Functional Genomics of Novel Secondary Metabolites from Diverse Cyanobacteria Using Untargeted Metabolomics
Abstract
:1. Introduction
| Taxonomic Subsection | Species | Strain | Abbreviation | Medium |
|---|---|---|---|---|
| 1 (Chroococcales) | Halothece sp. | PCC 7418 | 7418 | ASNIII/Tu4X |
| 1 (Chroococcales) | Synechococcus elongatus | PCC 6301 | 6301 | BG11 |
| 1 (Chroococcales) | Synechococcus sp. | PCC 7002 | 7002 | ASNIII/BG11 + vit. B12 |
| 2 (Pleurocapsales) | Chroococcidiopsis sp. | PCC 6712 | 6712 | ASNIII/BG11 |
| 2 (Pleurocapsales) | Pleurocapsa sp. | PCC 7327 | 7327 | BG11 |
| 3 (Oscillatoriales) | Geitlerinema sp. | PCC 7407 | 7407 | BG11 |
| 3 (Oscillatoriales) | Leptolyngbya sp. | PCC 7376 | 7376 | ASNIII + vit. B12 |
| 3 (Oscillatoriales) | Microcoleus vaginatus | PCC 9802 | 9802 | BG11 |
| 4 (Nostocales) | Calothrix sp. | PCC 7507 | 7507 | BG11o |
| 4 (Nostocales) | Nostoc sp. | PCC 7107 | 7107 | BG11o |
2. Results and Discussion
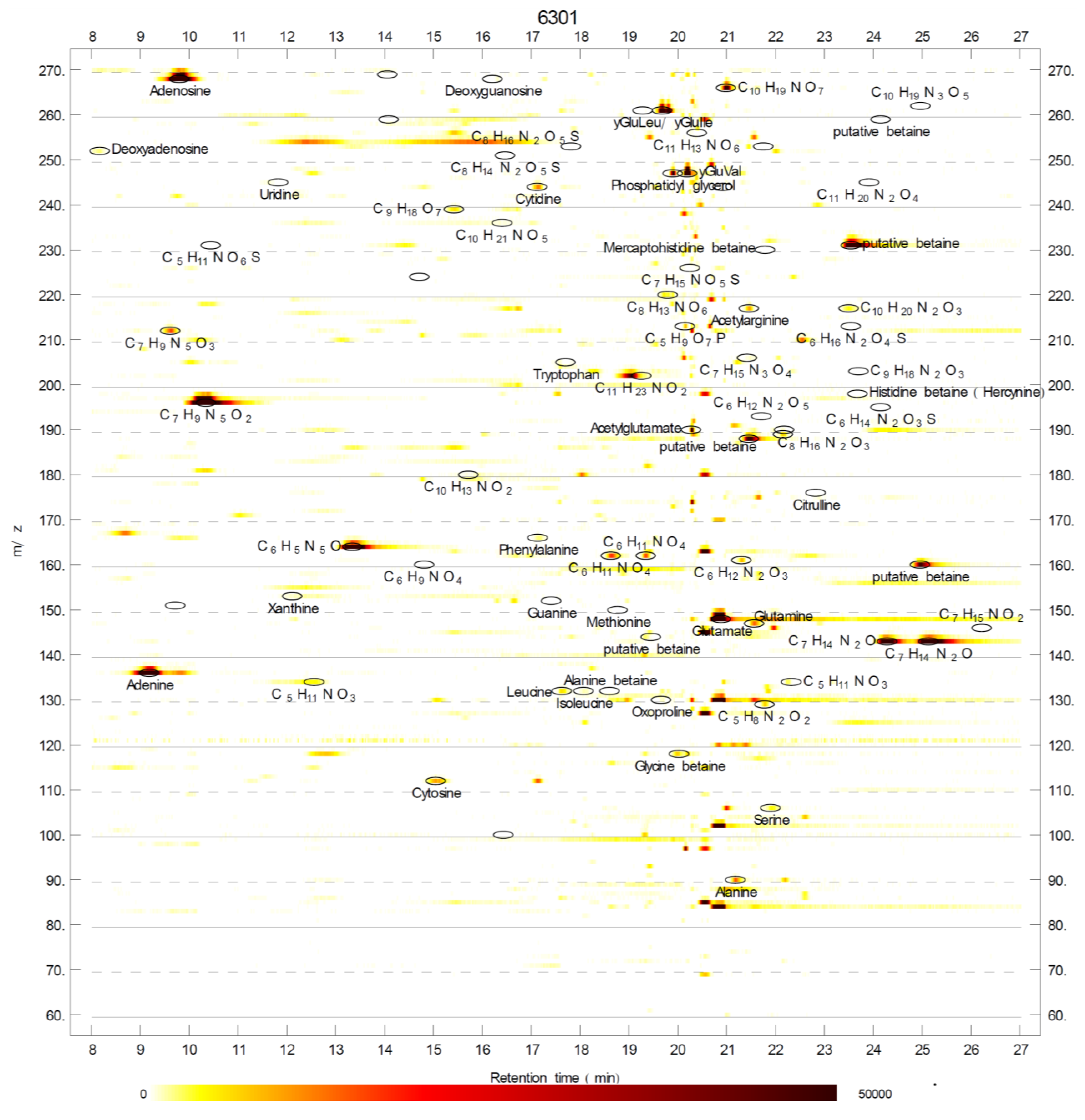
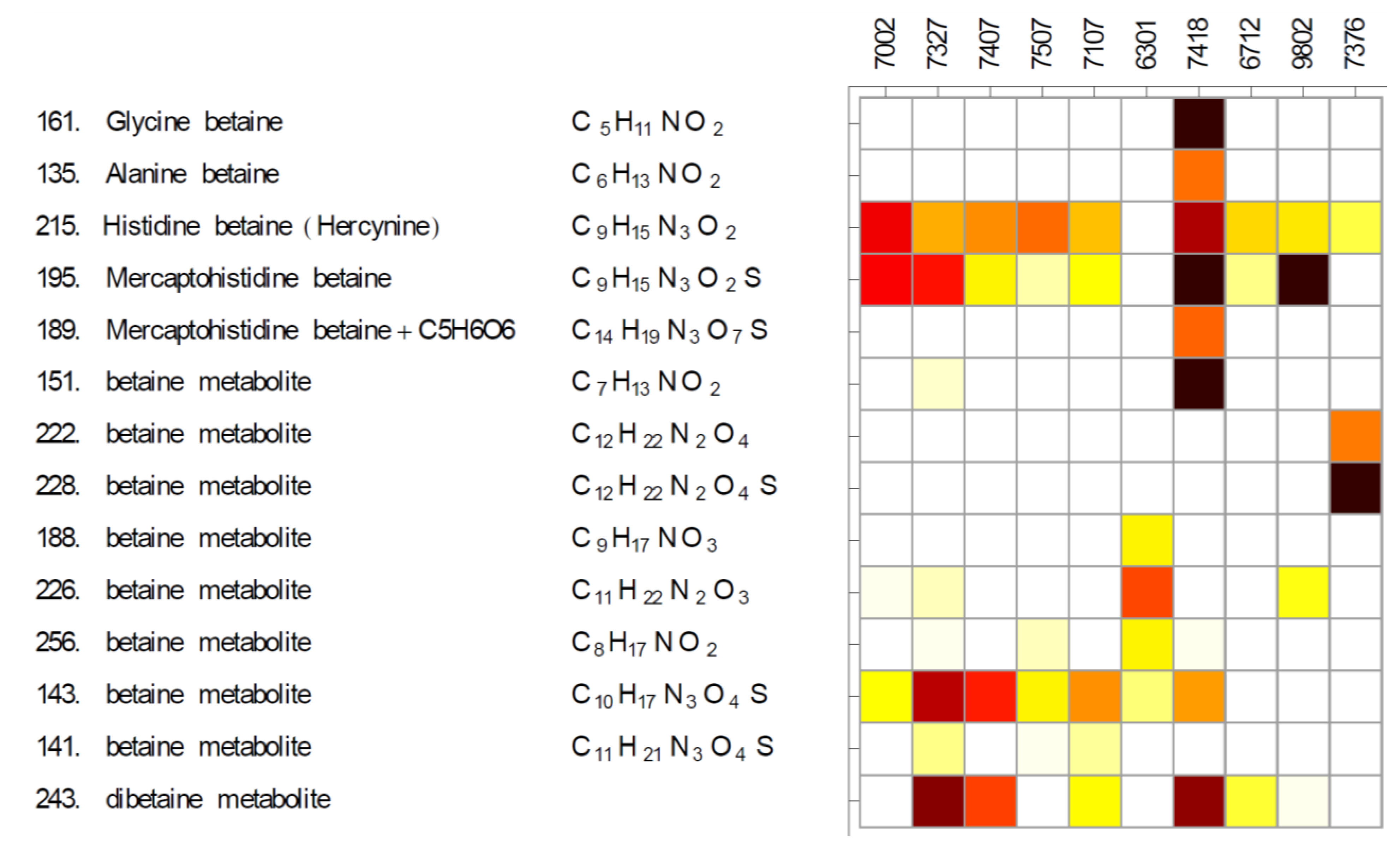
2.1. Betaines and Their Biosynthetic Genes
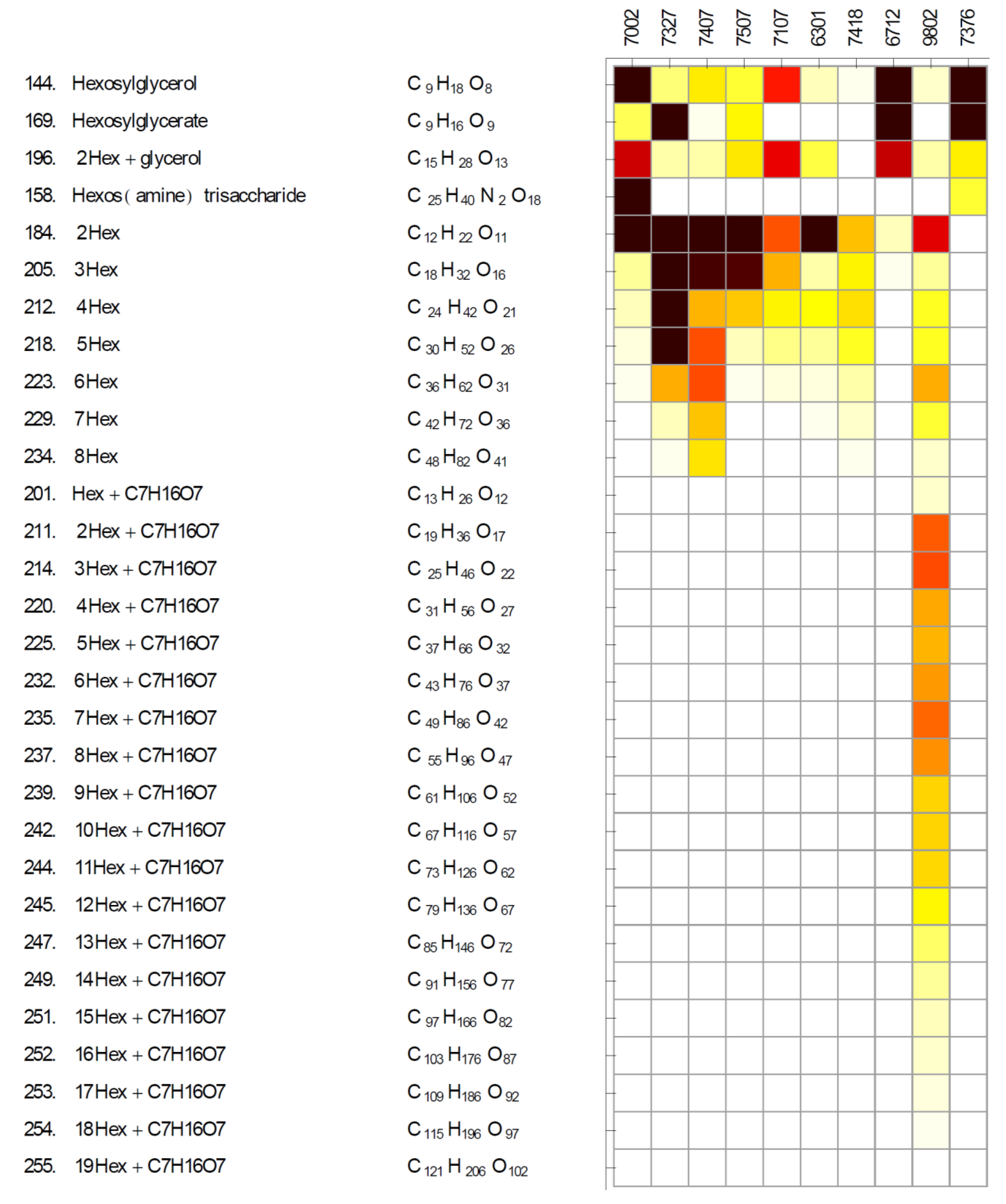
2.2. Diversity of Glycosides and Oligosaccharides
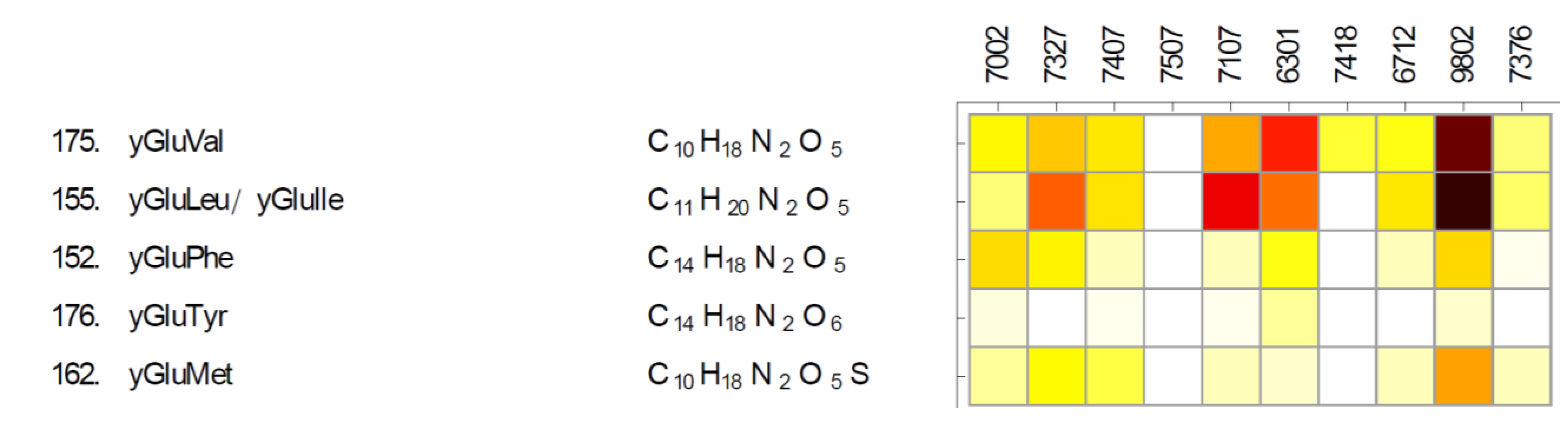
2.3. Gamma-Glutamyl Dipeptides and Gamma-Glutamyltransferase (ggt)
2.4. Other Metabolites
3. Experimental Section
3.1. Strains and Culture Conditions
3.2. Metabolite Extraction
3.3. LC-MS Analysis
3.4. Data Analysis
3.5. Genome Analysis
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Seckbach, J.; Oren, A. Oxygenic Photosynthetic Microorganisms in Extreme Environments. In Algae and Cyanobacteria in Extreme Environments; Seckbach, J., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 5–25. [Google Scholar]
- Whitton, B.A.; Potts, M. Introduction to the Cyanobacteria. In Ecology of Cyanobacteria II; Whitton, B.A., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 1–13. [Google Scholar]
- Garcia-Pichel, F.; Loza, V.; Marusenko, Y.; Mateo, P.; Potrafka, R.M. Temperature drives the continental-scale distribution of key microbes in topsoil communities. Science 2013, 340, 1574–1577. [Google Scholar] [CrossRef]
- Garcia-Pichel, F.; Belnap, J.; Neuer, S.; Schanz, F. Estimates of global cyanobacterial biomass and its distribution. Algol. Stud. 2003, 109, 213–227. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive Earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef]
- Gao, Q.; Garcia-Pichel, F. Microbial ultraviolet sunscreens. Nat. Rev. Microbiol. 2011, 9, 791–802. [Google Scholar] [CrossRef]
- Atsumi, S.; Higashide, W.; Liao, J.C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 2009, 27, 1177–1180. [Google Scholar] [CrossRef]
- McNeely, K.; Xu, Y.; Bennette, N.; Bryant, D.A.; Dismukes, G.C. Redirecting reductant flux into hydrogen production via metabolic engineering of fermentative carbon metabolism in a cyanobacterium. Appl. Environ. Microbiol. 2010, 76, 5032–5038. [Google Scholar] [CrossRef]
- Ducat, D.C.; Way, J.C.; Silver, P.A. Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 2011, 29, 95–103. [Google Scholar]
- Lan, E.I.; Liao, J.C. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 6018–6023. [Google Scholar] [CrossRef]
- Xu, Y.; Guerra, L.T.; Li, Z.; Ludwig, M.; Dismukes, G.C.; Bryant, D.A. Altered carbohydrate metabolism in glycogen synthase mutants of Synechococcus sp. strain PCC 7002: Cell factories for soluble sugars. Metab. Eng. 2013, 16, 56–67. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Koonin, E.V. From complete genome sequence to “complete” understanding? Trends Biotechnol. 2010, 28, 398–406. [Google Scholar] [CrossRef]
- Hanson, A.D.; Pribat, A.; Waller, J.C.; de Crécy-Lagard, V. “Unknown” proteins and “orphan” enzymes: The missing half of the engineering parts list—and how to find it. Biochem. J. 2009, 425, 1–11. [Google Scholar]
- Gerdes, S.; El Yacoubi, B.; Bailly, M.; Blaby, I.K.; Blaby-Haas, C.E.; Jeanguenin, L.; Lara-Núñez, A.; Pribat, A.; Waller, J.C.; Wilke, A.; et al. Synergistic use of plant-prokaryote comparative genomics for functional annotations. BMC Genomics 2011, 12 (Suppl. 1), S2. [Google Scholar]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005, 12, 291–299. [Google Scholar]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar]
- Nichols, R.J.; Sen, S.; Choo, Y.J.; Beltrao, P.; Zietek, M.; Chaba, R.; Lee, S.; Kazmierczak, K.M.; Lee, K.J.; Wong, A.; et al. Phenotypic landscape of a bacterial cell. Cell 2011, 144, 143–156. [Google Scholar] [CrossRef]
- Deutschbauer, A.; Price, M.N.; Wetmore, K.M.; Shao, W.; Baumohl, J.K.; Xu, Z.; Nguyen, M.; Tamse, R.; Davis, R.W.; Arkin, A.P. Evidence-based annotation of gene function in Shewanella oneidensis MR-1 using genome-wide fitness profiling across 121 conditions. PLoS Genet. 2011, 7, e1002385. [Google Scholar] [CrossRef]
- Reed, J.L.; Patel, T.R.; Chen, K.H.; Joyce, A.R.; Applebee, M.K.; Herring, C.D.; Bui, O.T.; Knight, E.M.; Fong, S.S.; Palsson, B.O. Systems approach to refining genome annotation. Proc. Natl. Acad. Sci. USA 2006, 103, 17480–17484. [Google Scholar] [CrossRef]
- Baran, R.; Reindl, W.; Northen, T.R. Mass spectrometry based metabolomics and enzymatic assays for functional genomics. Curr. Opin. Microbiol. 2009, 12, 547–552. [Google Scholar] [CrossRef]
- Saito, N.; Ohashi, Y.; Soga, T.; Tomita, M. Unveiling cellular biochemical reactions via metabolomics-driven approaches. Curr. Opin. Microbiol. 2010, 13, 358–362. [Google Scholar] [CrossRef]
- Saito, N.; Robert, M.; Kitamura, S.; Baran, R.; Soga, T.; Mori, H.; Nishioka, T.; Tomita, M. Metabolomics approach for enzyme discovery. J. Proteome Res. 2006, 5, 1979–1987. [Google Scholar] [CrossRef]
- Saito, N.; Robert, M.; Kochi, H.; Matsuo, G.; Kakazu, Y.; Soga, T.; Tomita, M. Metabolite profiling reveals YihU as a novel hydroxybutyrate dehydrogenase for alternative succinic semialdehyde metabolism in Escherichia coli. J. Biol. Chem. 2009, 284, 16442–16451. [Google Scholar]
- Baran, R.; Bowen, B.P.; Price, M.N.; Arkin, A.P.; Deutschbauer, A.M.; Northen, T.R. Metabolic footprinting of mutant libraries to map metabolite utilization to genotype. ACS Chem. Biol. 2013, 8, 189–199. [Google Scholar] [CrossRef]
- Kind, T.; Fiehn, O. Advances in structure elucidation of small molecules using mass spectrometry. Bioanal. Rev. 2010, 2, 23–60. [Google Scholar] [CrossRef]
- Bowen, B.P.; Northen, T.R. Dealing with the unknown: metabolomics and metabolite atlases. J. Am. Soc. Mass Spectrom. 2010, 21, 1471–1476. [Google Scholar] [CrossRef]
- Baran, R; Bowen, B.P.; Bouskill, N.J.; Brodie, E.L.; Yannone, S.M.; Northen, T.R. Metabolite identification in Synechococcus sp. PCC 7002 using untargeted stable isotope assisted metabolite profiling. Anal. Chem. 2010, 82, 9034–9042. [Google Scholar] [CrossRef]
- Baran, R.; Bowen, B.P.; Northen, T.R. Untargeted metabolic footprinting reveals a surprising breadth of metabolite uptake and release by Synechococcus sp. PCC 7002. Mol. Biosyst. 2011, 7, 3200–3206. [Google Scholar] [CrossRef]
- Sugita, C.; Ogata, K.; Shikata, M.; Jikuya, H.; Takano, J.; Furumichi, M.; Kanehisa, M.; Omata, T.; Sugiura, M.; Sugita, M. Complete nucleotide sequence of the freshwater unicellular cyanobacterium Synechococcus elongatus PCC 6301 chromosome: gene content and organization. Photosynth. Res. 2007, 93, 55–67. [Google Scholar] [CrossRef]
- Starkenburg, S.R.; Reitenga, K.G.; Freitas, T.; Johnson, S.; Chain, P.S.; Garcia-Pichel, F.; Kuske, C.R. Genome of the cyanobacterium Microcoleus vaginatus FGP-2, a photosynthetic ecosystem engineer of arid land soil biocrusts worldwide. J. Bacteriol. 2011, 193, 4569–4570. [Google Scholar] [CrossRef]
- Shih, P.M.; Wu, D.; Latifi, A.; Axen, S.D.; Fewer, D.P.; Talla, E.; Calteau, A.; Cai, F.; Tandeau de Marsac, N.; Rippka, R.; et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 1053–1058. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stainer, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Baran, R.; Kochi, H.; Saito, N.; Suematsu, M.; Soga, T.; Nishioka, T.; Robert, M.; Tomita, M. MathDAMP: A package for differential analysis of metabolite profiles. BMC Bioinforma. 2006, 7, 530. [Google Scholar] [CrossRef]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Scheubert, K.; Hufsky, F.; Böcker, S. Computational mass spectrometry for small molecules. J. Cheminforma. 2013, 5, 12. [Google Scholar] [CrossRef]
- Warren, C.R. Quaternary ammonium compounds can be abundant in some soils and are taken up as intact molecules by plants. New Phytol. 2013, 198, 476–485. [Google Scholar] [CrossRef]
- Seebeck, F.P. In vitro reconstitution of mycobacterial ergothioneine biosynthesis. J. Am. Chem. Soc. 2010, 132, 6632–6633. [Google Scholar] [CrossRef]
- Pfeiffer, C.; Bauer, T.; Surek, B.; Schomig, E.; Grundemann, D. Cyanobacteria produce high levels of ergothioneine. Food Chem. 2011, 129, 4. [Google Scholar]
- Nyyssola, A.; Kerovuo, J.; Kaukinen, P.; von Weymarn, N.; Reinikainen, T. Extreme halophiles synthesize betaine from glycine by methylation. J. Biol. Chem. 2000, 275, 22196–22201. [Google Scholar] [CrossRef]
- Garcia-Pichel, F.; Nübel, U.; Muyzer, G. The phylogeny of unicellular, extremely halotolerant cyanobacteria. Arch. Microbiol. 1998, 169, 469–482. [Google Scholar] [CrossRef]
- Bello, M.H.; Barrera-Perez, V.; Morin, D.; Epstein, L. The Neurospora crassa mutant NcΔEgt-1 identifies an ergothioneine biosynthetic gene and demonstrates that ergothioneine enhances conidial survival and protects against peroxide toxicity during conidial germination. Fungal Genet. Biol. 2012, 49, 160–172. [Google Scholar] [CrossRef]
- Hagemann, M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 2011, 35, 87–123. [Google Scholar] [CrossRef]
- Pereira, S.; Zille, A.; Micheletti, E.; Moradas-Ferreira, P.; De Philippis, R.; Tamagnini, P. Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol. Rev. 2009, 33, 917–941. [Google Scholar] [CrossRef]
- Luley-Goedl, C.; Nidetzky, B. Glycosides as compatible solutes: Biosynthesis and applications. Nat. Prod. Rep. 2011, 28, 875–896. [Google Scholar] [CrossRef]
- Ojima, T.; Saburi, W.; Yamamoto, T.; Kudo, T. Characterization of Halomonas sp. strain H11 α-glucosidase activated by monovalent cations and its application for efficient synthesis of α-d-glucosylglycerol. Appl. Environ. Microbiol. 2012, 78, 1836–1845. [Google Scholar] [CrossRef]
- Wieneke, R.; Klein, S.; Geyer, A.; Loos, E. Structural and functional characterization of galactooligosaccharides in Nostoc commune: β-d-galactofuranosyl-(1→6)-[β-d-galactofuranosyl-(1→6)]2-β-d-1,4-anhydrogalactitol and β-(1→6)-galactofuranosylated homologues. Carbohydr. Res. 2007, 342, 2757–2765. [Google Scholar] [CrossRef]
- Pontis, H.G.; Vargas, W.A.; Salerno, G.L. Structural characterization of the members of a polymer series, compatible solutes in Anabaena cells exposed to salt stress. Plant Sci. 2007, 172, 29–35. [Google Scholar] [CrossRef]
- Oss, M.; Kruve, A.; Herodes, K.; Leito, I. Electrospray ionization efficiency scale of organic compounds. Anal. Chem. 2010, 82, 2865–2872. [Google Scholar] [CrossRef]
- Hoiczyk, E.; Baumeister, W. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr. Biol. 1998, 8, 1161–1168. [Google Scholar] [CrossRef]
- Garcia-Pichel, F.; Pringault, O. Microbiology: Cyanobacteria track water in desert soils. Nature 2001, 413, 380–381. [Google Scholar] [CrossRef]
- Rajeev, L.; da Rocha, U.N.; Klitgord, N.; Luning, E.G.; Fortney, J.; Axen, S.D.; Shih, P.M.; Bouskill, N.J.; Bowen, B.P.; Kerfeld, C.A.; et al. Dynamic cyanobacterial response to hydration and dehydration in a desert biological soil crust. ISME J. 2013. [Google Scholar] [CrossRef]
- Suzuki, H.; Yamada, C.; Kato, K. Gamma-glutamyl compounds and their enzymatic production using bacterial gamma-glutamyltranspeptidase. Amino Acids 2007, 32, 333–340. [Google Scholar] [CrossRef]
- Boanca, G.; Sand, A.; Okada, T.; Suzuki, H.; Kumagai, H.; Fukuyama, K.; Barycki, J.J. Autoprocessing of Helicobacter pylori gamma-glutamyltranspeptidase leads to the formation of a threonine-threonine catalytic dyad. J. Biol. Chem. 2007, 282, 534–541. [Google Scholar]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Caspi, R.; Altman, T.; Dreher, K.; Fulcher, C.A.; Subhraveti, P.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012, 40, D742–D753. [Google Scholar] [CrossRef]
- The Pasteur Culture Collection of Cyanobacteria (PCC). Available online: http://www.pasteur.fr/pcc_cyanobacteria (accessed on 22 September 2013).
- Markowitz, V.M.; Chen, I.-M.A.; Palaniappan, K.; Chu, K.; Szeto, E.; Grechkin, Y.; Ratner, A.; Jacob, B.; Huang, J.; Williams, P.; et al. IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012, 40, D115–D122. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Braunshausen, A.; Seebeck, F.P. Identification and characterization of the first ovothiol biosynthetic enzyme. J. Am. Chem. Soc. 2011, 133, 1757–1759. [Google Scholar] [CrossRef]
- Schoor, A.; Hagemann, M.; Erdmann, N. Glucosylglycerol-phosphate synthase: target for ion-mediated regulation of osmolyte synthesis in the cyanobacterium Synechocystis sp. strain PCC 6803. Arch. Microbiol. 1999, 171, 101–106. [Google Scholar] [CrossRef]
- Haft, D.H.; Selengut, J.D.; White, O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003, 31, 371–373. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Anderson, J.B.; Derbyshire, M.K.; DeWeese-Scott, C.; Gonzales, N.R.; Gwadz, M.; Hao, L.; He, S.; Hurwitz, D.I.; Jackson, J.D.; et al. CDD: A conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007, 35, D237–D240. [Google Scholar] [CrossRef]
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Baran, R.; Ivanova, N.N.; Jose, N.; Garcia-Pichel, F.; Kyrpides, N.C.; Gugger, M.; Northen, T.R. Functional Genomics of Novel Secondary Metabolites from Diverse Cyanobacteria Using Untargeted Metabolomics. Mar. Drugs 2013, 11, 3617-3631. https://doi.org/10.3390/md11103617
Baran R, Ivanova NN, Jose N, Garcia-Pichel F, Kyrpides NC, Gugger M, Northen TR. Functional Genomics of Novel Secondary Metabolites from Diverse Cyanobacteria Using Untargeted Metabolomics. Marine Drugs. 2013; 11(10):3617-3631. https://doi.org/10.3390/md11103617
Chicago/Turabian StyleBaran, Richard, Natalia N. Ivanova, Nick Jose, Ferran Garcia-Pichel, Nikos C. Kyrpides, Muriel Gugger, and Trent R. Northen. 2013. "Functional Genomics of Novel Secondary Metabolites from Diverse Cyanobacteria Using Untargeted Metabolomics" Marine Drugs 11, no. 10: 3617-3631. https://doi.org/10.3390/md11103617
APA StyleBaran, R., Ivanova, N. N., Jose, N., Garcia-Pichel, F., Kyrpides, N. C., Gugger, M., & Northen, T. R. (2013). Functional Genomics of Novel Secondary Metabolites from Diverse Cyanobacteria Using Untargeted Metabolomics. Marine Drugs, 11(10), 3617-3631. https://doi.org/10.3390/md11103617




