Fucoxanthin Enhances the Level of Reduced Glutathione via the Nrf2-Mediated Pathway in Human Keratinocytes
Abstract
:1. Introduction
2. Results
2.1. Fucoxanthin Increases Expression of GCLC and GSS at the mRNA and Protein Levels
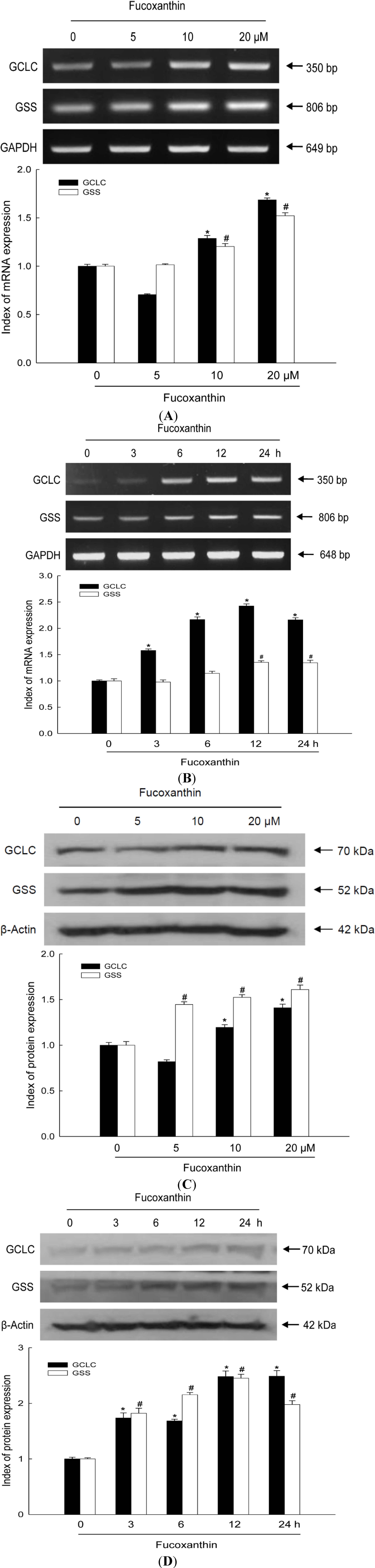
2.2. Fucoxanthin Induces Activation of Nrf2 and Enhances Binding of Nrf2 to the ARE in the Promoters of the GCLC and GSS Genes
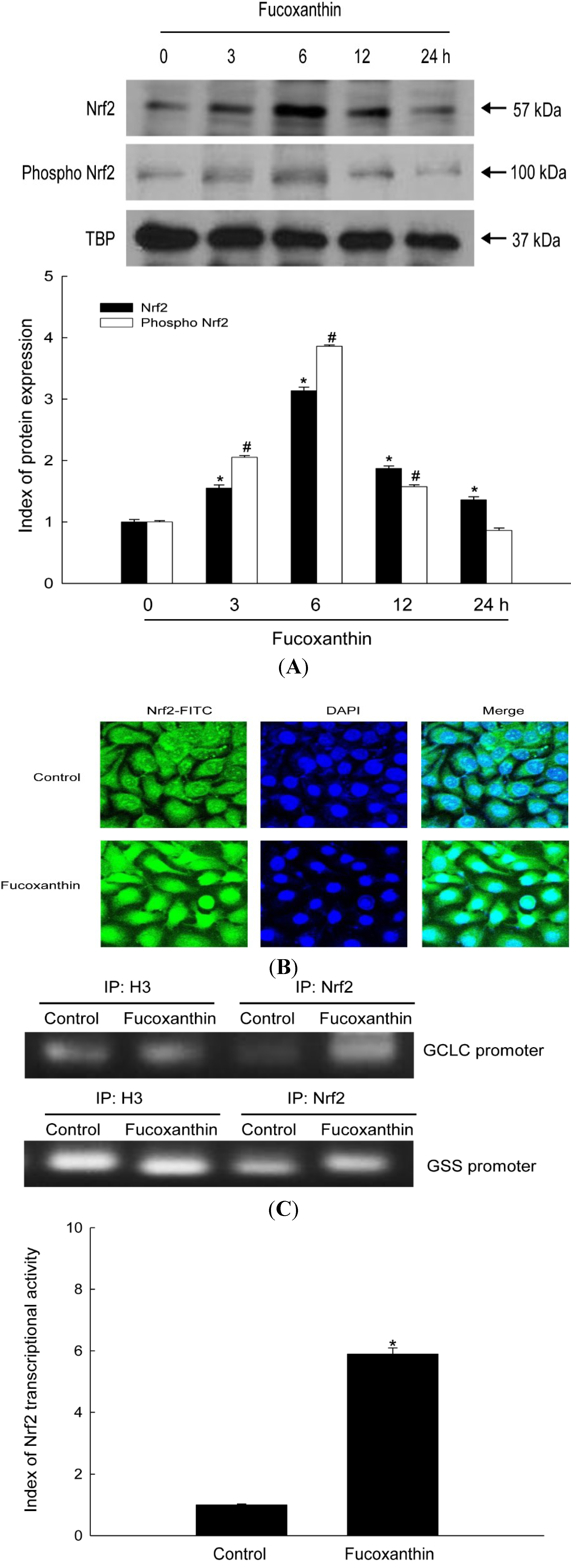
2.3. Fucoxanthin Involves Nrf2-Driven GCLC and GSS via Phosphorylation of Akt
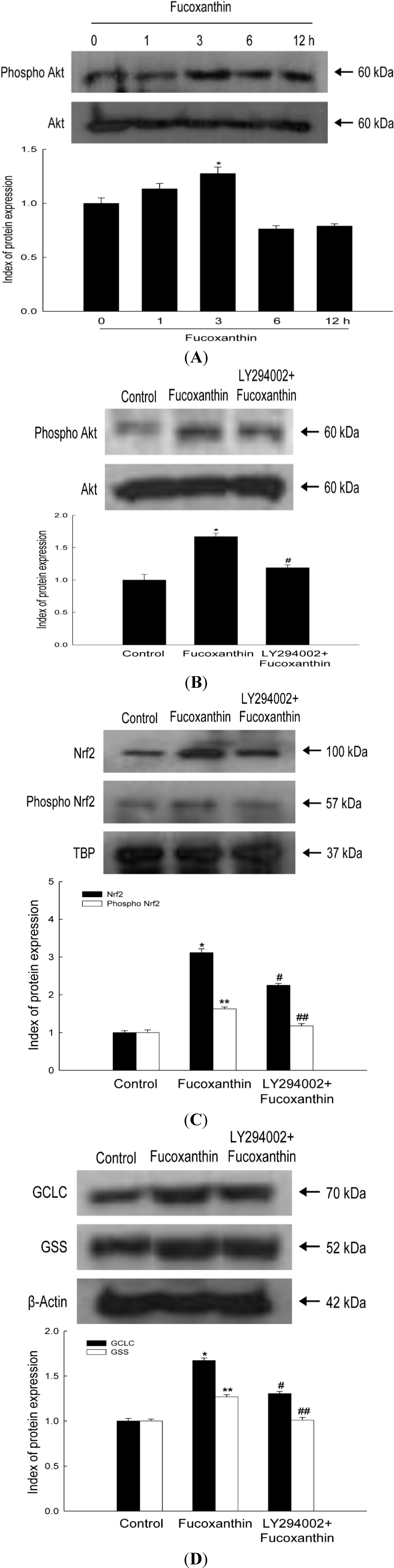
2.4. Fucoxanthin Promotes the Synthesis of GSH Catalyzed by GCLC and GSS
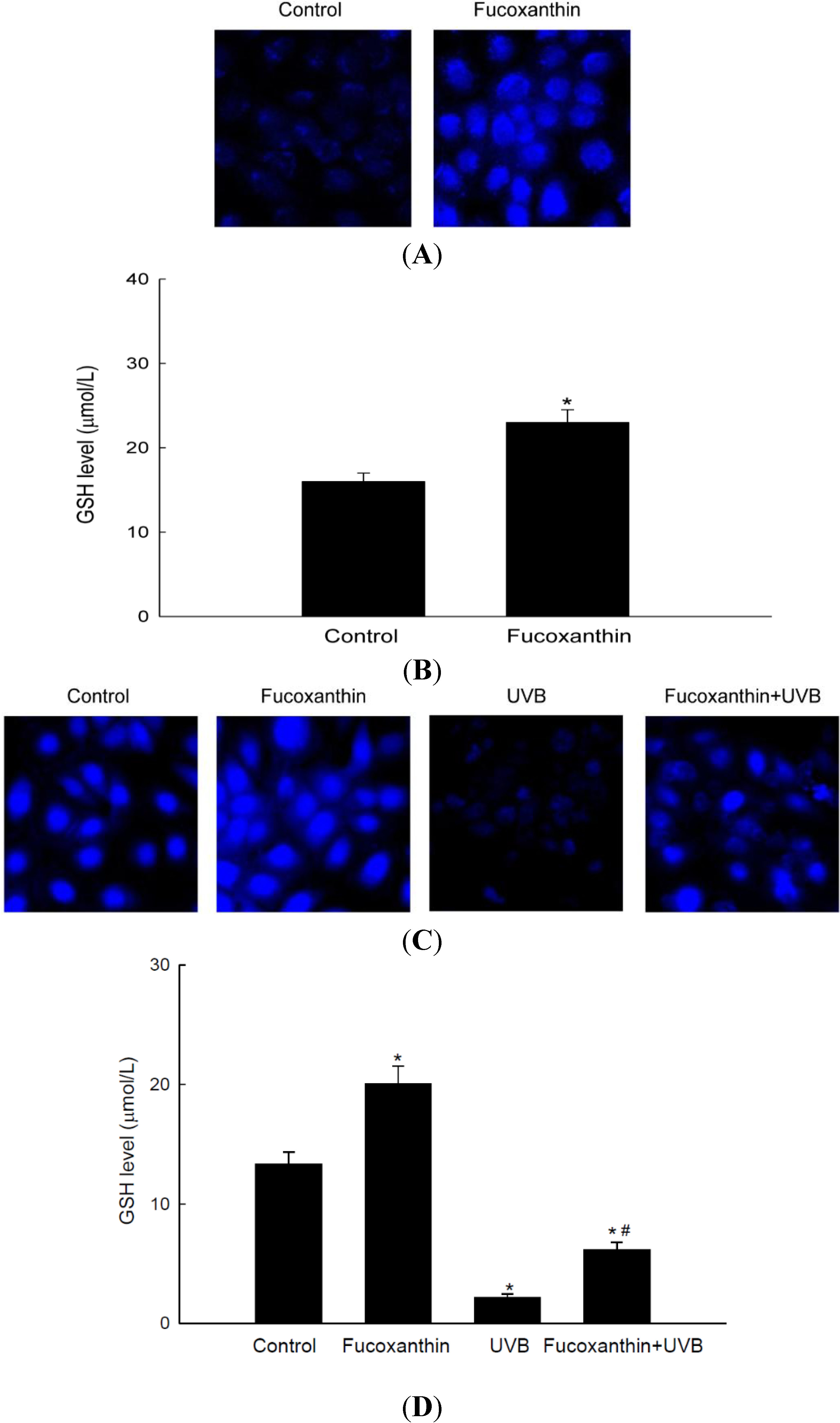
3. Materials and Methods
3.1. Materials
3.2. Cell Culture
3.3. Reverse Transcription-PCR (RT-PCR)
3.4. Western Blot Analysis
3.5. Immunocytochemistry
3.6. ChIP Assay
3.7. Luciferase Reporter Assay
3.8. Detection of GSH
3.9. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Halliwell, B. Biochemistry of oxidative stress. Biochem. Soc. Trans. 2007, 35, 1147–1150. [Google Scholar] [CrossRef]
- Surh, Y.J.; Kundu, J.K.; Na, H.K. Nrf2 as a master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes by some chemopreventive phytochemicals. Planta Med. 2008, 74, 1526–1539. [Google Scholar] [CrossRef]
- Wu, K.C.; Liu, J.J.; Klaassen, C.D. Nrf2 activation prevents cadmium-induced acute liver injury. Toxicol. Appl. Pharmacol. 2012, 263, 14–20. [Google Scholar] [CrossRef]
- Harvey, C.J.; Thimmulappa, R.K.; Singh, A.; Blake, D.J.; Ling, G.; Wakabayashi, N.; Fujii, J.; Myers, A.; Biswal, S. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic. Biol. Med. 2009, 46, 443–453. [Google Scholar]
- Yang, Y.; Chen, Y.; Johansson, E.; Schneider, S.N.; Shertzer, H.G.; Nebert, D.W.; Dalton, T.P. Interaction between the catalytic and modifier subunits of glutamate-cysteine ligase. Biochem. Pharmacol. 2007, 74, 372–381. [Google Scholar] [CrossRef]
- Wu, G.; Fang, Y.Z.; Yang, S.; Lupton, J.; Turner, N.D. Glutathione metabolism and its implications for health. J. Nutr. 2004, 134, 489–492. [Google Scholar]
- Fang, Y.Z.; Yang, S.; Wu, G. Free radicals, antioxidants, and nutrition. Nutrition 2002, 18, 872–879. [Google Scholar] [CrossRef]
- Filomeni, G.; Rotilio, G.; Ciriolo, M.R. Cell signalling and the glutathione redox system. Biochem. Pharmacol. 2002, 64, 1057–1064. [Google Scholar] [CrossRef]
- Clément, M.V.; Ponton, A.; Pervaiz, S. Apoptosis induced by hydrogen peroxide is mediated by decreased superoxide anion concentration and reduction of intracellular milieu. FEBS Lett. 1998, 440, 13–18. [Google Scholar] [CrossRef]
- Pi, J.; Zhang, Q.; Woods, C.G.; Wong, V.; Collins, S.; Andersen, M.E. Activation of Nrf2-mediated oxidative stress response in macrophages by hypochlorous acid. Toxicol. Appl. Pharmacol. 2008, 226, 236–243. [Google Scholar] [CrossRef]
- Marrot, L.; Jones, C.; Perez, P.; Meunier, J.R. The significance of Nrf2 pathway in (photo)-oxidative stress response in melanocytes and keratinocytes of the human epidermis. Pigment Cell Melanoma Res. 2008, 21, 79–88. [Google Scholar]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Eggler, A.L.; Gay, K.A.; Mesecar, A.D. Molecular mechanisms of natural products in chemoprevention: Induction of cytoprotective enzymes by Nrf2. Mol. Nutr. Food Res. 2008, 52, 84–94. [Google Scholar]
- Sugawara, T.; Baskaran, V.; Tsuzuki, W.; Nagao, A. Brown algae fucoxanthin is hydrolyzed to fucoxanthinol during absorption by Caco-2 human intestinal cells and mice. J. Nutr. 2002, 132, 946–951. [Google Scholar]
- Yan, X.; Chuda, Y.; Suzuki, M.; Nagatat, T. Fucoxanthin as the Major Antioxidant in Hifikia fusiformis, a Common Edible Seaweed. Biosci. Biotechnol. Biochem. 1999, 63, 605–607. [Google Scholar] [CrossRef]
- Hu, T.; Liu, D.; Chen, Y.; Wu, J.; Wang, S. Antioxidant activity of sulfated polysaccharide fractions extracted from Undaria pinnitafida in vitro. Int. J. Biol. Macromol. 2010, 46, 193–198. [Google Scholar] [CrossRef]
- Das, S.K.; Hashimoto, T.; Kanazawa, K. Growth inhibition of human hepatic carcinoma HepG2 cells by fucoxanthin is associated with down-regulation of cyclin D. Biochim. Biophys. Acta 2008, 1780, 743–749. [Google Scholar]
- Woo, M.N.; Jeon, S.M.; Shin, Y.C.; Lee, M.K.; Kang, M.; Choi, M.S. Anti-obese property of fucoxanthin is partly mediated by altering lipid-regulating enzymes and uncoupling proteins of visceral adipose tissue in mice. Mol. Nutr. Food Res. 2009, 53, 1603–1611. [Google Scholar] [CrossRef]
- Maeda, H.; Hosokawa, M.; Sashima, T.; Miyashita, K. Dietary combination of fucoxanthin and fish oil attenuates the weight gain of white adipose tissue and decreases blood glucose in obese/diabetic KK-Ay mice. J. Agric. Food Chem. 2007, 55, 7701–7706. [Google Scholar] [CrossRef]
- D’Orazio, N.; Gemello, E.; Gammone, M.A.; de Girolamo, M.; Ficoneri, C.; Riccioni, G. Fucoxantin: A treasure from the sea. Mar. Drugs 2012, 10, 604–616. [Google Scholar] [CrossRef]
- Zheng, J.; Piao, M.J.; Keum, Y.S.; Kim, H.S.; Hyun, J.W. Fucoxanthin protects cultured human keratinocytes against oxidative stress by blocking free radicals and inhibiting apoptosis. Biomol. Ther. 2013, 21, 270–276. [Google Scholar] [CrossRef]
- Nguyen, T.; Huang, H.C.; Pickett, C.B. Transcriptional regulation of the antioxidant response element Activation by Nrf2 and repression by MafK. J. Biol. Chem. 2000, 275, 15466–15473. [Google Scholar] [CrossRef]
- Nakaso, K.; Yano, H.; Fukuhara, Y.; Takeshima, T.; Wada-Isoe, K.; Nakashima, K. PI3K is a key molecule in the Nrf2-mediated regulation of antioxidative proteins by hemin in human neuroblastoma cells. FEBS Lett. 2003, 546, 181–184. [Google Scholar] [CrossRef]
- Vlahos, C.J.; Matter, W.F.; Hui, K.Y.; Brown, R.F. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J. Biol. Chem. 1994, 269, 5241–5248. [Google Scholar]
- Chen, J.S.; Huang, P.H.; Wang, C.H.; Lin, F.Y.; Tsai, H.Y.; Wu, T.C.; Lin, S.J.; Chen, J.W. Nrf-2 mediated heme oxygenase-1 expression, an antioxidant-independent mechanism, contributes to anti-atherogenesis and vascular protective effects of Ginkgo biloba extract. Atherosclerosis 2011, 214, 301–309. [Google Scholar] [CrossRef]
- Ogasawara, Y.; Takeda, Y.; Takayama, H.; Nishimoto, S.; Ichikawa, K.; Ueki, M.; Suzuki, T.; Ishii, K. Significance of the rapid increase in GSH levels in the protective response to cadmium exposure through phosphorylated Nrf2 signaling in Jurkat T-cells. Free Radic. Biol. Med. 2014, 69, 58–66. [Google Scholar] [CrossRef]
- Sies, H. Glutathione and its role in cellular functions. Free Radic. Biol. Med. 1999, 27, 916–921. [Google Scholar] [CrossRef]
- Kudugunti, S.K.; Vad, N.M.; Ekogbo, E.; Moridani, M.Y. Efficacy of caffeic acid phenethyl ester (CAPE) in skin B16-F0 melanoma tumor bearing C57BL/6 mice. Investig. New Drugs 2011, 29, 52–62. [Google Scholar] [CrossRef]
- Zhou, B.R.; Yin, H.B.; Xu, Y.; Wu, D.; Zhang, Z.H.; Yin, Z.Q.; Permatasari, F.; Luo, D. Baicalin protects human skin fibroblasts from ultraviolet A radiation-induced oxidative damage and apoptosis. Free Radic. Res. 2012, 46, 1458–1471. [Google Scholar] [CrossRef]
- Bolfa, P.; Vidrighinescu, R.; Petruta, A.; Dezmirean, D.; Stan, L.; Vlase, L.; Damiane, G.; Catoia, C.; Filip, A.; Clichici, S. Photoprotective effects of Romanian propolis on skin of mice exposed to UVB irradiation. Food Chem. Toxicol. 2013, 62, 329–342. [Google Scholar]
- Cullinan, S.B.; Gordan, J.D.; Jin, J.; Harper, J.W.; Diehl, J.A. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 2004, 24, 8477–8486. [Google Scholar] [CrossRef]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 2004, 279, 31556–31567. [Google Scholar] [CrossRef]
- Apopa, P.L.; He, X.; Ma, Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J. Biochem. Mol. Toxicol. 2008, 22, 63–76. [Google Scholar] [CrossRef]
- Jaiswal, A.K. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic. Biol. Med. 2004, 36, 1199–1207. [Google Scholar] [CrossRef]
- Liu, C.L.; Chiu, Y.T.; Hu, M.L. Fucoxanthin enhances HO-1 and NQO1 expression in murine hepatic BNL CL. 2 cells through activation of the Nrf2/ARE system partially by its pro-oxidant activity. J. Agric. Food Chem. 2011, 59, 11344–11351. [Google Scholar] [CrossRef]
- Martin, D.; Rojo, A.I.; Salinas, M.; Diaz, R.; Gallardo, G.; Alam, J.; Ruiz de Galarreta, C.M.; Cuadrado, A. Regulation of heme oxygenase-1 expression through the phosphatidylinositol 3-kinase/Akt pathway and the Nrf2 transcription factor in response to the antioxidant phytochemical carnosol. J. Biol. Chem. 2004, 279, 8919–8929. [Google Scholar] [CrossRef]
- Rodriguez, K.J.; Wong, H.K.; Oddos, T.; Southall, M.; Frei, B.; Kaur, S. A purified Feverfew extract protects from oxidative damage by inducing DNA repair in skin cells via a PI3-kinase-dependent Nrf2/ARE pathway. J. Dermatol. Sci. 2013, 72, 304–310. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zheng, J.; Piao, M.J.; Kim, K.C.; Yao, C.W.; Cha, J.W.; Hyun, J.W. Fucoxanthin Enhances the Level of Reduced Glutathione via the Nrf2-Mediated Pathway in Human Keratinocytes. Mar. Drugs 2014, 12, 4214-4230. https://doi.org/10.3390/md12074214
Zheng J, Piao MJ, Kim KC, Yao CW, Cha JW, Hyun JW. Fucoxanthin Enhances the Level of Reduced Glutathione via the Nrf2-Mediated Pathway in Human Keratinocytes. Marine Drugs. 2014; 12(7):4214-4230. https://doi.org/10.3390/md12074214
Chicago/Turabian StyleZheng, Jian, Mei Jing Piao, Ki Cheon Kim, Cheng Wen Yao, Ji Won Cha, and Jin Won Hyun. 2014. "Fucoxanthin Enhances the Level of Reduced Glutathione via the Nrf2-Mediated Pathway in Human Keratinocytes" Marine Drugs 12, no. 7: 4214-4230. https://doi.org/10.3390/md12074214
APA StyleZheng, J., Piao, M. J., Kim, K. C., Yao, C. W., Cha, J. W., & Hyun, J. W. (2014). Fucoxanthin Enhances the Level of Reduced Glutathione via the Nrf2-Mediated Pathway in Human Keratinocytes. Marine Drugs, 12(7), 4214-4230. https://doi.org/10.3390/md12074214



