4.02, m | 56.3 | 4.02, m |
| 6 | 34.9 | 2.97, br d (10.6) | 34.9 | 2.97, dd (14.5, 3.5) | 34.9 | 2.96, dd (14.3, 3.4) |
| 2.75, dd (14.1, 10.6) | 2.74, dd (14.5, 11.4) | 2.75, dd (14.3, 10.5) | ||||
| 7 | 128.0 | 128.0 | 128.0 | |||
| 8 | 129.8 | 7.03, d (8.4) | 129.8 | 7.03, d (8.8) | 129.8 | 7.03, d (8.4) |
| 9 | 115.1 | 6.66, d (8.3) | 115.1 | 6.66, d (8.7) | 115.1 | 6.66, d (8.4) |
| 10 | 155.8 | 155.8 | 155.8 | |||
| 11 | 115.1 | 6.66, d (8.3) | 115.1 | 6.66, d (8.7) | 115.1 | 6.66, d (8.4) |
| 12 | 129.8 | 7.03, d (8.4) | 129.8 | 7.03, d (8.8) | 129.8 | 7.03, d (8.4) |
| 13 | 171.3 | 171.3 | 171.2 | |||
| 5-NH | 8.69, s | 8.69, d (6.3) | 8.72, ovl | |||
| 10-OH | 9.21, s | 9.21, s | 9.19, s | |||
| 14 | 50.9 | 4.48, m | 50.9 | 4.48, m | 49.6 | 4.43, ovl |
| 15 | 36.2 | 2.59, dd (15.5, 10.0) | 36.2 | 2.59, dd (15.6, 10.6) | 36.0 | 2.59, dd (15.5, 9.6) |
| 2.50, ovl | 2.50, ovl | 2.49, ovl | ||||
| 16 | 171.1 | 171.1 | 171.2 | |||
| 17 | 171.2 | 171.2 | 171.1 | |||
| 14-NH | 8.07, d (7.1) | 8.07, d (8.2) | 8.06, d (6.6) | |||
| 16-NH2 | 7.21, ovl | 7.21, s | 7.22, s | |||
| 6.89, ovl | 6.89, ovl | 6.88, s | ||||
| 18 | 49.6 | 4.51, dd (14.1, 8.6) | 49.6 | 4.51, m | 49.6 | 4.51, dd (14.1, 8.5) |
| 19 | 26.7 | 2.00, m | 26.7 | 1.99, ovl | 26.5 | 2.02, ovl |
| 1.71, m | 1.72, m | 1.74, ovl | ||||
| 20 | 30.5 | 2.09, ovl | 30.5 | 2.09, ovl | 30.6 | 2.10, ovl |
| 21 | 174.0 | 174.0 | 174.1 | |||
| 22 | 171.2 | 171.1 | 170.9 | |||
| 18-NH | 6.92, ovl | 6.92, ovl | 6.98, d (7.8) | |||
| 21-NH2 | 7.10, s | 7.11, s | 7.13, s | |||
| 6.85, ovl | 6.85, ovl | 6.86, s | ||||
| 23 | 59.7 | 4.28, t (7.9) | 59.7 | 4.28, t (8.7) | 60.1 | 4.17, ovl |
| 24 | 37.5 | 2.03, m | 37.5 | 2.03, m | 29.0 | 2.13, ovl |
| 1.86, m | 1.86, m | 1.77, m | ||||
| 25 | 68.7 | 4.39, ovl | 68.7 | 4.39, ovl | 24.6 | 2.00, ovl |
| 1.88, m | ||||||
| 26 | 55.7 | 3.82, br d (6.9) | 55.6 | 3.82, dd (10.1, 3.4) | 47.2 | 3.78, m |
| 3.69, ovl | 3.69, ovl | 3.74, m | ||||
| 27 | 172.6 | 172.6 | 172.7 | |||
| 25-OH | 5.22, s | 5.23, d (2.9) | ||||
| 28 | 49.7 | 4.43, m | 49.7 | 4.43, m | 49.7 | 4.43, ovl |
| 29 | 35.2 | 2.70, dd (15.5, 5.0) | 35.2 | 2.70, dd (16.5, 5.7) | 35.2 | 2.71, dd (15.7, 5.4) |
| 2.43, dd (15.5, 7.5) | 2.43, dd (16.5, 8.1) | 2.47, ovl | ||||
| 30 | 171.8 | 171.8 | 171.8 | |||
| 31 | 170.8 | 170.8 | 170.8 | |||
| 28-NH | 8.79, d (6.8) | 8.80, d (8.0) | 8.72, ovl | |||
| 30-NH2 | 7.37, s | 7.38, s | 7.38, s | |||
| 6.85, ovl | 6.85, ovl | 6.85, s | ||||
| 32 | 56.3 | 4.17, dd (11.7, 7.0) | 56.3 | 4.17, dd (12.3, 7.3) | 56.2 | 4.17, ovl |
| 33 | 61.4 | 3.69, ovl | 61.4 | 3.69, ovl | 61.4 | 3.66, t (5.5) |
| 3.65, m | 3.65, ovl | |||||
| 34 | 170.5 | 170.5 | 170.3 | |||
| 32-NH | 7.40, s | 7.40, ovl | 7.33, ovl | |||
| 33-OH | 4.87, t (5.5) | 4.88, t (6.0) | 4.87, t (5.9) | |||
| 35 | 45.2 | 3.99, m | 45.2 | 3.99, m | 45.3 | 3.98, m |
| 36 | 41.9 | 2.35, m | 41.9 | 2.35, m | 41.8 | 2.34, ovl |
| 37 | 171.3 | 171.3 | 171.2 | |||
| 38 | 34.6 | 1.43, m | 34.6 | 1.40, m | 34.6 | 1.41, m |
| 1.40, m | ||||||
| 39 | 25.3 | 1.24, ovl | 25.3 | 1.24, ovl | 25.4 | 1.24, ovl |
| 1.13, ovl | 1.11, ovl | 1.13, ovl | ||||
| 40–46 | 28.6–29.3 | 1.24, ovl | 28.6–26.5 | 1.24, ovl | 28.6–29.3 | 1.24, ovl |
| 47 | 26.8 | 1.24, ovl | 36.0 | 1.26, ovl | 26.8 | 1.24, ovl |
| 1.13, ovl | 1.08, m | 1.13, ovl | ||||
| 48 | 38.5 | 1.24, ovl | 33.7 | 1.27, ovl | 38.5 | 1.24, ovl |
| 1.13, ovl | 1.13, ovl | |||||
| 49 | 27.4 | 1.49, m | 28.9 | 1.24, ovl | 27.4 | 1.49, m |
| 1.09, ovl | ||||||
| 50 | 22.5 | 0.84, d (6.6) | 11.2 | 0.82, ovl | 22.6 | 0.84, d (6.6) |
| 51 | 22.5 | 0.84, d (6.6) | 19.1 | 0.82, ovl | 22.6 | 0.84, d (6.6) |
| 35-NH | 7.21, ovl | 7.21, s | 7.14, ovl | |||
| Fungi | 1 | 2 | 3 | 4 | Amphotericin B |
|---|---|---|---|---|---|
| Aspergillus flavus | 3.125 | 3.125 | 25 | 12.5 | 0.195 |
| Neurospora crassa | 6.25 | 6.25 | 12.5 | 12.5 | 0.098 |
| Candida tropicalis | 6.25 | 6.25 | 6.25 | 6.25 | 0.781 |
| Candida albicans | 6.25 | 6.25 | 12.5 | 12.5 | 0.098 |
| Fusarium oxysporum | 25 | 25 | 25 | 25 | 12.5 |
| Alternaria brassicicola | 25 | 25 | 25 | 25 | 0.391 |
| Penicillium griseofulvum | 3.125 | 3.125 | 6.25 | 6.25 | 3.125 |
| Cell Line | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| HeLa | 8.9 | 7.8 | 4.6 | 5.6 |
| srcts-NRK | 8.9 | 7.4 | 5.2 | 9.0 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, S.; Ko, S.-K.; Jang, M.; Kim, J.W.; Kim, G.S.; Lee, J.K.; Jeon, E.S.; Futamura, Y.; Ryoo, I.-J.; Lee, J.-S.; et al. New Cyclic Lipopeptides of the Iturin Class Produced by Saltern-Derived Bacillus sp. KCB14S006. Mar. Drugs 2016, 14, 72. https://doi.org/10.3390/md14040072
Son S, Ko S-K, Jang M, Kim JW, Kim GS, Lee JK, Jeon ES, Futamura Y, Ryoo I-J, Lee J-S, et al. New Cyclic Lipopeptides of the Iturin Class Produced by Saltern-Derived Bacillus sp. KCB14S006. Marine Drugs. 2016; 14(4):72. https://doi.org/10.3390/md14040072
Chicago/Turabian StyleSon, Sangkeun, Sung-Kyun Ko, Mina Jang, Jong Won Kim, Gil Soo Kim, Jae Kyoung Lee, Eun Soo Jeon, Yushi Futamura, In-Ja Ryoo, Jung-Sook Lee, and et al. 2016. "New Cyclic Lipopeptides of the Iturin Class Produced by Saltern-Derived Bacillus sp. KCB14S006" Marine Drugs 14, no. 4: 72. https://doi.org/10.3390/md14040072
APA StyleSon, S., Ko, S.-K., Jang, M., Kim, J. W., Kim, G. S., Lee, J. K., Jeon, E. S., Futamura, Y., Ryoo, I.-J., Lee, J.-S., Oh, H., Hong, Y.-S., Kim, B. Y., Takahashi, S., Osada, H., Jang, J.-H., & Ahn, J. S. (2016). New Cyclic Lipopeptides of the Iturin Class Produced by Saltern-Derived Bacillus sp. KCB14S006. Marine Drugs, 14(4), 72. https://doi.org/10.3390/md14040072