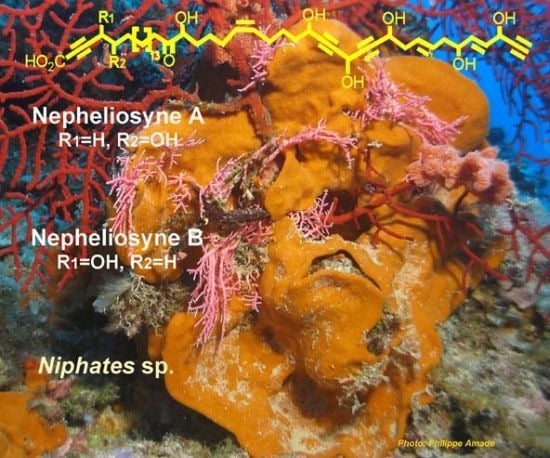
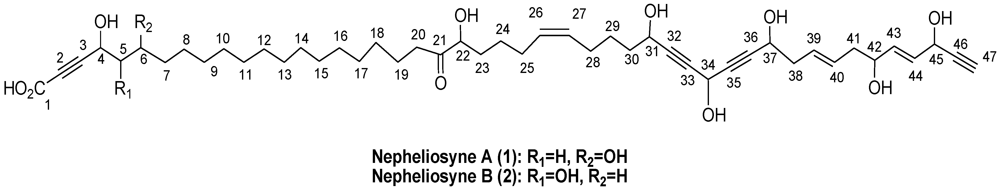
Nepheliosyne B, a New Polyacetylenic Acid from the New Caledonian Marine Sponge Niphates sp.
Abstract
:1. Introduction
2. Results and Discussion
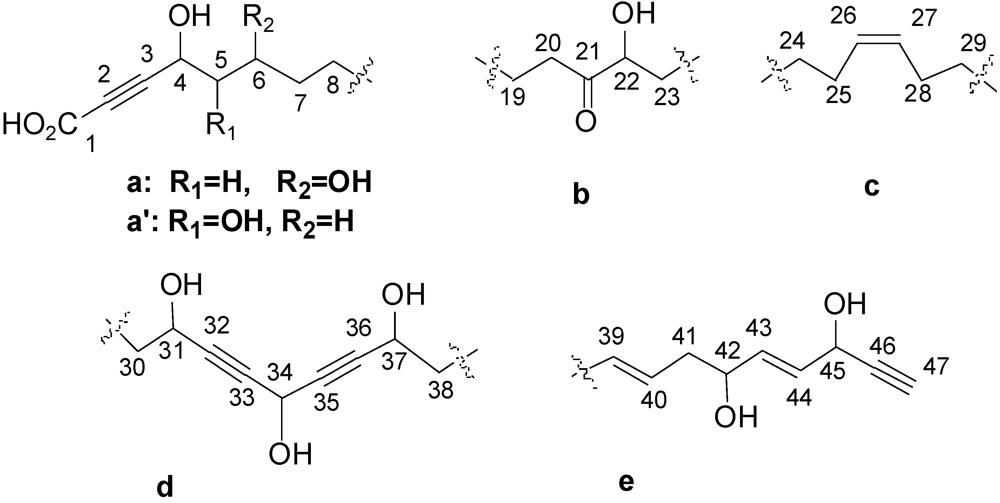
| Position | δC (ppm)/Multiplicity | δH (ppm)/ J (Hz)/Multiplicity | 1H-1H COSY/TOCSY | 1H-13C HMBC |
|---|---|---|---|---|
| 1 | 157.7, qC | |||
| 2 | 78.0, qC | |||
| 3 | 87.3, qC | |||
| 4 | 59.3, CH | 4.64, dd, 9.3, 3.5 | 5a, 6, 7 | 2, 3, 5, 6 |
| 5a | 45.6, CH2 | 1.74–1.71, m | 4, 5b, 6, 7 | 3, 4, 7 |
| 5b | 45.6, CH2 | 1.86–1.81, m | 4, 5a, 6, 7, 8 | 3, 4, 6, 7 |
| 6 | 68.4, CH | 3.84–3.79, m | 4, 5a, 5b, 7, 8 | 4, 5b |
| 7 | 38.7, CH2 | 1.45, m | 4, 6, 5a, 5b, 7, 8 | 8 |
| 8–18 | 30.8–30.4, CH2 | 1.30, m | 5a, 5b, 7, 17, 19 | |
| 19 | 24.1, CH2 | 1.59–1.55, m | 18, 20, 24, 25 | 20, 24, 25 |
| 20 | 38.6, CH2 | 2.61–2.49, m | 18, 19, 25, 26 | 18, 19, 21 |
| 21 | 215.4, qC | |||
| 22 | 78.0, CH | 4.05, dd, 8.1, 4.2 | 19, 23, 24 | 21, 23, 24 |
| 23a | 34.6, CH2 | 1.74–1.69, m | 22, 24, 25, 26 | 20 |
| 23b | 34.6, CH2 | 1.61–1.49, m | 22, 24, 25, 26 | 20 |
| 24 | 26.2, CH2 | 1.40–1.35, m | 20, 22, 23, 25, 26 | 19, 23, 25, 26, 27 |
| 25 | 28.0, CH2 | 2.14–2.05, m | 20, 23, 24, 26, 27 | 18, 19, 24, 26, 27 |
| 26 | 130.7, CH | 5.38, m | 20, 23, 24, 25, 28 | 25 |
| 27 | 130.9, CH | 5.38, m | 28, 29, 30, 31 | 28 |
| 28 | 27.8, CH2 | 2.14–2.05, m | 26, 27, 29, 30, 31 | 26, 27, 29, 30 |
| 29 | 26.7, CH2 | 1.56–1.49, m | 26, 27, 28, 30, 31 | 28, 30, 31 |
| 30 | 38.3, CH2 | 1.70–1.64, m | 26, 27, 28, 29, 31 | 28, 29, 31 |
| 31 | 62.6, CH | 4.33, dd, 6.7, 1.4 | 26, 27, 28, 29, 30, 34 | 30, 32, 33 |
| 32 | 83.1, qC | |||
| 33 | 82.9, qC | |||
| 34 | 52.3, CH | 5.17, t, 1.6 | 29, 30, 31, 37, 38 | 32, 33, 35, 36 |
| 35 | 85.6, qC | |||
| 36 | 85.9, qC | |||
| 37 | 62.6, CH | 4.36, dd, 6.6, 1.6 | 34, 38, 39, 40 | 35, 38 |
| 38 | 36.8, CH2 | 2.45, t, 6.1 | 31, 37, 39, 40 | 36, 37, 39, 40 |
| 39 | 129.2, CH | 5.64–5.54, m | 37, 38, 40, 41, 42 | 38, 42, 44 |
| 40 | 127.5, CH | 5.64–5.54, m | 37, 38, 39, 41, 42 | 41, 42, 44 |
| 41 | 36.3, CH2 | 2.38–2.25, m | 39, 40, 42 | 39, 40, 42, 43 |
| 42 | 72.2, CH | 4.15, dd, 6.3 | 39, 40, 41, 44 | 39, 40, 41, 43, 44 |
| 43 | 135.7, CH | 5.91, ddd, 15.4, 5.9, 1.3 | 41, 42, 44, 45 | 42, 44, 45 |
| 44 | 130.7, CH | 5.78, ddd, 15.4, 5.7, 1.2 | 41, 42, 43, 45 | 42, 43, 45, 46 |
| 45 | 62.5, CH | 4.83, d, 5.5 | 37, 42, 43, 44, 47 | 43, 44, 46, 47 |
| 46 | 84.4, qC | |||
| 47 | 74.4, CH | 2.90, d, 2.0 | 45 | 44, 45 |
| Position | δC (ppm)/Multiplicity | δH (ppm)/ J (Hz)/Multiplicity | 1H-1H COSY/TOCSY | 1H-13C HMBC |
|---|---|---|---|---|
| 1 | 157.7, qC | |||
| 2 | 80.1, qC | |||
| 3 | 85.3, qC | |||
| 4 | 67.1, CH | 4.26, d, 5.02 | 5, 6, 7 | 1, 5 |
| 5 | 74.9, CH | 3.58, m | 4, 6, 7 | 4 |
| 6a | 33.5, CH2 | 1.69–1.64, m | 4, 5, 7, 8 | 4, 5 |
| 6b | 33.5, CH2 | 1.52–1.44, m | 4, 5, 7, 8 | 4, 5 |
| 7 | 30.8–30.4, CH2 | 1.29, m | 4, 5, 6, 8 | 8 |
| 8–18 | 30.8–30.4, CH2 | 1.29, m | 5a, 5b, 7, 17, 19 | |
| 19 | 24.1, CH2 | 1.59–1.55, m | 18, 20, 24, 25 | 20, 24, 25 |
| 20 | 38.6, CH2 | 2.61–2.49, m | 18, 19, 25, 26 | 18, 19, 21 |
| 21 | 215.0, qC | |||
| 22 | 78.0, CH | 4.05, dd, 8.1, 4.2 | 19, 23, 24 | 21, 23, 24 |
| 23a | 34.6, CH2 | 1.57–1.50, m | 22, 24, 25, 26 | 20 |
| 23b | 34.6, CH2 | 1.73–1.67, m | 22, 24, 25, 26 | 20 |
| 24 | 25.9, CH2 | 1.42–1.34, m | 20, 22, 23, 25, 26 | 19, 23, 25, 26, 27 |
| 25 | 28.0, CH2 | 2.14–2.05, m | 20, 23, 24, 26, 27 | 18, 19, 24, 26, 27 |
| 26 | 130.7, CH | 5.38, m | 20, 23, 24, 25, 28 | 25 |
| 27 | 130.9, CH | 5.38, m | 28, 29, 30, 31 | 28 |
| 28 | 27.8, CH2 | 2.14–2.05, m | 26, 27, 29, 30, 31 | 26, 27, 29, 30 |
| 29 | 26.2, CH2 | 1.58–1.49, m | 26, 27, 28, 30, 31 | 28, 30, 31 |
| 30 | 38.6, CH2 | 1.70–1.62, m | 26, 27, 28, 29, 31 | 28, 29, 31 |
| 31 | 62.6, CH | 4.33, dd, 6.7, 1.4 | 26, 27, 28, 29, 30, 34 | 30, 32, 33 |
| 32 | 83.1, qC | |||
| 33 | 82.9, qC | |||
| 34 | 52.4, CH | 5.17, t, 1.6 | 29, 30, 31, 37, 38 | 32, 33, 35, 36 |
| 35 | 85.6, qC | |||
| 36 | 85.9, qC | |||
| 37 | 62.6, CH | 4.36, dd, 6.6, 1.6 | 34, 38, 39, 40 | 35, 38 |
| 38 | 36.9, CH2 | 2.45, t, 6.1 | 31, 37, 39, 40 | 36, 37, 39, 40 |
| 39 | 127.6, CH | 5.64–5.54, m | 37, 38, 40, 41, 42 | 38, 42, 44 |
| 40 | 129.2, CH | 5.64–5.54, m | 37, 38, 39, 41, 42 | 41, 42, 44 |
| 41 | 36.4, CH2 | 2.38–2.25, m | 39, 40, 42 | 39, 40, 42, 43 |
| 42 | 72.2, CH | 4.15, dd, 6.3 | 39, 40, 41, 44 | 39, 40, 41, 43, 44 |
| 43 | 135.7, CH | 5.91, ddd, 15.4, 5.9, 1.3 | 41, 42, 44, 45 | 42, 44, 45 |
| 44 | 130.7, CH | 5.78, ddd, 15.4, 5.7, 1.2 | 41, 42, 43, 45 | 42, 43, 45, 46 |
| 45 | 62.6, CH | 4.83, d, 5.5 | 37, 42, 43, 44, 47 | 43, 44, 46, 47 |
| 46 | 84.4, qC | |||
| 47 | 75.1, CH | 2.90, d, 2.0 | 45 | 44, 45 |
| Compounds | SKM1 (IC50) | U266 (IC50) | Kasumi (IC50) | K562 (IC50) |
|---|---|---|---|---|
| 1 | 250 | 170 | 200 | 200 |
| 2 | >250 | 200 | 150 | 150 |
3. Experimental Section
3.1. General
3.2. Sponge Material

3.3. Extraction and Isolation
3.4. Characterization Data
3.5. Biological Activity
4. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Minto, R.E.; Blacklock, B. Biosynthesis and function of polyacetylenes and allied natural products. J. Prog. Lipid Res. 2008, 47, 233–306. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.G.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2011, 28, 196–268. [Google Scholar]
- Kobayashi, J.; Naitoh, K.; Ishida, K.; Shigemori, H.; Ishibashi, M. Nepheliosyne A, new C47 acetylenic acid from the Okinawan marine sponge Xestospongia sp. J. Nat. Prod. 1994, 57, 1300–1303. [Google Scholar] [CrossRef]
- Brantley, S.E.; Molinski, T.F.; Preston, C.M.; DeLong, E.F. Brominated acetylenic fatty acids from Xestospongia sp., a marine sponge-bacteria association. Tetrahedron 1995, 51, 7667–7672. [Google Scholar]
- Jung, S.-K.; Young, J.-L.; Kwang, S.-I.; Jee, H.-J.; Chung, J.-S.; Chong, O.-L.; Jongki, H.; Hongkum, L. Cytotoxic polyacetylenes from the marine sponge Petrosia sp. J. Nat. Prod. 1999, 62, 554–559. [Google Scholar] [CrossRef]
- Isaacs, S.; Kashman, Y.; Loya, S.; Hizi, A.; Loya, Y. Petrosynol and petrosolic acid, two novel natural inhibitors of the reverse transcriptase of human immunodeficiency virus from Petrosia sp. Tetrahedron 1993, 49, 10435–10438. [Google Scholar] [CrossRef]
- Li, H.-Y.; Matsunaga, S.; Fusetani, N. Corticatic acids A–C, antifungal acetylenic acids from the marine sponge, Petrosia corticata. J. Nat. Prod. 1994, 57, 1464–1467. [Google Scholar] [CrossRef]
- Ueoka, R.; Ise, Y.; Matsunaga, S. Cytotoxic polyacetylenes related to petroformyne-1 from the marine sponge Petrosia sp. Tetrahedron 2009, 65, 5204–5208. [Google Scholar] [CrossRef]
- Okamoto, C.; Nakao, Y.; Fujita, T.; Iwashita, T.; van Soest, R.W.M.; Fusetani, N.; Matsunaga, S. Cytotoxic C47-polyacetylene carboxylic acids from a marine sponge Petrosia sp. J. Nat. Prod. 2007, 70, 1816–1819. [Google Scholar] [CrossRef]
- Shin, J.; Seo, Y.; Cho, K.W.; Rho, J.-R.; Paul, V.J. Osirisynes A–F, highly oxygenated polyacetylenes from the sponge Haliclona osiris. Tetrahedron 1998, 54, 8711–8720. [Google Scholar]
- Chill, L.; Miroz, A.; Kashmann, Y. Haliclonyne, a new highly oxygenated polyacetylene from the marine sponge Haliclona species. J. Nat. Prod. 2000, 63, 523–526. [Google Scholar] [CrossRef]
- Nuzzo, G.; Ciavatta, M.L.; Villani, G.; Manzo, E.; Zanfardino, A.; Varcamonti, M.; Gavagnin, M. Fulvynes, antimicrobial polyoxygenated acetylenes from the Mediterranean sponge Haliclona fulva. Tetrahedron 2012, 68, 754–760. [Google Scholar] [CrossRef]
- Nakao, Y.; Uehara, T.; Matsunaga, S.; Fusetani, N.; van Soest, R.W.M. Callyspongynic acid, a polyacetylenic acid which inhibits α-glucosidase, from the marine sponge Callyspongia truncata. J. Nat. Prod. 2002, 65, 922–924. [Google Scholar] [CrossRef]
- Sullivan, G.R.; Dale, J.A.; Mosher, H.S. Correlation of configuration and fluorine-19 chemical shifts of alpha-methoxy-alpha-trifluoromethylphenyl acetate derivatives. J. Org. Chem. 1973, 38, 2143–2147. [Google Scholar]
- Ohtani, I.; Kusumi, T.; Kashman, Y.; Kakisawa, H. High-field FT NMR application of Mosher’s method. The absolute configurations of marine terpenoids. J. Am. Chem. Soc. 1991, 113, 4092–4096. [Google Scholar]
- Any attempt to obtain the absolute configuration of C-22 by circular dichroism was also unsuccessful.
- Natural History Museum of Marseille (France). Available online: http://www.museum-marseille.org/ (accessed on 17 June 2013).
- Puissant, A.; Grosso, S.; Jacquel, A.; Belhacene, N.; Colosetti, P.; Cassuto, J.P.; Auberger, P. Imatinib mesylate-resistant human chronic myelogenous leukemia cell lines exhibit high sensitivity to the phytoalexin resveratrol. FASEB J. 2008, 22, 1894–1904. [Google Scholar] [CrossRef]
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Legrave, N.; Hamrouni-Buonomo, S.; Dufies, M.; Guérineau, V.; Vacelet, J.; Auberger, P.; Amade, P.; Mehiri, M. Nepheliosyne B, a New Polyacetylenic Acid from the New Caledonian Marine Sponge Niphates sp. Mar. Drugs 2013, 11, 2282-2292. https://doi.org/10.3390/md11072282
Legrave N, Hamrouni-Buonomo S, Dufies M, Guérineau V, Vacelet J, Auberger P, Amade P, Mehiri M. Nepheliosyne B, a New Polyacetylenic Acid from the New Caledonian Marine Sponge Niphates sp. Marine Drugs. 2013; 11(7):2282-2292. https://doi.org/10.3390/md11072282
Chicago/Turabian StyleLegrave, Nathalie, Souhir Hamrouni-Buonomo, Maeva Dufies, Vincent Guérineau, Jean Vacelet, Patrick Auberger, Philippe Amade, and Mohamed Mehiri. 2013. "Nepheliosyne B, a New Polyacetylenic Acid from the New Caledonian Marine Sponge Niphates sp." Marine Drugs 11, no. 7: 2282-2292. https://doi.org/10.3390/md11072282
APA StyleLegrave, N., Hamrouni-Buonomo, S., Dufies, M., Guérineau, V., Vacelet, J., Auberger, P., Amade, P., & Mehiri, M. (2013). Nepheliosyne B, a New Polyacetylenic Acid from the New Caledonian Marine Sponge Niphates sp. Marine Drugs, 11(7), 2282-2292. https://doi.org/10.3390/md11072282