Characterization of ACE Inhibitory Peptides from Mactra veneriformis Hydrolysate by Nano-Liquid Chromatography Electrospray Ionization Mass Spectrometry (Nano-LC-ESI-MS) and Molecular Docking
Abstract
:1. Introduction
2. Results and Discussion
2.1. ACE Inhibitory Activity of Fractions
2.2. Identification of Peptides and Evaluation of Their ACE Inhibitory Activity
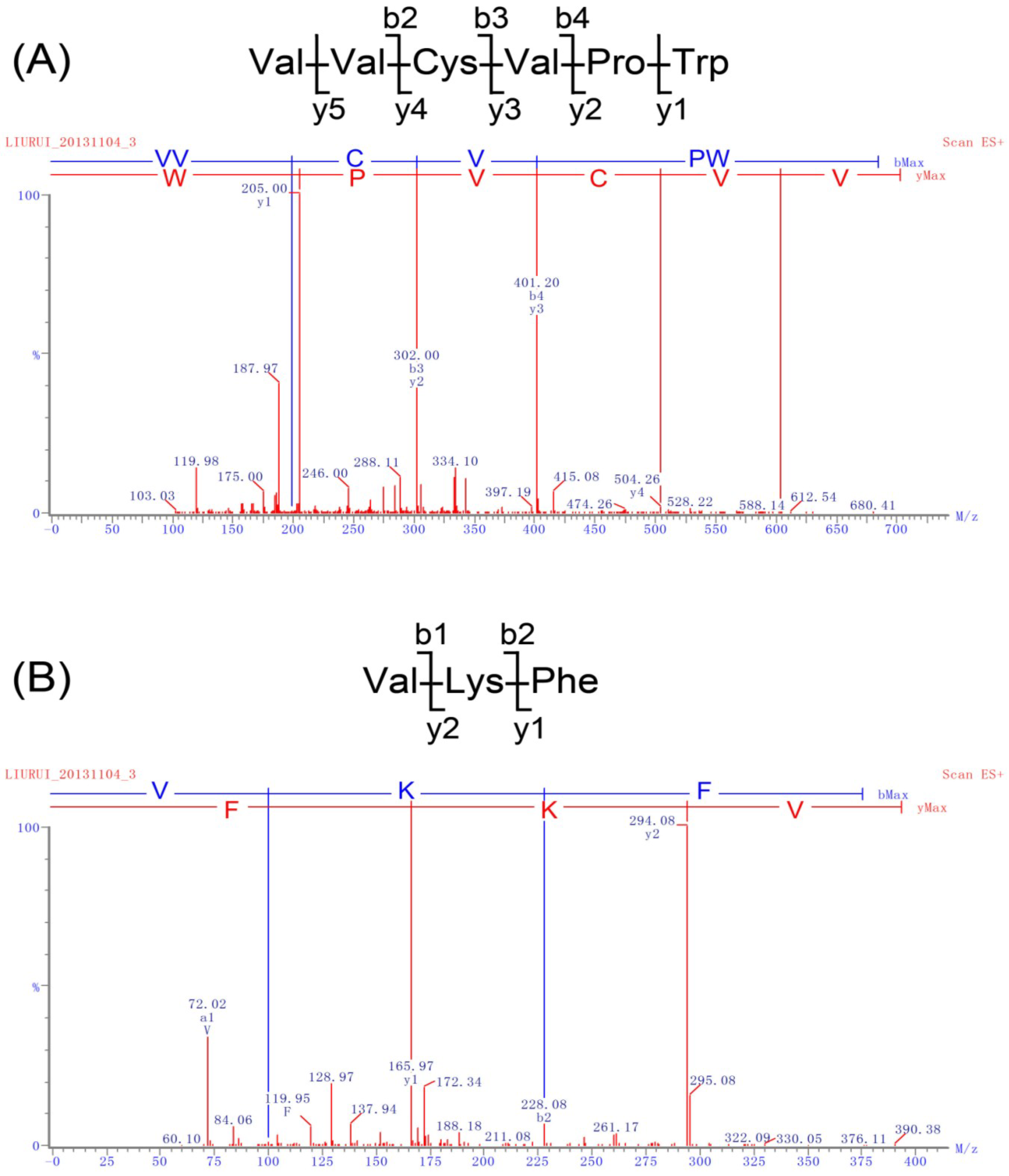
| Peptide Sequences | IC50 (μM) | Peptide Sequences | IC50 (μM) | Peptide Sequences | IC50 (μM) |
|---|---|---|---|---|---|
| VVCVPW | 4.1 | MPFLFK | 92.4 | LLMPK | 686.2 |
| VKF | 10.4 | VFKAF | 100.1 | LLLR | 1005 |
| LYHVL | 17.5 | LR | 158.0 | LEGR | 1090 |
| LVKF | 24.6 | HFEAMR | 192.3 | LALR | 1268 |
| LFR | 31.8 | LLHSPP | 222.0 | LGALPF | 2690 |
| PLFPK | 41.3 | LGECGGR | 232.8 | NKPGDML | - |
| LASPTM | 59.8 | LLRH | 307.5 | LLLLR | - |
| LFVAAP | 70.2 | LKLP | 367.1 | VGGPR | - |
| FKR | 77.9 | RR | 482.2 | LK | - |
2.3. Molecular Docking
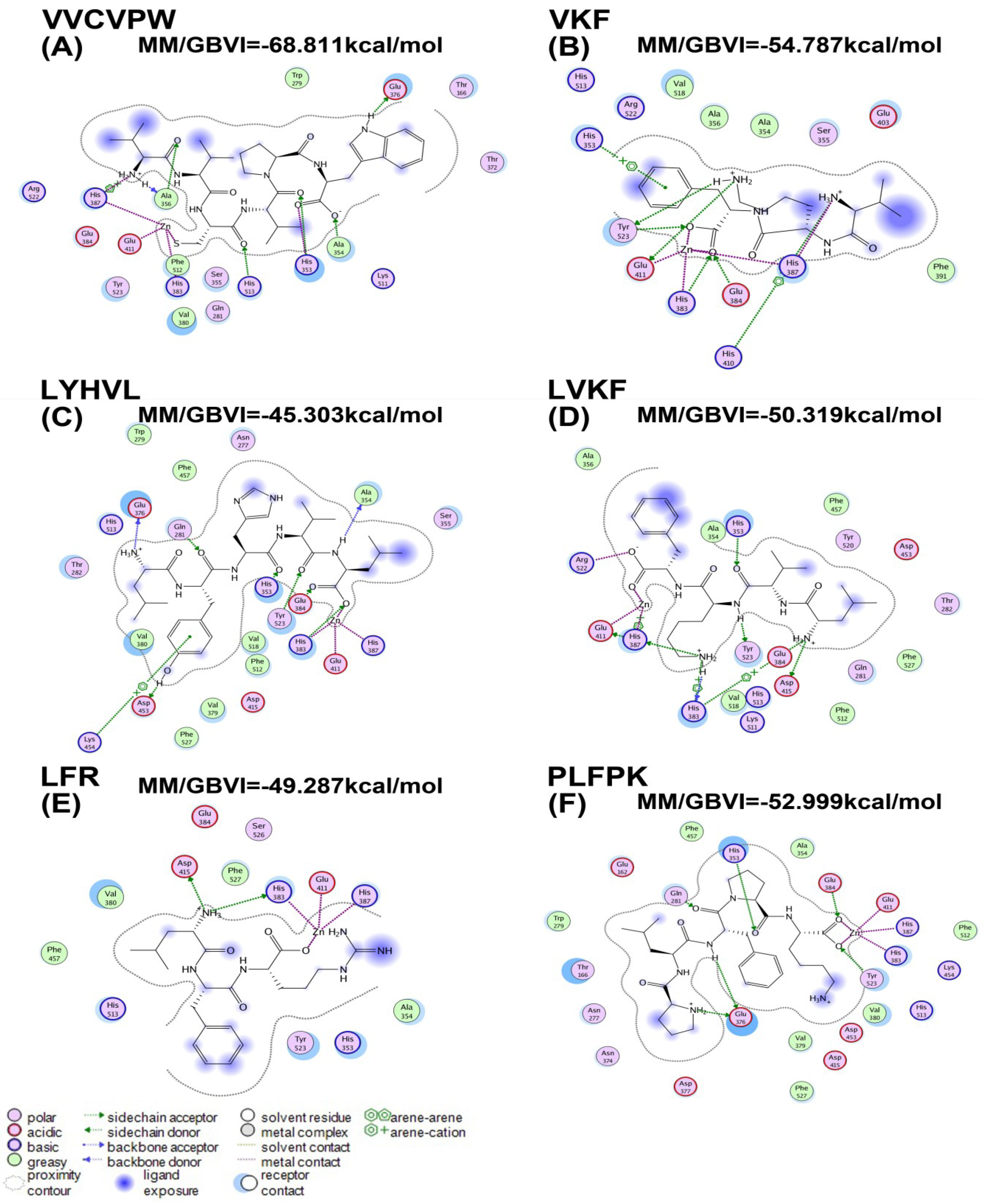
| Peptides | MM/GBVI (kcal/mol) |
|---|---|
| VVCVPW | −68.81 |
| VKF | −54.79 |
| LYHVL | −45.30 |
| LVKF | −50.32 |
| LFR | −49.29 |
| PLFPK | −52.99 |
3. Experimental Section
3.1. Materials
3.2. Preparation of Hydrolysates
3.3. Assay for ACE Inhibitory Activity
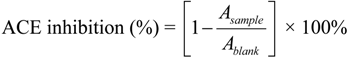
3.4. Ultrafiltration
3.5. Peptide Characterization by Nano-LC-ESI-MS/MS
3.6. Peptide Synthesis and ACE Inhibitory Activity Determination
3.7. Molecular Docking
3.8. Statistical Analysis
4. Conclusions
Supplementary Files
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Murray, B.A.; FitzGerald, R.J. Angiotensin converting enzyme inhibitory peptides derived from food proteins: Biochemistry, bioactivity and production. Curr. Pharm. Des. 2007, 13, 773–791. [Google Scholar] [CrossRef]
- Liu, R.; Wang, M.; Duan, J.A.; Guo, J.M.; Tang, Y.P. Purification and identification of three novel antioxidant peptides from Cornu Bubali (water buffalo horn). Peptides 2010, 31, 786–793. [Google Scholar] [CrossRef]
- Shahidi, F.; Han, X.Q.; Synowiecki, J. Production and characteristics of protein hydrolysates from capelin (Mallotus villosus). Food Chem. 1995, 53, 285–293. [Google Scholar] [CrossRef]
- Salampessy, J.; Phillips, M.; Seneweera, S.; Kailasapathy, K. Release of antimicrobial peptides through bromelain hydrolysis of leatherjacket (Meuchenia sp.) insoluble proteins. Food Chem. 2010, 120, 556–560. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, X.; Tang, L.; Xu, W.; Gong, M. GLP-1 analogs containing disulfide bond exhibited prolonged half-life in vivo than GLP-1. Peptides 2011, 32, 1303–1312. [Google Scholar] [CrossRef]
- Wijesekara, I.; Kim, S.K. Angiotensin-I-converting enzyme (ACE) inhibitors from marine resources: Prospects in the pharmaceutical industry. Mar. Drugs 2010, 8, 1080–1093. [Google Scholar] [CrossRef]
- Jimsheena, V.K.; Gowda, L.R. Arachin derived peptides as selective angiotensin I-converting enzyme (ACE) inhibitors: Structure-activity relationship. Peptides 2010, 31, 1165–1176. [Google Scholar] [CrossRef]
- Ferreira, S.H. A bradykinin-potentiating factor (BPF) present in the venom of Bothrops jararaca. Br. J. Pharmacol. Chemother. 1965, 24, 163–169. [Google Scholar] [CrossRef]
- Rao, S.; Sun, J.; Liu, Y.; Zeng, H.; Su, Y.; Yang, Y. ACE inhibitory peptides and antioxidant peptides derived from in vitro digestion hydrolysate of hen egg white lysozyme. Food Chem. 2012, 135, 1245–1252. [Google Scholar] [CrossRef]
- Wu, S.; Sun, J.; Tong, Z.; Lan, X.; Zhao, Z.; Liao, D. Optimization of hydrolysis conditions for the production of angiotensin-I converting enzyme-inhibitory peptides and isolation of a novel peptide from lizard fish (Saurida elongata) muscle protein hydrolysate. Mar. Drugs 2012, 10, 1066–1080. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y.; Zhong, Q.; Wu, Y.; Xia, W. Purification and characterization of a novel angiotensin-I converting enzyme (ACE) inhibitory peptide derived from enzymatic hydrolysate of grass carp protein. Peptides 2012, 33, 52–58. [Google Scholar] [CrossRef]
- Tomatsu, M.; Shimakage, A.; Shinbo, M.; Yamada, S.; Takahashi, S. Novel angiotensin I-converting enzyme inhibitory peptides derived from soya milk. Food Chem. 2013, 136, 612–616. [Google Scholar] [CrossRef]
- Liu, R.; Ji, J.; Wang, L.C.; Chen, S.Y.; Guo, S.; Wu, H. Characterisation of nucleosides and nucleobases in Mactra veneriformis by high performance liquid chromatography coupled with diode array detector-mass spectrometry (HPLC-DAD-MS). Food Chem. 2012, 135, 548–554. [Google Scholar] [CrossRef]
- Wang, L.C.; Zhang, K.; Di, L.Q.; Liu, R.; Wu, H. Isolation and structural elucidation of novel homogenous polysaccharide from Mactra veneriformis. Carbohydr. Polym. 2011, 86, 982–987. [Google Scholar] [CrossRef]
- Luan, H.M.; Wang, L.C.; Wu, H.; Jin, Y.; Ji, J. Antioxidant activities and antioxidative components in the surf clam, Mactra veneriformis. Nat. Prod. Res. 2011, 25, 1838–1848. [Google Scholar] [CrossRef]
- Wolters, D.A.; Washburn, M.P.; Yates, J.R., III. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 2001, 73, 5683–5690. [Google Scholar] [CrossRef]
- Köcher, T.; Swart, R.; Mechtler, K. Ultra-high-pressure RPLC hyphenated to an LTQ-Orbitrap Velos reveals a linear relation between peak capacity and number of identified peptides. Anal. Chem. 2011, 83, 2699–2704. [Google Scholar] [CrossRef]
- Van Elswijk, D.A.; Diefenbach, O.; van der Berg, S.; Irth, H.; Tjaden, U.R.; van der Greef, J. Rapid detection and identification of angiotensin-converting enzyme inhibitors by on-line liquid chromatography-biochemical detection, coupled to electrospray mass spectrometry. J. Chromatogr. A 2003, 1020, 45–58. [Google Scholar]
- Banerjeel, P.; Shanthi, C. Isolation of novel bioactive regions from bovine Achilles tendon collagen having angiotensin I-converting enzyme-inhibitory properties. Process Biochem. 2012, 47, 2335–2346. [Google Scholar] [CrossRef]
- Yuriev, E.; Ramsland, P.A. Latest developments in molecular docking: 2010–2011 in review. J. Mol. Recognit. 2013, 26, 215–239. [Google Scholar] [CrossRef]
- Gentilucci, L.; Tolomelli, A.; de Marco, R.; Artali, R. Molecular docking of opiates and opioid peptides, a tool for the design of selective agonists and antagonists, and for the investigation of atypical ligand-receptor interactions. Curr. Med. Chem. 2012, 19, 1587–1601. [Google Scholar] [CrossRef]
- Wang, J.P.; Hu, J.E.; Cui, J.Z.; Bai, X.F.; Du, Y.G.; Miyaguchi, Y.; Lin, B.C. Purification and identification of a ACE inhibitory peptide from oyster proteins hydrolysate and the antihypertensive effect of hydrolysate in spontaneously hypertensive rats. Food Chem. 2008, 111, 302–308. [Google Scholar] [CrossRef]
- Ko, S.C.; Kang, N.; Kim, E.A.; Kang, M.C.; Lee, S.H.; Kang, S.M.; Lee, J.B.; Jeon, B.T.; Kim, S.K.; Park, S.J.; et al. novel angiotensin I-converting enzyme (ACE) inhibitory peptide from a marine Chlorella ellipsoidea and its antihypertensive effect in spontaneously hypertensive rats. Process Biochem. 2012, 47, 2005–2011. [Google Scholar] [CrossRef]
- Fujita, H.; Yoshikawa, M. LKPNM: A prodrug-type ACE-inhibitory peptide derived from fish protein. Immunopharmacology 1999, 44, 123–127. [Google Scholar] [CrossRef]
- Balti, R.; Nedjar-Arroume, N.; Adjé, E.Y.; Guillochon, D.; Nasri, M. Analysis of novel angiotensin I-converting enzyme inhibitory peptides from enzymatic hydrolysates of cuttlefish (Sepia officinalis) muscle proteins. J. Agric. Food Chem. 2010, 58, 3840–3846. [Google Scholar] [CrossRef]
- Tavares, T.; Sevilla, M.Á.; Montero, M.J.; Carrón, R.; Malcata, F.X. Acute effect of whey peptides upon blood pressure of hypertensive rats, and relationship with their angiotensin-converting enzyme inhibitory activity. Mol. Nutr. Food Res. 2012, 56, 316–324. [Google Scholar] [CrossRef]
- Pan, D.D.; Cao, J.X.; Guo, H.Q.; Zhao, B. Studies on purification and the molecular mechanism of a novel ACE inhibitory peptide from whey protein hydrolysate. Food Chem. 2012, 130, 121–126. [Google Scholar] [CrossRef]
- Cushman, D.W.; Cheung, H.S. Spectrophotometric assay and properties of the angiotensin-converting enzyme of rabbit lung. Biochem. Pharmacol. 1971, 20, 1637–1648. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, R.; Zhu, Y.; Chen, J.; Wu, H.; Shi, L.; Wang, X.; Wang, L. Characterization of ACE Inhibitory Peptides from Mactra veneriformis Hydrolysate by Nano-Liquid Chromatography Electrospray Ionization Mass Spectrometry (Nano-LC-ESI-MS) and Molecular Docking. Mar. Drugs 2014, 12, 3917-3928. https://doi.org/10.3390/md12073917
Liu R, Zhu Y, Chen J, Wu H, Shi L, Wang X, Wang L. Characterization of ACE Inhibitory Peptides from Mactra veneriformis Hydrolysate by Nano-Liquid Chromatography Electrospray Ionization Mass Spectrometry (Nano-LC-ESI-MS) and Molecular Docking. Marine Drugs. 2014; 12(7):3917-3928. https://doi.org/10.3390/md12073917
Chicago/Turabian StyleLiu, Rui, Yunhan Zhu, Jiao Chen, Hao Wu, Lei Shi, Xinzhi Wang, and Lingchong Wang. 2014. "Characterization of ACE Inhibitory Peptides from Mactra veneriformis Hydrolysate by Nano-Liquid Chromatography Electrospray Ionization Mass Spectrometry (Nano-LC-ESI-MS) and Molecular Docking" Marine Drugs 12, no. 7: 3917-3928. https://doi.org/10.3390/md12073917
APA StyleLiu, R., Zhu, Y., Chen, J., Wu, H., Shi, L., Wang, X., & Wang, L. (2014). Characterization of ACE Inhibitory Peptides from Mactra veneriformis Hydrolysate by Nano-Liquid Chromatography Electrospray Ionization Mass Spectrometry (Nano-LC-ESI-MS) and Molecular Docking. Marine Drugs, 12(7), 3917-3928. https://doi.org/10.3390/md12073917




