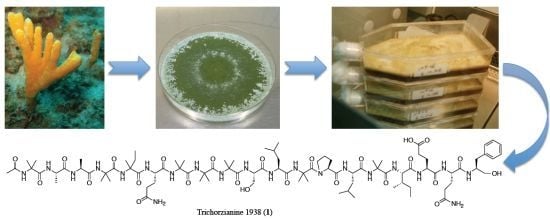
Eight New Peptaibols from Sponge-Associated Trichoderma atroviride
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Elucidation of Trichorzianine 1938 (TA1938) (1)
| Compound name | New/known | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA1938 (1) | new | AcAib | Ala | Ala | Aib | Iva | Gln | Aib | Aib | Aib | Ser | Leu | Aib | Pro | Leu | Aib | Ile | Glu | Gln | Pheol |
| TA1909 (2) | new | AcAib | Ala | Ala | Aib | Aib | Gln | Aib | Aib | Ala | Ser | Leu | Aib | Pro | Leu | Aib | Ile | Gln | Gln | Pheol |
| TA1895 (3) | new | AcAib | Ala | Ala | Aib | Aib | Gln | Aib | Aib | Ala | Ser | Leu | Aib | Pro | Val | Aib | Ile | Gln | Gln | Pheol |
| TA1896 (4) | new | AcAib | Ala | Ala | Aib | Aib | Gln | Aib | Aib | Ala | Ser | Leu | Aib | Pro | Val | Aib | Ile | Glu-OMe | Gln | Pheol |
| TA1924 (5) | new | AcAib | Ala | Ala | Aib | Iva | Gln | Aib | Aib | Aib | Ser | Leu | Aib | Pro | Val | Aib | Ile | Glu-OMe | Gln | Pheol |
| TA1910 (6) | new | AcAib | Ala | Ala | Aib | Iva | Gln | Aib | Aib | Ala | Ser | Leu | Aib | Pro | Val | Aib | Ile | Glu-OMe | Gln | Pheol |
| TA1924a (7) | new | AcAib | Ala | Ala | Aib | Aib | Gln | Aib | Aib | Aib | Ser | Leu | Aib | Pro | Leu | Aib | Ile | Glu-OMe | Gln | Pheol |
| TA1909a (8) | new | AcAib | Ala | Ala | Aib | Iva | Gln | Aib | Aib | Ala | Ser | Leu | Aib | Pro | Val | Aib | Ile | Gln | Gln | Pheol |
| TA.VIb (9) | known | AcAib | Ala | Ala | Aib | Aib | Gln | Aib | Aib | Aib | Ser | Leu | Aib | Pro | Val | Aib | Ile | Gln | Gln | Pheol |
| TA.VIa (10) | known | AcAib | Ala | Ala | Aib | Iva | Gln | Aib | Aib | Aib | Ser | Leu | Aib | Pro | Leu | Aib | Ile | Gln | Gln | Pheol |
| TA.VII (11) | known | AcAib | Ala | Ala | Aib | Iva | Gln | Aib | Aib | Aib | Ser | Leu | Aib | Pro | Val | Aib | Ile | Gln | Gln | Pheol |
| TA.Vb (12) | known | AcAib | Ala | Ala | Aib | Aib | Gln | Aib | Aib | Aib | Ser | Leu | Aib | Pro | Leu | Aib | Ile | Gln | Gln | Pheol |

| Position | δC, mult. b | δH, mult., J (Hz) | LR C–H correlations c | NOE correlations d | |
|---|---|---|---|---|---|
| Ac | 1 | 171.1 s | Ac-2, 1Aib-NH,4 | ||
| 2 | 23.1 q | 1.93 s | 1Aib-NH,3,4 | ||
| 1Aib | 1 | 175.7 e s | 2Ala-NH, 1Aib-NH,3,4 | ||
| 2 | 55.8 s | 1Aib-NH,3,4 | |||
| 3 | 26.2 g q | 1.35 s | 1Aib-NH | 1Aib-NH | |
| 4 | 24.0 h q | 1.32 s | 1Aib-3 | 1Aib-NH | |
| NH | 8.56 s | Ac-2, 1Aib-3,4 | |||
| 2Ala | 1 | 174.5 s | 3Ala-NH, 2Ala-2,3 | ||
| 2 | 50.9 d | 4.02 m | 2Ala-NH,3 | 2Ala-NH,3 | |
| 3 | 16.8 q | 1.31 d | 2Ala-NH,2 | 2Ala-NH,2 | |
| NH | 8.25 d (5.2) | 3Ala-NH, 2Ala-2,3 | |||
| 3Ala | 1 | 174.5 s | 4Aib-NH, 3Ala-2,3 | ||
| 2 | 51.0 d | 3.99 m | 3Ala-NH,3 | 3Ala-NH,3, 4Aib-NH | |
| 3 | 16.2 q | 1.33 d | 3Ala-NH,2 | 3Ala-NH,2 | |
| NH | 7.68 d (5.6) | 4Aib-NH, 3Ala-2,3, 2Ala-NH | |||
| 4Aib | 1 | 175.0 s | 5Iva-NH | ||
| 2 | 56.0 f s | 4Aib-NH,3,4 | |||
| 3 | 26.1 g q | 1.43 k s | 4Aib-NH,4 | 4Aib-NH | |
| 4 | 23.2 h q | 1.35 l s | 4Aib-NH,3 | 4Aib-NH | |
| NH | 7.87 s | 4Aib-2,3, 3Ala-NH,2 | |||
| 5Iva | 1 | 176.4 s | 6Gln-NH, 5Iva-3,NH | ||
| 2 | 58.6 s | 5Iva-NH,3a,3b,4,5 | |||
| 3a | 25.6 t | 2.20 m | 5Iva-4 | 5Iva-4,3b | |
| 3b | 1.62 m | 5Iva-4,3a | |||
| 4 | 7.4 q | 0.74 t | 5Iva-3a,3b | 5Iva-NH,3a,3b | |
| 5 | 22.7 q | 1.35 l s | 5Iva-NH,3a | 5Iva-NH, | |
| NH | 7.47 s | 5Iva-4,5 | |||
| 6Gln | 1 | 174.0 s | 7Aib-NH, 6Gln-2 | ||
| 2 | 56.0 d | 3.78 m | 6Gln-NH,3,4a,4b | 7Aib-NH, 6Gln-NH,3,4a,4b | |
| 3 | 26.2 t | 1.97 m | 6Gln-2,4a,4b,NH | 6Gln-2,4a,4b,NH, 7Aib-NH | |
| 4a | 31.6 i t | 2.15 m | 6Gln-3,NH2b | 6Gln-2,3,4b,NH,NH2a | |
| 4b | 2.23 m | 6Gln-2,3,4a | |||
| 5 | 173.6 s | 6Gln-3,4a,4b,NH2a,b | |||
| NH2a | 7.17 s | 6Gln-4a,NH2b | |||
| NH2b | 6.76 s | 6Gln-NH2a | |||
| NH | 7.74 m | 6Gln-2,3,4a | |||
| 7Aib | 1 | 175.8 e s | 8Aib-NH, 7Aib-NH,3,4 | ||
| 2 | 56.1 f s | 7Aib-NH,3,4 | |||
| 3 | 25.8 g q | 1.42 k s | 7Aib-NH,4 | 7Aib-NH | |
| 4 | 22.6 h q | 1.34 s | 7Aib-NH | 7Aib-NH | |
| NH | 7.86 s | 6Gln-2,3, 7Aib-3,4, 8Aib-NH | |||
| 8Aib | 1 | 175.9 e s | 9Aib-NH, 8Aib-3,4 | ||
| 2 | 56.1 f s | 8Aib-NH,3,4 | |||
| 3 | 26.3 g q | 1.42 k s | 8Aib-NH,4 | 8Aib-NH | |
| 4 | 23.3 h q | 1.39 l s | 8Aib-3 | 8Aib-NH | |
| NH | 7.55 | 8Aib-3,4, 7Aib-NH, 9Aib-NH | |||
| 9Aib | 1 | 176.4 s | 10Ser-NH, 9Aib-3,4 | ||
| 2 | 56.1 f s | 9Aib-NH,3,4 | |||
| 3 | 26.5 g q | 1.46 k s | 9Aib-NH,4 | ||
| 4 | 23.0 h q | 1.42 l s | 9Aib-3 | 9Aib-NH | |
| NH | 7.77 | 9Aib-4, 8Aib-NH | |||
| 10Ser | 1 | 170.6 s | 11Leu-NH, 10Ser-2,3a | - | |
| 2 | 58.9 d | 4.02 m | 10Ser-NH,3a,3b, 11Leu -NH | ||
| 3a | 61.2 t | 3.73 m | 10Ser-NH,2 | 10Ser-NH,2 | |
| 3b | 3.78 m | 10Ser-NH,2 | |||
| OH | |||||
| NH | 7.74 m | 10Ser-2,3a,3b | |||
| 11Leu | 1 | 173.7 j s | 12Aib-NH, 11Leu-2 | ||
| 2 | 51.5 d | 4.27 m | 11Leu-NH,4 | 12Aib-NH, 11Leu-NH,3a,3b,5, 13Pro-5a | |
| 3a | 39.2 t | 1.56 m | 11Leu-2,5,6 | 11Leu-2,3b,5 | |
| 3b | 1.70 m | 11Leu-NH,2,3a,5 | |||
| 4 | 23.9 d | 1.73 m | 11Leu-5,6 | 11Leu-NH,6 | |
| 5 | 20.5 q | 0.76 d | 11Leu-6,3b | 11Leu-2,3a,3b | |
| 6 | 23.0 q | 0.82 d | 11Leu-5,3b | 11Leu-4 | |
| NH | 7.59 d (8.0) | 10Ser-2, 11Leu-2,3b,4 | |||
| 12Aib | 1 | 173.0 s | 12Aib-NH,3,4 | ||
| 2 | 56.2 f s | 12Aib-NH,3,4 | |||
| 3 | 25.8 g q | 1.39 s | 12Aib-NH | 12Aib-NH | |
| 4 | 23.2 h q | 1.49 s | 12Aib-NH | 12Aib-NH, 13Pro-5b | |
| NH | 7.89 s | 13Pro-5a,5b, 11Leu-2, 12Aib-3,4 | |||
| 13Pro | 1 | 173.4 s | 14Leu-NH, 13Pro-2,3a | ||
| 2 | 63.1 d | 4.21 t (8.0) | 13Pro-3a,4 | 14Leu-NH,5, 13Pro-3b,4 | |
| 3a | 28.7 t | 1.59 m | 13Pro-2,5b | 13Pro-3b,5a | |
| 3b | 2.22 m | 13Pro-3a,2,4,5b | |||
| 4 | 26.2 t | 1.86 m | 13Pro-2 | 13Pro-2,3b,5a,5b | |
| 5a | 48.7 t | 3.44 m | 13Pro-3b | 13Pro-2,3a,4,5b, 11Leu-2, 14Leu-NH, 12Aib-NH | |
| 5b | 3.69 m | 13Pro-4,5a, 12Aib-NH,3 | |||
| 14Leu | 1 | 173.7 j s | 15Aib-NH, 14Leu-2 | ||
| 2 | 53.4 d | 3.91 m | 14Leu-NH,4 | 14Leu-3a,3b,4,5,6,NH, 15Aib-NH | |
| 3a | 38.7 t | 1.51 m | 14Leu-2,5,6 | 14Leu-2,3b,6,NH | |
| 3b | 1.78 m | 14Leu-2,3a,6,NH | |||
| 4 | 24.8 d | 1.68 m | 14Leu-5,6 | 14Leu-2,5,NH | |
| 5 | 23.0 q | 0.92 d (6.0) | 14Leu-6 | 14Leu-2,4, 13Pro-2 | |
| 6 | 21.0 q | 0.82 m | 14Leu-5 | 14Leu-2,3a,3b | |
| NH | 7.72 m | 14Leu-2,3a,3b,4, 13Pro-2,5a | |||
| 15Aib | 1 | 175.4 s | 16Ile-NH, 15Aib-NH,3,4 | ||
| 2 | 56.3 f s | 15Aib-NH,3,4 | |||
| 3 | 26.1 g q | 1.44 k s | 15Aib-4 | 15Aib -NH | |
| 4 | 23.4 h q | 1.37 l s | 15Aib-NH,3 | 15Aib-NH | |
| NH | 7.64 s | 16Ile-NH, 15Aib-3,4, 14Leu-2 | |||
| 16Ile | 1 | 172.0 s | 17Glu-NH, 16Ile-2 | - | |
| 2 | 58.9 d | 3.93 m | 16Ile-NH,4b,6 | 16Ile-NH,3,4b,6 | |
| 3 | 35.6 d | 1.88 m | 16Ile-2,4a,4b,5,6 | 16Ile-NH,2,6 | |
| 4a | 25.0 t | 1.20 m | 16Ile-2,3,5,6 | 16Ile-NH,4b | |
| 4b | 1.47 m | 16Ile-NH,2,4a,5 | |||
| 5 | 11.5 q | 0.82 m | 16Ile-4a | 16Ile-4b | |
| 6 | 15.6 q | 0.86 d | 16Ile-4a,2 | 16Ile-NH,2,3 | |
| NH | 6.96 s | 16Ile-2,3,4a,4b,6, 15Aib-NH | |||
| 17Glu | 1 | 171.3 s | 18Gln-NH, 17Glu-2,3a,3b | ||
| 2 | 53.6 d | 4.06 m | 17Glu-NH,4a,4b | 18Gln-NH, 17Glu-NH,3a,3b,4a,4b | |
| 3a | 26.5 t | 1.87 m | 17Glu-2,4a,4b | 17Glu-NH,2,4b | |
| 3b | 1.97 m | 17Glu-NH,2.4a | |||
| 4a | 30.4 t | 2.25 m | 17Glu-2,3a,3b | 17Glu-NH,2,3b | |
| 4b | 2.37 m | 17Glu-NH,2,3a | |||
| 5 | 173.8 j s | 17Glu-4b | |||
| NH | 7.68 d (5.6) | 17Glu-2,3a,3b,4a,4b | |||
| 18Gln | 1 | 170.8 s | 19PheOH-NH, 18Gln-2,3a,3b | ||
| 2 | 53.1 d | 4.06 m | 18Gln-NH,3a,3b,4a,4b | 19PheOH-NH, 18Gln-NH,3a,3b,,4b | |
| 3a | 27.7 d | 1.83 m | 18Gln-2,4a,4b,NH | 18Gln-NH,2,3b,4a | |
| 3b | 1.70 m | 18Gln-NH,2,3a,4b | |||
| 4a | 31.7 i t | 2.05 m | 18Gln-NH2a,2,3a,3b | 18Gln-3a | |
| 4b | 1.97 m | 18Gln-NH,NH2b,2,3b | |||
| 5 | 173.6 | 18Gln-NH2a,b,3a,3b,4a,4b | |||
| NH2a | 6.65 s | 18Gln-NH2b | |||
| NH2b | 7.10 s | 18Gln-NH2a,4b | |||
| NH | 7.46 d | 19PheOH-NH, 18Gln-2,3a,3b,4b | |||
| 19PheOH | 1a | 62.7 t | 3.29 m | 19PheOH-2,3a,3b | 19PheOH-NH,2,3a,3b,5 |
| 1b | 3.32 m | 19PheOH-NH,2,3a,3b | |||
| 2 | 52.5 d | 3.86 m | 19PheOH-1a,1b,3a,3b,NH | 19PheOH-NH,1a,1b,3a,3b,5 | |
| 3a | 36.6 t | 2.61 dd (13.4, 8.2) | 19PheOH-1a,1b,2,5 | 19PheOH-NH,1a,1b,2,5 | |
| 3b | 2.84 dd (13.2,4.8) | 19PheOH-1a,1b,2,5 | |||
| 4 | 139.2 s | 19PheOH-2,3a,3b,5 | |||
| 5 | 129.4 d | 7.21 m | 19PheOH-3a,3b,6,7 | 19PheOH-1a,2,3a,3b,7,NH | |
| 6 | 128.2 d | 7.19 m | 19PheOH-5,7 | ||
| 7 | 126.0 d | 7.12 m | 19PheOH-5,6 | 19PheOH-5,NH | |
| OH | |||||
| NH | 7.27 d (8) | 18Gln-NH,2, 19PheOH-1a,1b,2,3a,5,7 |
 ) and ROESY (
) and ROESY (  ) correlations.
) correlations.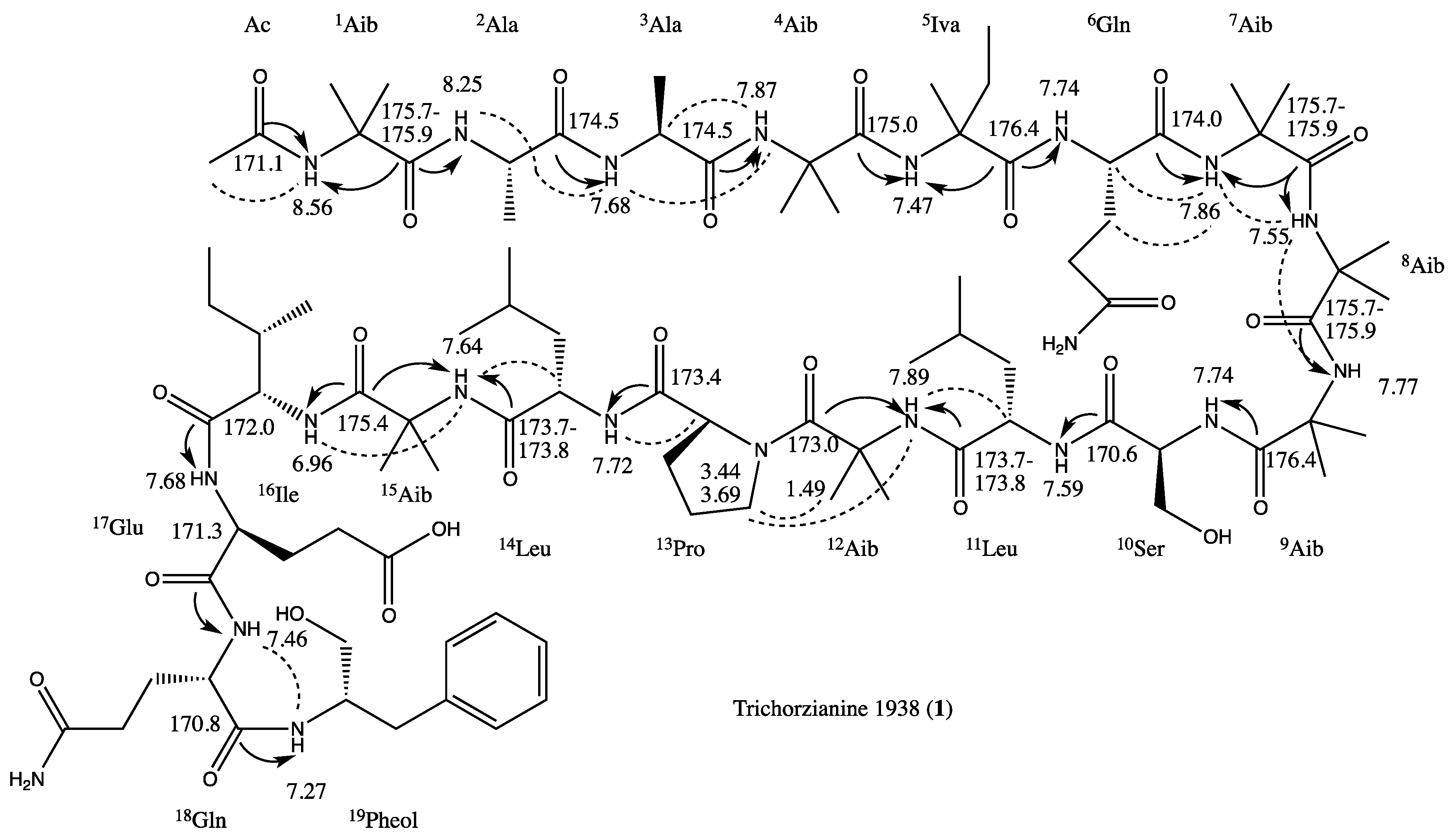
2.2. Structure Elucidation of 2–12
| Position | TA1909 (2) a | TA1895 (3) a | TA1896 (4) b | TA1924 (5) b | TA1910 (6) a | TA1924a (7) a | TA1909a (8) a | |
|---|---|---|---|---|---|---|---|---|
| Ac | 2 | 1.93 s | 1.92 s | 1.92 s | 1.92 s | 1.92 s | 1.93 s | 1.93 s |
| 1Aib | 3 | 1.35 s | 1.35 s | 1.35 s | 1.35 s | 1.35 s | 1.36 s | 1.36 s |
| 4 | 1.32 s | 1.32 s | 1.32 s | 1.32 s | 1.33 s | 1.32 s | 1.33 s | |
| NH | 8.57 s | 8.57 s | 8.58 s | 8.54 s | 8.57 s | 8.57 s | 8.56 s | |
| 2Ala | 2 | 3.99 m | 3.98 m | 3.98 m | 4.00 m | 4.01 m | 4.01 m | 4.01 m |
| 3 | 1.31 d | 1.31 d | 1.31 d | 1.30 d | 1.31 d | 1.31 d | 1.31 d | |
| NH | 8.27 d | 8.27 d | 8.27 d | 8.24 d | 8.26 d | 8.27 d | 8.25 d | |
| 3Ala | 2 | 3.99 m | 3.99 m | 3.98 m | 4.02 m | 4.01 m | 4.01 m | 4.01 m |
| 3 | 1.33 d | 1.33 d | 1.33 d | 1.33 d | 1.33 d | 1.33 d | 1.33 d | |
| NH | 7.69 d | 7.69 d | 7.70 d | 7.68 d | 7.68 d | 7.71 m | 7.68 d | |
| 4Aib | 3 | 1.41 d s | 1.39 c s | 1.43 c s | 1.43 s | 1.45 c s | 1.45 s | 1.46 s |
| 4 | 1.36 c s | 1.38 s | 1.38 d s | 1.35 s | 1.35 d s | 1.38 c s | 1.35 c s | |
| NH | 7.81 s | 7.81 s | 7.82 s | 7.86 s | 7.87 s | 7.83 s | 7.87 s | |
| 5Iva/5Aib | 3a | 1.44 s | 1.42 e s | 1.43 c s | 2.20 m | 2.21 m | 1.44 s | 2.21 m |
| 3b | 1.62 m | 1.62 m | 1.62 m | |||||
| 4 | 1.39 s | 1.37 d s | 1.35 s | 0.73 t | 0.73 m | 1.36 c s | 0.73 t | |
| 5 | 1.35 c s | 1.35 s | 1.36 s | |||||
| NH | 7.54 s | 7.53 s | 7.53 s | 7.46 s | 7.47 s | 7.55 s | 7.47 s | |
| 6Gln | 2 | 3.74 m | 3.73 m | 3.74 m | 3.78 m | 3.78 m | 3.77 m | 3.78 m |
| 3 | 1.98 m | 1.99 m | 1.99 m | 1.95 m | 1.97 m | 1.97 m | 1.97 m | |
| 4a | 2.13 m | 2.13 m | 2.13 m | 2.14 m | 2.14 m | 2.15 m | 2.14 m | |
| 4b | 2.23 m | 2.23 m | 2.23 m | 2.23 m | 2.23 m | 2.23 m | 2.23 m | |
| NH2a | 7.13 s | 7.13 s | 7.17 s | 7.16 s | 7.16 s | 7.16 s | 7.16 s | |
| NH2b | 6.75 s | 6.75 s | 6.75 s | 6.74 s | 6.77 s | 6.76 s | 6.76 s | |
| NH | 7.74 m | 7.74 m | 7.74 m | 7.72 m | 7.73 m | 7.77 m | 7.73 m | |
| 7Aib | 3 | 1.41 d s | 1.41 c s | 1.44 s | 1.42 d s | 1.43 c s | 1.45 d s | 1.43 d s |
| 4 | 1.34 c s | 1.37 d s | 1.39 d s | 1.35 c s | 1.36 d s | 1.35 c s | 1.36 c s | |
| NH | 7.90 s | 7.89 s | 7.89 s | 7.84 s | 7.90 s | 7.86 s | 7.90 s | |
| 8Aib | 3 | 1.46 c s | 1.45 s | 1.46 s | 1.42 d s | 1.45 c s | 1.43 d s | 1.43 d s |
| 4 | 1.38 d s | 1.35 d s | 1.39 d s | 1.38 c s | 1.35 d s | 1.35 c s | 1.39 c s | |
| NH | 7.59 | 7.58 s | 7.58 s | 7.54 s | 7.55 s | 7.59 s | 7.55 s | |
| 9Ala/9Aib | 2 | 3.94 m | 3.93 m | 3.93 m | 3.96 m | 3.96 m | ||
| 3 | 1.40 d | 1.40 d | 1.40 d | 1.46 s | 1.40 d | 1.48 s | 1.40 d | |
| 4 | 1.42 s | 1.43 s | ||||||
| NH | 7.73 m | 7.73 m | 7.73 m | 7.75 s | 7.72 s | 7.79 s | 7.72 s | |
| 10Ser | 2 | 4.09 m | 4.09 m | 4.09 m | 4.02 m | 4.10 m | 4.03 m | 4.11 m |
| 3a | 3.72 m | 3.72 m | 3.73 m | 3.76 m | 3.73 m | 3.75 m | 3.72 m | |
| 3b | 3.77 m | 3.77 m | 3.77 m | 3.78 m | 3.76 m | 3.79 m | 3.76 | |
| OH | 4.86 t | 4.85 t | 4.85 t | - | - | - | 4.87 t | |
| NH | 7.77 m | 7.77 m | 7.76 m | 7.73 m | 7.76 d | 7.74 m | 7.76 m | |
| 11Leu | 2 | 4.27 m | 4.27 m | 4.27 m | 4.28 m | 4.28 m | 4.27 m | 4.28 m |
| 3a | 1.54 m | 1.52 m | 1.53 m | 1.55 m | 1.55 m | 1.56 m | 1.55 m | |
| 3b | 1.67 m | 1.65 m | 1.66 m | 1.68 m | 1.67 m | 1.70 m | 1.67 m | |
| 4 | 1.73 m | 1.68 m | 1.69 m | 1.69 m | 1.69 m | 1.73 m | 1.69 m | |
| 5 | 0.76 d | 0.77 d | 0.77 m | 0.76 d | 0.78 d | 0.76 m | 0.78 d | |
| 6 | 0.81 d | 0.83 d | 0.83 d | 0.83 d | 0.84 d | 0.82 d | 0.84 d | |
| NH | 7.43 d | 7.44 d | 7.43 m | 7.59 d | 7.44 d | 7.59 d | 7.44 m | |
| 12Aib | 3 | 1.40 s | 1.38 e s | 1.36 s | 1.38 s | 1.39 s | 1.37 s | 1.40 s |
| 4 | 1.46 s | 1.45 s | 1.47 s | 1.48 s | 1.46 s | 1.49 s | 1.49 s | |
| NH | 7.78 s | 7.91 s | 7.89 s | 7.93 s | 7.91 s | 7.92 s | 7.93 s | |
| 13Pro | 2 | 4.21 t | 4.23 t | 4.21 t | 4.22 t | 4.22 t | 4.21 t | 4.24 t |
| 3a | 1.58 m | 1.64 m | 1.64 m | 1.65 m | 1.65 m | 1.60 m | 1.65 m | |
| 3b | 2.22 m | 2.22 m | 2.22 m | 2.22 m | 2.22 m | 2.24 m | 2.22 m | |
| 4 | 1.84 m | 1.84 m | 1.84 m | 1.86 m | 1.86 m | 1.86 m | 1.86 m | |
| 5a | 3.38 m | 3.47 m | 3.47 m | 3.52 m | 3.48 m | 3.45 m | 3.48 m | |
| 5b | 3.71 m | 3.69 m | 3.69 m | 3.67 m | 3.70 m | 3.70 m | 3.70 m | |
| 14Leu/14Val | 2 | 3.94 m | 3.76 m | 3.68 m | 3.72 m | 3.68 m | 3.90 m | 3.77 m |
| 3a | 1.51 m | 2.22 m | 2.22 m | 2.21 m | 2.21 m | 1.52 m | 2.21 m | |
| 3b | 1.78 m | 1.78 m | ||||||
| 4 | 1.67 m | 0.93 d | 0.93 d | 0.93 d | 0.94 d | 1.68 m | 0.94 d | |
| 5 | 0.91 d | 0.86 d | 0.86 d | 0.87 d | 0.89 d | 0.92 d | 0.88 d | |
| 6 | 0.82 d | 0.82 d | ||||||
| NH | 7.74 m | 7.59 m | 7.59 m | 7.58 m | 7.59 d | 7.72 m | 7.60 d | |
| 15Aib | 3 | 1.42 d s | 1.43 s | 1.42 c s | 1.42 d s | 1.43 c s | 1.42 d s | 1.45 d s |
| 4 | 1.35 c s | 1.37 d s | 1.35 s | 1.35 c s | 1.38 d s | 1.36 c s | 1.39 c s | |
| NH | 7.63 s | 7.48 s | 7.45 s | 7.43 s | 7.43 s | 7.63 s | 7.43 s | |
| 16Ile | 2 | 3.91 m | 3.84 t | 3.87 t | 3.88 m | 3.88 m | 3.93 m | 3.87 m |
| 3 | 1.87 m | 1.88 m | 1.88 m | 1.88 m | 1.88 m | 1.88 m | 1.88 m | |
| 4a | 1.20 m | 1.20 m | 1.20 m | 1.20 m | 1.20 m | 1.20 m | 1.20 m | |
| 4b | 1.47 m | 1.47 m | 1.47 m | 1.50 m | 1.50 m | 1.47 m | 1.50 m | |
| 5 | 0.81 t | 0.81 t | 0.80 t | 0.80 t | 0.81 t | 0.82 t | 0.81 t | |
| 6 | 0.85 d | 0.85 d | 0.85 d | 0.84 d | 0.85 d | 0.86 d | 0.86 d | |
| NH | 6.96 d | 7.22 m | 7.20 m | 7.20 m | 7.21 m | 6.94 d | 7.22 m | |
| 17Gln/17Glu | 2 | 4.03 m | 3.95 m | 4.01 m | 4.01 m | 4.02 m | 4.06 m | 3.97 m |
| 3a | 1.85 m | 1.90 m | 1.98 m | 1.95 m | 1.95 m | 1.87 m | 1.90 m | |
| 3b | 1.94 m | 1.97 m | ||||||
| 4a | 2.08 m | 2.13 m | 2.41 m | 2.40 m | 2.49 m | 2.37 m | 2.13 m | |
| 4b | 2.18 m | 2.23 m | 2.47 m | 2.30 m | 2.40 m | 2.44 m | 2.23 m | |
| NH | 7.69 d | 7.69 d | 7.67 d | 7.68 d | 7.68 d | 7.68 m | 7.69 d | |
| OMe | 3.55 s | 3.55 s | 3.55 s | 3.54 s | ||||
| NH2a | 6.72 s | 6.72 s | 6.72 s | |||||
| NH2b | 7.13 s | 7.13 s | 7.13 s | |||||
| 18Gln | 2 | 4.04 m | 4.00 m | 4.01 m | 4.03 m | 4.03 m | 4.06 m | 4.03 m |
| 3a | 1.83 m | 1.81 m | 1.83 m | 1.81 m | 1.83 m | 1.83 m | 1.85 m | |
| 3b | 1.70 m | 1.70 m | 1.70 m | 1.70 m | 1.72 m | 1.70 m | 1.72 m | |
| 4a | 2.03 m | 2.04 m | 2.05 m | 2.05 m | 2.05 m | 2.05 m | 2.05 m | |
| 4b | 1.97 m | 1.97 m | 1.97 m | 1.97 m | 1.98 m | 1.97 m | 1.98 m | |
| 5 | ||||||||
| NH2a | 6.64 s | 6.62 s | 6.51 s | 6.61 s | 6.63 s | 6.65 s | 6.63 s | |
| NH2b | 7.10 s | 7.07 s | 7.05 s | 7.04 s | 7.07 s | 7.10 s | 7.07 s | |
| NH | 7.47 d | 7.44 d | 7.45 d | 7.44 d | 7.46 d | 7.48 d | 7.46 d | |
| 19Pheol | 1a | 3.28 m | 3.30 m | 3.30 m | 3.30 m | 3.30 m | 3.28 m | 3.30 m |
| 1b | 3.32 m | 3.32 m | 3.33 m | 3.33 m | 3.33 m | 3.32 m | 3.33 m | |
| 2 | 3.86 m | 3.84 t | 3.86 m | 3.86 m | 3.86 m | 3.87 m | 3.86 m | |
| 3a | 2.61 dd | 2.61 dd | 2.61 dd | 2.61 dd | 2.62 dd | 2.61 dd | 2.63 dd | |
| 3b | 2.83 dd | 2.83 dd | 2.83 dd | 2.84 dd | 2.84 dd | 2.84 dd | 2.84 dd | |
| 5 | 7.20 m | 7.22 m | 7.21 m | 7.21 m | 7.22 m | 7.21 m | 7.22 m | |
| 6 | 7.19 m | 7.18 m | 7.19 m | 7.19 m | 7.20 m | 7.19 m | 7.20 m | |
| 7 | 7.11 m | 7.11 m | 7.11 m | 7.11 m | 7.12 m | 7.13 m | 7.12 m | |
| OH | 4.70 t | 4.69 t | 4.67 t | 4.69 t | ||||
| NH | 7.27 d | 7.15 m | 7.18 m | 7.18 m | 7.19 m | 7.30 d | 7.18 m |
| Position | TA1909 (2) a | TA1895 (3) a | TA1896 (4) b | TA1924 (5) b | TA1910 (6) a | TA1924a (7) a | TA1909a (8) a | |
|---|---|---|---|---|---|---|---|---|
| Ac | 1 | 171.1 s | 171.2 s | 171.1s | 171.0 s | 171.1 s | 171.1 s | 171.1 s |
| 2 | 23.1 q | 23.3 q | 23.2 q | 23.0 q | 23.0 q | 23.1 q | 23.1 q | |
| 1Aib | 1 | 176.0 e s | 176.0 e s | 175.9 e s | 175.6 e s | 175.8 s | 175.9 e s | 175.8 e s |
| 2 | 55.7 d s | 55.8 d s | 55.8 s | 55.8 s | 55.8 s | 55.7 d s | 55.8 d s | |
| 3 | 26.5 g q | 26.8 q | 26.5 g q | 26.5 q | 26.5 q | 26.5 q | 26.3 q | |
| 4 | 24.1 q | 24.2 q | 24.1 q | 24.1 q | 24.1 q | 24.1 q | 23.7 q | |
| 2Ala | 1 | 174.8 s | 174.6 s | 174.6 s | 174.5 s | 174.6 s | 174.6 s | |
| 2 | 51.0 d | 51.0 d | 51.0 d | 50.8 d | 50.9 d | 50.9 d | 50.9 d | |
| 3 | 16.8 q | 16.8 q | 16.8 q | 16.8 q | 16.9 q | 16.8 q | 16.9 q | |
| 3Ala | 1 | 174.8 s | 174.8 s | 174.8 s | 174.5 s | 174.6 s | 174.7 s | 174.6 s |
| 2 | 51.4 d | 51.3 d | 51.3 d | 51.0 d | 51.1 d | 51.2 d | 51.1 d | |
| 3 | 16.1 q | 16.1 q | 16.1 q | 16.2 q | 16.2 q | 16.1 q | 16.2 q | |
| 4Aib | 1 | 175.0 s | 175.0 s | 175.0 s | 175.0 s | 175.0 s | 175.0 s | 175.0 s |
| 2 | 56.0 s | 56.0 s | 55.7 d s | 55.9 d s | 56.4 s | 56.0 d s | 56.4 s | |
| 3 | 26.2q | 25.8 j q | 26.2 g q | 26.5 q | 26.0 g q | 26.1 q | 26.3 g q | |
| 4 | 23.3 f q | 22.6 f q | 23.4 h q | 22.9 h q | 23.1 f q | 22.5 f q | 23.3 f q | |
| 5Aib/5Iva | 1 | 176.0 e s | 176.0 e s | 175.9 e s | 176.4 s | 176.2 s | 176.0 e s | 176.2 s |
| 2 | 55.8 d s | 55.7 d s | 55.8 d s | 58.6 s | 58.5 s | 55.8 d s | 58.5 s | |
| 3 | 26.7 g q | 26.7 g s | 26.5 g q | 25.6 t | 25.8 t | 26.6 q | 25.9 t | |
| 4 | 22.6 q | 22.8 f q | 22.6 h q | 7.4 q | 7.4 q | 22.7 f q | 7.4 q | |
| 5 | 22.7 h q | 22.7 q | 22.6 q | |||||
| 6Gln | 1 | 174.1 s | 174.1 s | 174.1 s | 173.8 s | 174.1 s | 173.8 s | 174.1 s |
| 2 | 56.2 d d | 56.4 d | 56.4 d | 56.0 d | 56.0 d | 56.0 d | 56.0 d | |
| 3 | 26.2 t | 26.5 t | 26.2 t | 26.2 t | 26.2 t | 26.2 t | 26.2 t | |
| 4 | 31.5 c t | 31.5 c t | 31.5 c t | 31.6 c t | 31.6 c t | 31.5 c t | 31.7 c t | |
| 5 | 173.6 s | 173.6 k s | 173.6 f s | 173.8 s | 173.6 s | 173.7 s | 173.6 f s | |
| 7Aib | 1 | 176.0 e s | 176.0 e s | 176.0 e s | 175.7 e s | 176.0 e s | 176.0 e s | 176.0 e s |
| 2 | 56.0 s | 56.0 s | 55.9 d s | 55.9 d s | 56.4 d s | 55.9 d s | 56.2 d s | |
| 3 | 26.4 g q | 26.2 j q | 26.5 g q | 26.2 g q | 26.3 g q | 26.2 q | 26.2 g q | |
| 4 | 23.0 q | 23.0 f q | 22.9 h q | 22.6 h q | 22.7 e q | 22.8 f q | 22.9 f q | |
| 8Aib | 1 | 176.3 s | 176.2 s | 176.2 s | 175.9 e s | 176.4 s | 175.6 s | 176.4 s |
| 2 | 56.0 s | 56.0 s | 56.0 d s | 56.1 d s | 56.0 s | 56.0 d s | 56.1 d s | |
| 3 | 26.8 h q | 26.8 q | 26.8 g q | 25.8 g q | 26.8 g q | 26.6 g q | 26.4 g q | |
| 4 | 22.7 q | 23.2 h q | 23.3 h q | 23.0 h q | 22.9 e q | 22.8 f q | 23.2 e q | |
| 9Ala/9Aib | 1 | 174.7 s | 174.6 s | 174.6 s | 176.5 s | 174.6 s | 176.4 s | 174.6 s |
| 2 | 51.9 d | 51.8 d | 51.8 d | 56.0 d s | 51.7 d | 56.1 d s | 51.7 d | |
| 3 | 16.5 q | 16.5 q | 16.5 q | 26.2 g q | 16.5 q | 26.5 g q | 16.5 q | |
| 4 | 23.2 h q | 22.9 f q | ||||||
| 10Ser | 1 | 170.7 s | 170.7 s | 170.7 s | 170.6 s | 170.6 s | 170.6 s | 170.6 s |
| 2 | 58.3 d | 58.2 d | 58.2 d | 58.8 d | 58.1 d | 58.9 d | 58.1 d | |
| 3 | 61.1 t | 61.1 t | 61.1 t | 61.0 t | 61.1 t | 61.2 t | 61.0 h t | |
| 11Leu | 1 | 173.5 s | 173.3 s | 173.3 s | 173.5 s | 173.3 s | 173.7 s | 173.3 s |
| 2 | 51.6 d | 51.5 d | 51.5 d | 51.4 d | 51.4 d | 51.6 d | 51.4 d | |
| 3 | 39.5 t | 39.5 t | 39.5 t | 39.8 t | 39.8 t | 39.2 t | 39.8 t | |
| 4 | 24.0 d | 24.2 d | 24.2 d | 24.1 d | 24.2 d | 23.9 d | 24.0 d | |
| 5 | 20.6 q | 21.0 q | 20.8 q | 20.8 q | 20.9 q | 20.5 q | 21.0 q | |
| 6 | 23.0 q | 23.1 q | 23.1 q | 23.4 q | 22.9 q | 23.0 q | 22.9 q | |
| 12Aib | 1 | 173.0 s | 172.7 s | 172.8 s | 172.8 s | 172.8 s | 173.0 s | 172.7 s |
| 2 | 55.9 s | 56.4 s | 56.1 d s | 56.1 d s | 56.2 d s | 56.3 d s | 56.1 d s | |
| 3 | 25.6 q | 26.3 g q | 26.4 g q | 26.4 g q | 26.4 q | 25.8 q | 25.5 q | |
| 4 | 23.4 f q | 23.1 q | 23.2 h q | 22.6 q | 23.0 q | 23.1 q | 22.9 q | |
| 13Pro | 1 | 173.4 s | 173.8 s | 173.8 s | 173.7 s | 173.8 s | 173.3 s | 173.8 s |
| 2 | 63.0 d | 63.0 d | 63.0 d | 63.0 d | 63.0 d | 63.0 d | 63.0 d | |
| 3 | 28.8 t | 28.8 t | 28.8 t | 28.7 t | 28.7–28.8 t | 28.7 t | 29.1 t | |
| 4 | 25.9 t | 25.8 t | 25.8 t | 26.0 t | 25.9 t | 26.1 t | 26.0 t | |
| 5 | 48.7 t | 48.6 t | 48.6 t | 48.5 t | 48.6 t | 48.7 t | 48.6 t | |
| 14Leu/14Val | 1 | 173.7 | 172.7 | 172.6 | 172.7 s | 172.7 s | 173.8 s | 172.7 s |
| 2 | 53.1 d | 61.0 d | 61.1 d | 61.1 d | 61.1 d | 53.6 d | 61.1 h d | |
| 3 | 38.7 t | 28.8 d | 28.8 d | 28.8 d | 28.7–28.8 d | 38.7 t | 28.8 d | |
| 4 | 24.8 d | 19.1 q | 19.1 k q | 19.1 q | 19.2 q | 24.8 d | 19.2 q | |
| 5 | 23.0 q | 19.1 q | 19.2 k q | 19.2 q | 19.1 q | 23.0 q | 19.2 q | |
| 6 | 21.1 q | 20.9 q | ||||||
| 15Aib | 1 | 175.4 s | 175.7 s | 175.6 s | 175.6 s | 175.7 s | 175.3 s | 175.7 s |
| 2 | 56.3 s | 56.4 s | 56.2 d s | 55.9 d s | 56.1 d s | 56.2 d s | 56.1 d s | |
| 3 | 26.3 g q | 26.4 g q | 26.2 g q | 26.6 g q | 26.5 g q | 26.1 g q | 26.3 g q | |
| 4 | 23.4 f q | 23.4 f q | 22.9 h q | 23.3 h q | 23.3 f q | 23.4 f q | 23.2 f q | |
| 16Ile | 1 | 172.0 s | 172.4 s | 172.3 s | 172.2 s | 172.3 s | 171.9 s | 172.4 s |
| 2 | 58.9 d | 59.6 d | 59.5 d | 59.5 d | 59.5 d | 58.9 d | 59.6 d | |
| 3 | 35.6 d | 35.5 d | 35.5 d | 35.5 d | 35.5 d | 35.6 d | 35.5 d | |
| 4 | 25.0 t | 25.4 t | 25.3 t | 25.4 t | 25.4 t | 25.0 t | 25.4 t | |
| 5 | 11.4 q | 11.4 q | 11.4 q | 11.4 q | 11.4 q | 11.5 q | 11.4 q | |
| 6 | 15.7 q | 15.7 q | 15.7 q | 15.7 q | 15.7 q | 15.7 q | 15.7 q | |
| 17Glu/17Gln | 1 | 171.5 s | 171.8 s | 171.4 s | 171.6 s | 171.4 s | 171.1 s | 171.8 s |
| 2 | 53.8 d | 54.3 d | 53.7 d | 53.8 d | 53.7 d | 53.1 d | 54.3 d | |
| 3 | 27.1 t | 26.8 t | 26.0 t | 26.3 t | 26.3 t | 26.5 t | 26.8 t | |
| 4 | 31.7 c t | 31.8 c t | 30.2 t | 30.5 t | 30.2 t | 30.1 t | 31.7 c t | |
| 5 | 173.7 s | 173.7 k s | 173.0 s | 172.9 s | 173.0 s | 172.9 s | 173.7 f s | |
| OMe | 51.4 q | 51.4 q | 51.4 q | 51.4 q | ||||
| 18Gln | 1 | 170.9 s | 171.0 s | 170.9 s | 170.9 s | 170.9 s | 170.8 s | 171.0 s |
| 2 | 53.1 d | 53.4 d | 53.3 d | 53.3 d | 53.3 d | 53.0 d | 53.4 d | |
| 3 | 27.7 t | 27.5 t | 27.5 t | 27.5 t | 27.5 t | 27.7 t | 27.4 t | |
| 4 | 31.7 c t | 31.7 c t | 31.7 c t | 31.7 c t | 31.7 c t | 31.6 c t | 31.5 c t | |
| 5 | 173.7 | 173.8 k s | 173.5 f s | 173.6 s | 173.6 | 173.6 s | 173.8 f s | |
| 19PheOH | 1 | 62.7 t | 62.9 t | 62.8 t | 62.8 t | 62.8 t | 62.7 t | 62.9 t |
| 2 | 52.6 d | 52.6 d | 52.6 d | 52.6 d | 52.6 d | 52.5 d | 52.6 d | |
| 3 | 36.7 t | 36.7 t | 36.7 t | 36.7 t | 36.7 t | 36.6 t | 36.7 t | |
| 4 | 139.2 s | 139.3 s | 139.3 s | 139.3 s | 139.3 s | 139.2 s | 139.3 s | |
| 5 | 129.4 d | 129.4 d | 129.4 d | 129.4 d | 129.4 d | 129.4 d | 129.4 d | |
| 6 | 128.2 d | 128.1 d | 128.1 d | 128.1 d | 128.1 d | 128.1 d | 128.1 d | |
| 7 | 126.0 d | 126.0 d | 125.9 d | 126.0 d | 126.0 d | 126.0 d | 126.0 d |
2.3. Determination of the Absolute Stereochemistry
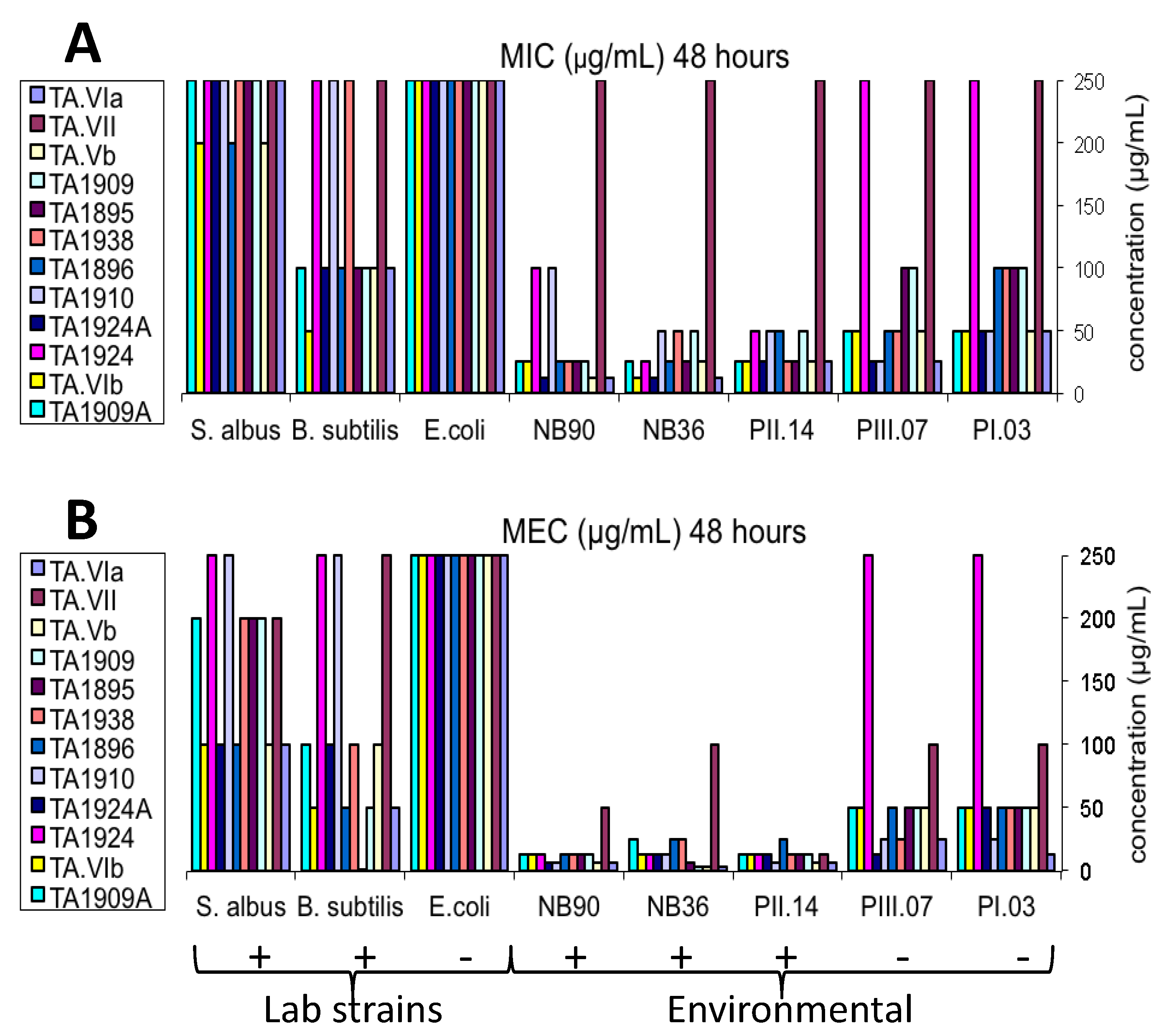
2.4. Antibacterial Bioassay
3. Experimental Section
3.1. General Experimental Procedures
3.2. Biological Material
3.3. Culture Procedure
3.4. Isolation Procedure
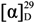 −25 (c 0.68, MeOH); UV (MeOH) λmax (log ε) 203 (4.61); IR (KBr) νmax 3320, 2960, 2360, 1661, 1541, 1204 cm−1; 1H and 13C NMR (see Table 2); HR TOF-MS-ES+ m/z 1961.1099 [M + Na]+ (calcd for C91H151N21NaO25, 1961.1088). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.5 min (d-Ile 47.1 min), l-Leu 45.3 min (d-Leu 47.7 min), l-Pro 35.8 min (d-Pro 36.4 min), l-Glu 32.7 min (d-Glu 33.6 min) and l-Ser 29.7 min (d-Ser 33.2 min). Advanced Marfey: l-Ala (l-Ala-l-FDAA, 3.40 min; l-Ala-d-FDAA, 3.99 min). Iva, not determined; Pheol, (l as in TA1909).
−25 (c 0.68, MeOH); UV (MeOH) λmax (log ε) 203 (4.61); IR (KBr) νmax 3320, 2960, 2360, 1661, 1541, 1204 cm−1; 1H and 13C NMR (see Table 2); HR TOF-MS-ES+ m/z 1961.1099 [M + Na]+ (calcd for C91H151N21NaO25, 1961.1088). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.5 min (d-Ile 47.1 min), l-Leu 45.3 min (d-Leu 47.7 min), l-Pro 35.8 min (d-Pro 36.4 min), l-Glu 32.7 min (d-Glu 33.6 min) and l-Ser 29.7 min (d-Ser 33.2 min). Advanced Marfey: l-Ala (l-Ala-l-FDAA, 3.40 min; l-Ala-d-FDAA, 3.99 min). Iva, not determined; Pheol, (l as in TA1909).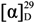 −23 (c 0.4, MeOH); UV (MeOH) λmax (log ε) 203 (4.46); IR (ATR probe) νmax 3309, 1647, 1541, 1201 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S3); HR TOF-MS-ES+ m/z 1932.0946 [M + Na]+ (calcd for C89H148N22NaO24, 1932.0935). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.2 min (d-Ile 47.8 min), l-Leu 45.0 min (d-Leu 47.7 min), l-Pro 35.5 min (d-Pro 36.4 min), l-Glu 32.7 min (d-Glu 33.6 min), l-Pheol: as l-Phe 45.6 min (D-Phe 47.7 min), l-Ser 29.4 min (d-Ser 33.2 min) and Ala, (l as in TA1938).
−23 (c 0.4, MeOH); UV (MeOH) λmax (log ε) 203 (4.46); IR (ATR probe) νmax 3309, 1647, 1541, 1201 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S3); HR TOF-MS-ES+ m/z 1932.0946 [M + Na]+ (calcd for C89H148N22NaO24, 1932.0935). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.2 min (d-Ile 47.8 min), l-Leu 45.0 min (d-Leu 47.7 min), l-Pro 35.5 min (d-Pro 36.4 min), l-Glu 32.7 min (d-Glu 33.6 min), l-Pheol: as l-Phe 45.6 min (D-Phe 47.7 min), l-Ser 29.4 min (d-Ser 33.2 min) and Ala, (l as in TA1938).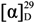 −32 (c 0.26, MeOH); UV (MeOH) λmax (log ε) 203 (4.48); IR (ATR probe) νmax 3308, 2936, 1638, 1537, 1202, 1047 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S4); HR TOF-MS-ES+ m/z 1918.0864 [M + Na]+ (calcd for C88H146N22NaO24, 1918.0778). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.3 min (d-Ile 47.8 min), l-Leu 45.0 min (d-Leu 47.7 min), l-Pro 35.6 min (d-Pro 36.4 min), l-Glu 32.7 min (d-Glu 33.6 min), l-Ser 29.5 min (d-Ser 33.2 min) and l-Val 40.9 min (d-Val 43.7 min). Pheol, (l as in TA1909); Ala, (l as in TA1938).
−32 (c 0.26, MeOH); UV (MeOH) λmax (log ε) 203 (4.48); IR (ATR probe) νmax 3308, 2936, 1638, 1537, 1202, 1047 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S4); HR TOF-MS-ES+ m/z 1918.0864 [M + Na]+ (calcd for C88H146N22NaO24, 1918.0778). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.3 min (d-Ile 47.8 min), l-Leu 45.0 min (d-Leu 47.7 min), l-Pro 35.6 min (d-Pro 36.4 min), l-Glu 32.7 min (d-Glu 33.6 min), l-Ser 29.5 min (d-Ser 33.2 min) and l-Val 40.9 min (d-Val 43.7 min). Pheol, (l as in TA1909); Ala, (l as in TA1938).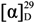 −18 (c 0.48, MeOH); UV (MeOH) λmax (log ε) 203 (4.56); IR (ATR probe) νmax 3312, 1653, 1540, 1201 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S5); HR TOF-MS-ES+ m/z 1933.0819 [M + Na]+ (calcd for C89H147N21NaO25, 1933.0775). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.3 min (d-Ile 47.8 min), l-Leu 45.1 min (d-Leu 47.7 min), l-Pro 35.7 min (d-Pro 36.4 min), l-Glu 32.7 min (d-Glu 33.6 min), l-Ser 29.6 min (d-Ser 33.2 min), l-Val 41.0 min (d-Val 43.7 min), Pheol, (l as in TA1909) and Ala, (l as in TA1938).
−18 (c 0.48, MeOH); UV (MeOH) λmax (log ε) 203 (4.56); IR (ATR probe) νmax 3312, 1653, 1540, 1201 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S5); HR TOF-MS-ES+ m/z 1933.0819 [M + Na]+ (calcd for C89H147N21NaO25, 1933.0775). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.3 min (d-Ile 47.8 min), l-Leu 45.1 min (d-Leu 47.7 min), l-Pro 35.7 min (d-Pro 36.4 min), l-Glu 32.7 min (d-Glu 33.6 min), l-Ser 29.6 min (d-Ser 33.2 min), l-Val 41.0 min (d-Val 43.7 min), Pheol, (l as in TA1909) and Ala, (l as in TA1938).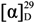 −17 (c 0.26, MeOH); UV (MeOH) λmax (log ε) 203 (4.48); IR (KBr) νmax 3335, 2966, 2363, 1663, 1541, 1207 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S6); HR TOF-MS-ES+ m/z 1961.1240 [M + Na]+ (calcd for C91H151N21NaO25, 1961.1088). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.4 min (d-Ile 47.8 min), l-Leu 45.1 min (d-Leu 47.7 min), L-Val 41.0 min (D-Val 43.7 min), l-Pro 35.7 min (d-Pro 36.4 min), l-Glu 32.7 min (d-Glu 33.6 min) and l-Ser 29.3 min (d-Ser 33.2 min). Iva, not determined; Pheol, (l as in TA1909); Ala, (l as in TA1938).
−17 (c 0.26, MeOH); UV (MeOH) λmax (log ε) 203 (4.48); IR (KBr) νmax 3335, 2966, 2363, 1663, 1541, 1207 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S6); HR TOF-MS-ES+ m/z 1961.1240 [M + Na]+ (calcd for C91H151N21NaO25, 1961.1088). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.4 min (d-Ile 47.8 min), l-Leu 45.1 min (d-Leu 47.7 min), L-Val 41.0 min (D-Val 43.7 min), l-Pro 35.7 min (d-Pro 36.4 min), l-Glu 32.7 min (d-Glu 33.6 min) and l-Ser 29.3 min (d-Ser 33.2 min). Iva, not determined; Pheol, (l as in TA1909); Ala, (l as in TA1938). 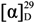 −36 (c 0.39, MeOH); UV (MeOH) λmax (log ε) 203 (4.60); IR (ATR probe) νmax 3310, 1654, 1541, 1217 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S7); HR TOF-MS-ES+ m/z 1947.0995 [M + Na]+ (calcd for C90H149N21NaO25, 1947.0931). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.5 min (d-Ile 47.8 min), l-Leu 45.3 min (d-Leu 48.1 min), l-Pro 35.8 min (d-Pro 37.0 min), l-Glu 33.4 min (d-Glu 34.1 min), l-Ser 29.8 min (d-Ser 33.6 min) and l-Val 41.2 min (d-Val 44.1 min). Iva, not determined; Pheol, (l as in TA1909); Ala, (l as in TA1938).
−36 (c 0.39, MeOH); UV (MeOH) λmax (log ε) 203 (4.60); IR (ATR probe) νmax 3310, 1654, 1541, 1217 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S7); HR TOF-MS-ES+ m/z 1947.0995 [M + Na]+ (calcd for C90H149N21NaO25, 1947.0931). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.5 min (d-Ile 47.8 min), l-Leu 45.3 min (d-Leu 48.1 min), l-Pro 35.8 min (d-Pro 37.0 min), l-Glu 33.4 min (d-Glu 34.1 min), l-Ser 29.8 min (d-Ser 33.6 min) and l-Val 41.2 min (d-Val 44.1 min). Iva, not determined; Pheol, (l as in TA1909); Ala, (l as in TA1938).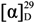 −30 (c 0.21, MeOH); UV (MeOH) λmax (log ε) 203 (4.67); IR (ATR probe) νmax 3310, 1655, 1541, 1204 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S8); HR TOF-MS-ES+ m/z 1961.1086 [M + Na]+ (calcd for C90H149N21NaO25, 1961.1088). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.4 min (d-Ile 47.8 min), l-Leu 45.2 min (d-Leu 48.1 min), l-Pro 35.7 min (d-Pro 37.0 min), l-Glu 32.7 min (d-Glu 34.1 min) and l-Ser 29.7 min (d-Ser 33.6 min), Pheol, (l as in TA1909); Ala, (l as in TA1938).
−30 (c 0.21, MeOH); UV (MeOH) λmax (log ε) 203 (4.67); IR (ATR probe) νmax 3310, 1655, 1541, 1204 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S8); HR TOF-MS-ES+ m/z 1961.1086 [M + Na]+ (calcd for C90H149N21NaO25, 1961.1088). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.4 min (d-Ile 47.8 min), l-Leu 45.2 min (d-Leu 48.1 min), l-Pro 35.7 min (d-Pro 37.0 min), l-Glu 32.7 min (d-Glu 34.1 min) and l-Ser 29.7 min (d-Ser 33.6 min), Pheol, (l as in TA1909); Ala, (l as in TA1938).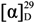 −49 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 203 (4.60); IR (ATR probe) νmax 3312, 2292, 1661, 1541, 1204 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S9); HR TOF-MS-ES+ m/z 1932.1022 [M + Na]+ (calcd for C89H148N22NaO24, 1,932.0935). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.5 min (d-Ile 47.8 min), l-Leu 45.2 min (d-Leu 48.1 min), l-Val 41.1 min (d-Val 44.1 min), l-Pro 35.9 min (d-Pro 37.0 min), l-Glu 32.7 min (d-Glu 34.1 min) and l-Ser 29.7 min (d-Ser 33.6 min). Iva, not determined; Pheol, (l as in TA1909); Ala, (l as in TA1938).
−49 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 203 (4.60); IR (ATR probe) νmax 3312, 2292, 1661, 1541, 1204 cm−1; 1H and 13C NMR (see Table 3 and Table 4 and Supplementary Table S9); HR TOF-MS-ES+ m/z 1932.1022 [M + Na]+ (calcd for C89H148N22NaO24, 1,932.0935). Retention time of amino acid (AA) Marfey’s derivatives: l-Ile 44.5 min (d-Ile 47.8 min), l-Leu 45.2 min (d-Leu 48.1 min), l-Val 41.1 min (d-Val 44.1 min), l-Pro 35.9 min (d-Pro 37.0 min), l-Glu 32.7 min (d-Glu 34.1 min) and l-Ser 29.7 min (d-Ser 33.6 min). Iva, not determined; Pheol, (l as in TA1909); Ala, (l as in TA1938).3.5. Antibacterial Bioassay
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Chugh, K.J.; Wallace, B.A. Peptaibols: Models for ion channels. Biochem. Soc. Trans. 2001, 29, 565–570. [Google Scholar] [CrossRef]
- Schuhmacher, R.; Stoppacher, N.; Zeilinger, S. Peptaibols of Trichoderma atroviride: Screening, Identification, and Structure Elucidation by Liquid Chromatography-Tandem Mass Spectrometry. In Communicating Current Research and Education Topics and Trends in Applied Microbiology; Méndez-Vilas, A., Ed.; Formatex: Badajoz, Spain, 2007; pp. 609–617. [Google Scholar]
- Kleinkauf, H.; Dohren, H. Nonribosomal biosynthesis of peptide antibiotics. Eur. J. Biochem. 1990, 192, 1–15. [Google Scholar]
- Peptaibol Database. Available online: http://www.cryst.bbk.ac.uk/peptaibol (accessed on 14 May 2012).
- Benitez, T.; Rincon, A.M.; Limon, M.C.; Codon, A.C. Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 2004, 7, 249–260. [Google Scholar]
- Bodo, B.; Rebuffat, S.; El Hajii, M.; Davoust, D. Structure of Trichorzianine A IIIC, an antifungal peptide from Trichoderma harzianum. J. Am. Chem. Soc. 1985, 107, 6011–6017. [Google Scholar] [CrossRef]
- El Hajii, M.; Rebuffat, S.; Lecommandeur, D.; Bodo, B. Isolation and sequence determination of trichorzianines A antifungal peptides from Trichoderma harzianum. Int. J. Peptide Prot. Res. 1987, 29, 207–215. [Google Scholar]
- Rebuffat, S.; El Hajii, M.; Hennig, P.; Davoust, D.; Bodo, B. Isolation, sequence and conformation of seven trichorzianines B from Trichoderma harzianum. Int. J. Peptide. Prot. Res. 1989, 34, 200–210. [Google Scholar]
- Pocsfalvi, G.; Scala, F.; Lorito, M.; Ritieni, A.; Randazzo, G.; Ferranti, P.; Vekey, K.; Malorni, A. Microheterogeneity characterization of a trichorzianine-A mixture from Trichoderma harzianum. J. Mass. Spectrom. 1998, 33, 154–163. [Google Scholar] [CrossRef]
- Carroux, A.; van Bohemen, A.-I.; Roullier, C.; du Pont, T.R.; Vansteelandt, M.; Bondon, A.; Zalouk-Vergnoux, A.; Pouchus, Y.F.; Ruiz, N. Unprecedented 17-residue peptaibiotics produced by marine-derived Trichoderma atroviride. Chem. Biodivers. 2013, 10, 772–786. [Google Scholar] [CrossRef]
- Oh, S.U.; Lee, S.J.; Kim, J.H.; Yoo, I.D. Structural elucidation of new antibiotic peptides, atroviridins A, B and C from Trichoderma atroviride. Tetrahedron Lett. 2000, 41, 61–64. [Google Scholar] [CrossRef]
- Oh, S.U.; Yun, B.S.; Lee, S.J.; Kim, J.H.; Yoo, I.D. Atroviridins A–C and neoatroviridins A–D, novel peptaibol antibiotics produced by Trichoderma atroviride F80317. I. Taxonomy, fermentation, isolation and biological activity. J. Antibiot. 2002, 55, 557–564. [Google Scholar] [CrossRef]
- Stoppacher, N.; Reithner, B.; Omann, M.; Zeilinger, S.; Krska, R.; Schuhmacher, R. Profiling of trichorzianines in culture samples of Trichoderma atroviride by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 3963–3970. [Google Scholar]
- Sabareesh, V.; Balaram, P. Tandem electrospray mass spectrometric studies of proton and sodium ion adducts of neutral peptides with modified N- and C-termini: Synthetic model peptides and microheterogeneous peptaibol antibiotics. Rapid Commun. Mass Spectrom. 2006, 20, 618–628. [Google Scholar] [CrossRef]
- Marfey, P. Determination of d-amino acids. II. Use of a bifunctional reagent, 1,5-fluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1987, 49, 591–596. [Google Scholar] [CrossRef]
- Fujii, K.; Shimoya, T.; Ikai, Y.; Oka, H.; Harada, K. Further application of Advanced Marfey’s method for determination of absolute configuration of primary amino compound. Tetrahedron Lett. 1998, 39, 2579–2582. [Google Scholar] [CrossRef]
- Bowden, K.; Heilbron, I.M.; Jones, E.R.H.; Weedon, B.C.L. Researches on acetylenic compounds. Part I. The preparation of acetylenic ketones by oxidation of acetylenic carbinols and glycols. J. Chem. Soc. 1946, 39–45. [Google Scholar]
- Gal-Hemed, I.; Atanasova, L.; Komon-Zelazowska, M.; Druzhinina, I.S.; Viterbo, A.; Yarden, O. Marine isolates of Trichoderma as potential halotolerant agents of biological control for arid-zone agriculture. Appl. Environ. Microbiol. 2011, 77, 5100–5109. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocol: A Guide to Methods and Applications; Innis, M.A., Gellfand, D.H., Sninisky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Paz, Z.; Komon-Zelazowska, M.; Druzhinina, I.S.; Aveskamp, M.M.; Shnaiderman, A.; Aluma, Y.; Carmeli, S.; Ilan, M.; Yarden, O. Diversity and potential antifungal properties of fungi associated with a Mediterranean sponge. Fungal Divers. 2010, 42, 17–26. [Google Scholar] [CrossRef]
- International Subcommission on Trichderma and Hypocrea Taxonomy. Available online: http://www.isth.info (accessed on 11 August 2011).
- National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov (accessed on 20 September 2012).
- Pitcher, D.J.; Saunders, N.A.; Owen, R.J. Rapid extraction of bacterial genomic DNA with guanidinium thiocyanate. Lett. Appl. Microbiol. 1989, 8, 151–156. [Google Scholar] [CrossRef]
- Oh, D.C.; Jensen, P.R.; Fenical, W. Zygosporamide, a cytotoxic cyclic depsipeptide from the marine-derived fungus Zygosporium masonii. Tetrahedron Lett. 2006, 47, 8625–8628. [Google Scholar] [CrossRef]
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Panizel, I.; Yarden, O.; Ilan, M.; Carmeli, S. Eight New Peptaibols from Sponge-Associated Trichoderma atroviride. Mar. Drugs 2013, 11, 4937-4960. https://doi.org/10.3390/md11124937
Panizel I, Yarden O, Ilan M, Carmeli S. Eight New Peptaibols from Sponge-Associated Trichoderma atroviride. Marine Drugs. 2013; 11(12):4937-4960. https://doi.org/10.3390/md11124937
Chicago/Turabian StylePanizel, Irina, Oded Yarden, Micha Ilan, and Shmuel Carmeli. 2013. "Eight New Peptaibols from Sponge-Associated Trichoderma atroviride" Marine Drugs 11, no. 12: 4937-4960. https://doi.org/10.3390/md11124937
APA StylePanizel, I., Yarden, O., Ilan, M., & Carmeli, S. (2013). Eight New Peptaibols from Sponge-Associated Trichoderma atroviride. Marine Drugs, 11(12), 4937-4960. https://doi.org/10.3390/md11124937