Cultivable Alginate Lyase-Excreting Bacteria Associated with the Arctic Brown Alga Laminaria
Abstract
:1. Introduction
2. Materials and Method
2.1. Materials
| Station | Location | |
|---|---|---|
| 0 | 78°55′3.0″N | 11°58′14.0″E |
| 1 | 78°55′19.7″N | 11°56′53.8″E |
| 2 | 78°55′17.0″N | 11°57′0.2″E |
| 3 | 78°55′3.8″N | 11°58′11.6″E |
| B | 78°55′20.5″N | 11°54′49.7″E |
| C | 78°55′3.8″N | 11°56′53.8″E |
2.2. Enrichment Cultivation and Isolation of the Bacteria Associated with the Arctic Brown Alga Laminaria
2.3. Screening of Alginate Lyase-Excreting Bacteria
2.4. Amplification, Cloning, and Sequencing of the 16S rRNA Gene
2.5. Assay for Alginate Lyase Activity
2.6. Analysis of the Optimal Temperatures for the Growth and Alginate Lyase Production of the Isolates and for the Activity of Alginate Lyase from Each Strain
3. Results
3.1. Diversity of Bacteria Associated with the Arctic Brown Alga Laminaria
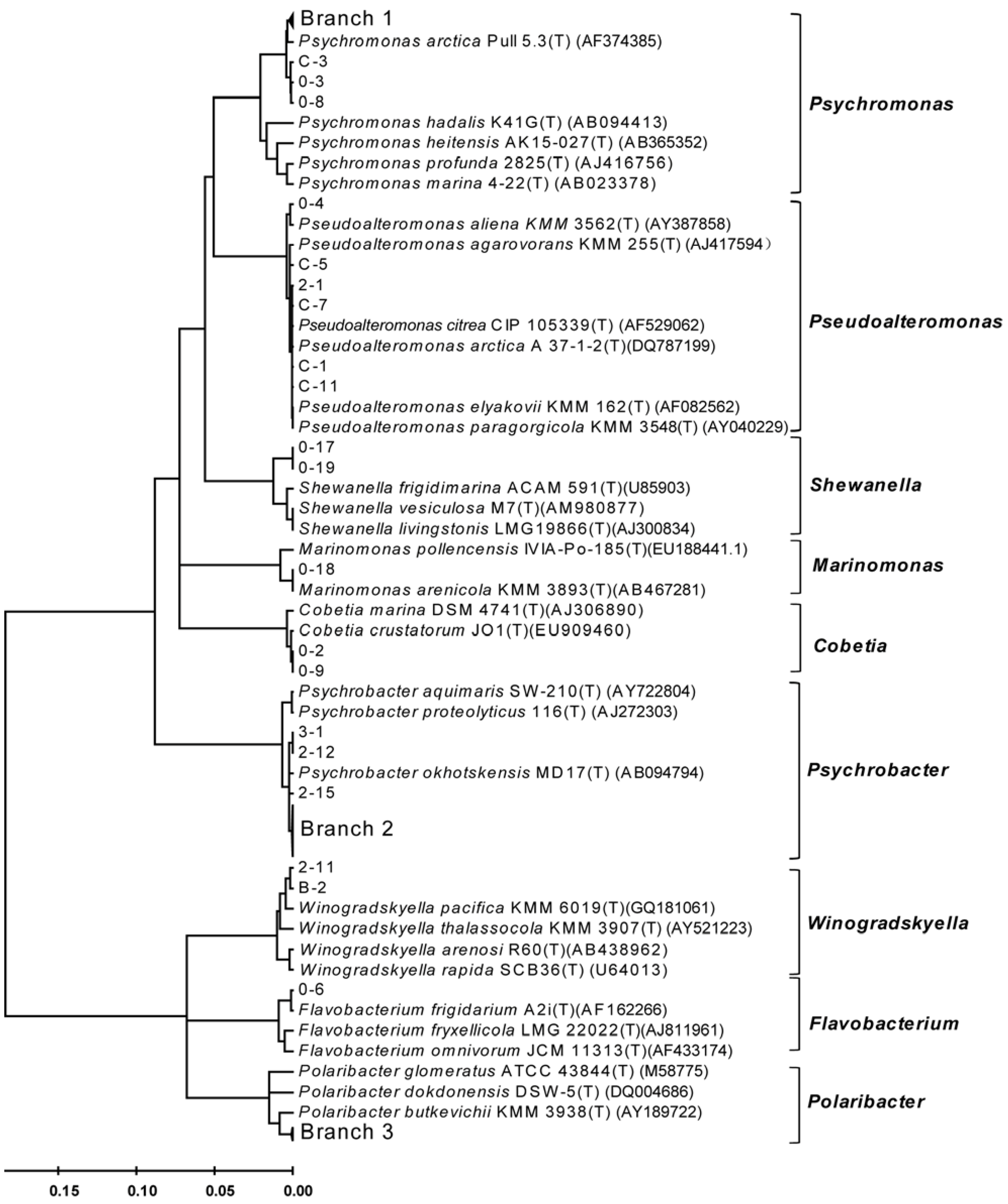
3.2. Diversity of Alginate Lyase-Excreting Bacteria from the Arctic Brown Alga Laminaria

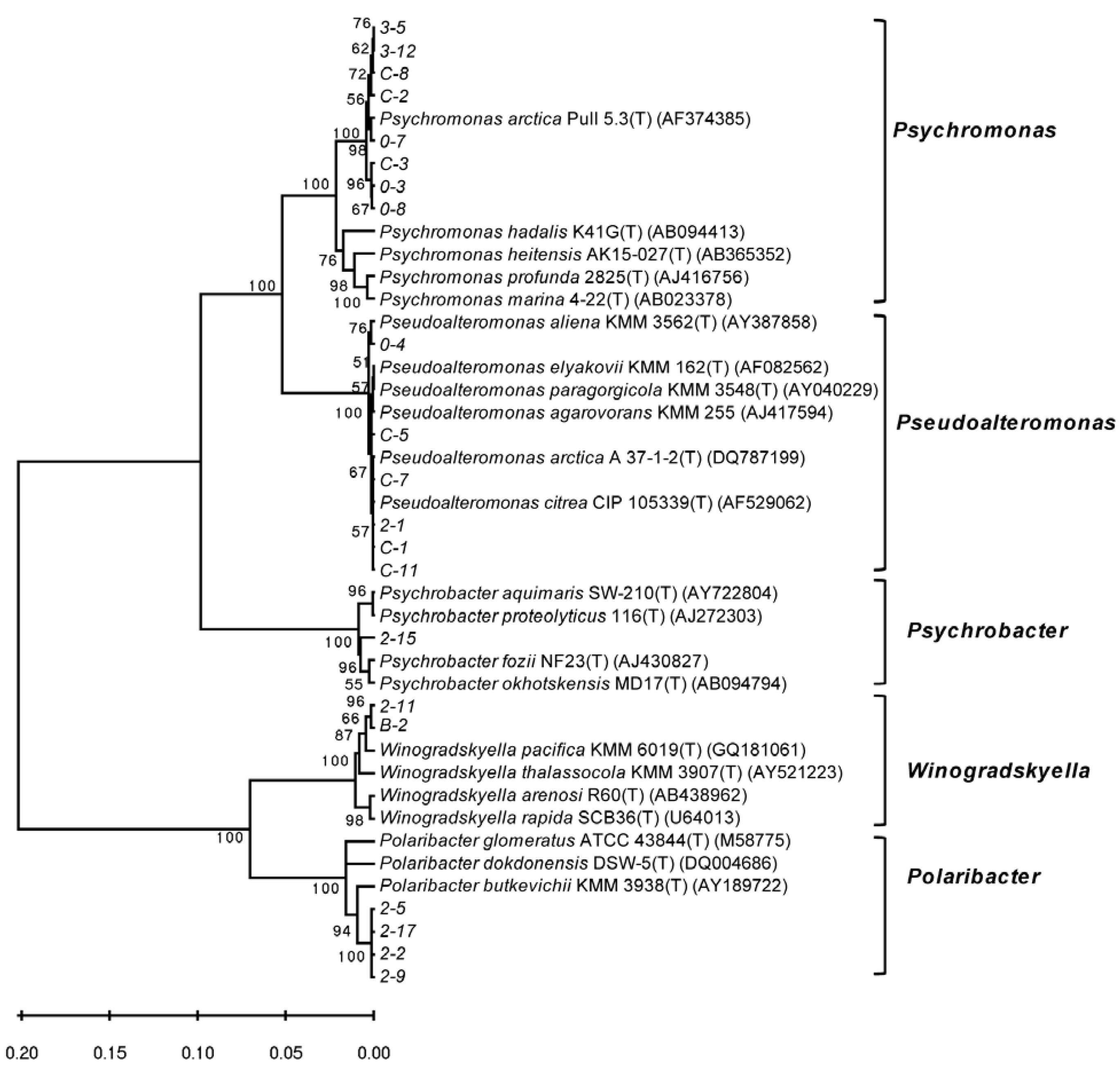
3.3. Optimal Temperatures for Growth and Alginate Lyase Production of the Strains
| Genera | Strains | Optimum Temperature( °C) | ||
|---|---|---|---|---|
| for Growth | for Enzyme Production | for Enzyme Activity | ||
| Polaribacter | 2-2 | 20 | 25 | 30 |
| 2-9 | 20 | 25 | 50 | |
| 2-5 | 25 | 25 | 50 | |
| 2-17 | 15 | 25 | 40 | |
| Pseudoalteromonas | 0-4 | 15 | 15 | 30 |
| 2-1 | 10 | 25 | 40 | |
| C-1 | 30 | 25 | 20 | |
| C-5 | 25 | 25 | 40 | |
| C-7 | 25 | 30 | 30 | |
| C-11 | 25 | 25 | 40 | |
| Psychrobacter | 2-15 | 20 | 20 | 50 |
| Psychromonas | 0-3 | 20 | 20 | 30 |
| 0-7 | 10 | 20 | 45 | |
| 0-8 | 20 | 15 | 30 | |
| 3-5 | 20 | 20 | 30 | |
| 3-12 | 15 | 10 | 30 | |
| C-2 | 20 | 20 | 20 | |
| C-3 | 20 | 25 | 20 | |
| C-8 | 25 | 25 | 30 | |
| Winogradskyella | 2-11 | 20 | 15 | 40 |
| B-2 | 25 | 25 | 40 | |
3.4. Optimal Temperatures for the Enzymatic Activity of the Alginate Lyases from the Strains
3.5. Levels of Extracellular Alginate Lyase Activity of the Strains
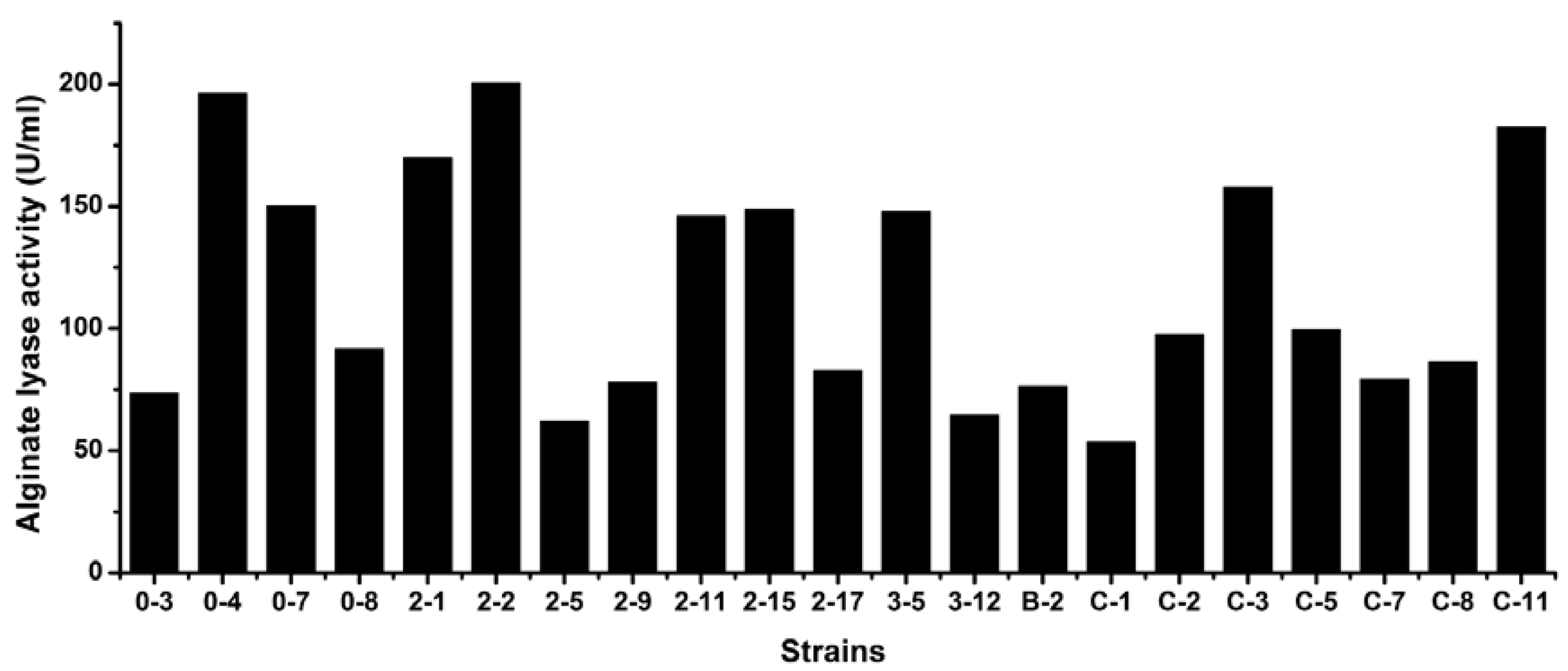
4. Discussion
Acknowledgments
Supplementary Files
References
- Gacesa, P. Alginates. Carbohydr. Polym. 1988, 8, 161–182. [Google Scholar] [CrossRef]
- Pawar, S.N.; Edgar, K.J. Alginate derivatization: A review of chemistry, properties and applications. Biomaterials 2012, 33, 3279–3305. [Google Scholar] [CrossRef]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties and biomedical applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar] [CrossRef]
- Preiss, J.; Ashwell, G. Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosac-charides and 4-deoxy-L-erythro-5-hexoseulose uronic acid. J. Biol. Chem. 1962, 237, 309–316. [Google Scholar]
- Kim, D.E.; Lee, E.Y.; Kim, H.S. Cloning and characterization of alginate lyase from a marine bacterium Streptomyces sp. ALG-5. Mar. Biotechnol. 2009, 11, 10–16. [Google Scholar]
- Gacesa, P. Enzymic degradation of alginates. Int. J. Biochem. 1992, 24, 545–552. [Google Scholar] [CrossRef]
- Osawa, T.; Matsubara, Y.; Muramatsu, T.; Kimura, M.; Kakuta, Y. Crystal structure of the alginate (poly alpha-L-guluronate) lyase from Corynebacterium sp. at 1.2 A resolution. J. Mol. Biol. 2005, 345, 1111–1118. [Google Scholar] [CrossRef]
- Caborhydrate-Active enZYmes (CAZy) database. Available online: http://www.cazy.org/fam/acc_PL.html (accessed on 24 September 2012).
- Wong, T.Y.; Preston, L.A.; Schiller, N.L. ALGINATE LYASE: Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000, 54, 289–340. [Google Scholar] [CrossRef]
- Sawabe, T.; Tanaka, R.; Iqbal, M.M.; Tajima, K.; Ezura, Y.; Ivanova, E.P.; Christen, R. Assignment of Alteromonas elyakovii KMM 162T and five strains isolated from spot-wounded fronds of Laminaria japonica to Pseudoalteromonas elyakovii comb. nov. and the extended description of the species. Int. J. Syst. Evol. Microbiol. 2000, 50, 265–271. [Google Scholar] [CrossRef]
- Li, J.W.; Dong, S.; Song, J.; Li, C.-B.; Chen, X.-L.; Xie, B.-B.; Zhang, Y.-Z. Purification and characterization of a bifunctional alginate lyase from Pseudoalteromonas sp. SM0524. Mar. Drugs 2011, 9, 109–123. [Google Scholar] [CrossRef]
- Lee, Y.K.; Jung, H.J.; Lee, H.K. Marine bacteria associated with the Korean brown alga, Undaria pinnatifida. J. Microbiol. 2006, 44, 694–698. [Google Scholar]
- Engel, A.S.; Porter, M.L.; Stern, L.A.; Quinlan, S.; Bennett, P.C. Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria”. FEMS Microbiol. Ecol. 2004, 51, 31–53. [Google Scholar] [CrossRef]
- Kumar, S.; Tamura, K.; Nei, M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 2004, 5, 150–163. [Google Scholar] [CrossRef]
- Green, F., III; Clausen, C.A.; Highley, T.L. Adaptation of the Nelson-Somogyi reducing-sugar assay to a microassay using microtiter plates. Anal. Biochem. 1989, 182, 197–199. [Google Scholar]
- Boyd, J.; Turvey, J.R. Structural studies of alginicacid, using abacterial poly-α-L-guluronate lyase. Carbohydr. Res. 1978, 66, 8. [Google Scholar]
- Ostgaard, K. Determination of alginate composition by a simple enzymatic assay. Hydrobiologia 1993, 260–261, 513–520. [Google Scholar]
- Declan, M.; Butler, K.Ø.; Catherine, L.V.; Evans, A.J.; Kloareg, B. Isolation conditions for high yields of protoplasts from Laminaria saccharina and L. digitata (Phaeophyceae). J. Exp. Bot. 1989, 40, 1237–1246. [Google Scholar]
- Quatrano, R.S.; Stevens, P.T. Cell wall assembly in Fucus zygotes: I. Characterization of the polysaccharide components. Plant Physiol. 1976, 58, 224–231. [Google Scholar] [CrossRef]
- Heyraud, A.; Colin-Morel, P.; Gey, C.; Chavagnat, F.; Guinand, M.; Wallach, J. An enzymatic method for preparation of homopolymannuronate blocks and strictly alternating sequences of mannuronic and guluronic units. Carbohydr. Res. 1998, 308, 417–422. [Google Scholar] [CrossRef]
- Iwamoto, M.; Kurachi, M.; Nakashima, T.; Kim, K.; Yamaguchi, K.; Oda, T.; Iwamoto, Y.; Muramatsu, T. Structure-activity relationship of alginate oligosaccharides in the induction of cytokine production from RAW264.7 cells. FEBS Lett. 2005, 579, 4423–4429. [Google Scholar] [CrossRef]
- Akiyama, H.; Endo, T.; Nakakita, R.; Murata, K.; Yonemoto, Y.; Okayama, K. Effect of depolymerized alginates on the growth of bifidobacteria. Biosci. Biotechnol. Biochem. 1992, 56, 355–356. [Google Scholar] [CrossRef]
- Mrsny, R.J.; Lazazzera, B.A.; Baughety, A.L.; Schiller, N.L.; Patapoff, T.W. Addition of a bacterial alginate lyase to purulent CF sputum in vitro can result in the disruption of alginate and modification of sputum viscoelasticity. Pulm. Pharmacol. 1994, 7, 357–366. [Google Scholar] [CrossRef]
- Wargacki, A.J.; Leonard, E.; Win, M.N.; Regitsky, D.D.; Santos, C.N.S.; Kim, P.B.; Cooper, S.R.; Raisner, R.M.; Herman, A.; Sivitz, A.B.; et al. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 2012, 335, 308–313. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Dong, S.; Yang, J.; Zhang, X.-Y.; Shi, M.; Song, X.-Y.; Chen, X.-L.; Zhang, Y.-Z. Cultivable Alginate Lyase-Excreting Bacteria Associated with the Arctic Brown Alga Laminaria. Mar. Drugs 2012, 10, 2481-2491. https://doi.org/10.3390/md10112481
Dong S, Yang J, Zhang X-Y, Shi M, Song X-Y, Chen X-L, Zhang Y-Z. Cultivable Alginate Lyase-Excreting Bacteria Associated with the Arctic Brown Alga Laminaria. Marine Drugs. 2012; 10(11):2481-2491. https://doi.org/10.3390/md10112481
Chicago/Turabian StyleDong, Sheng, Jie Yang, Xi-Ying Zhang, Mei Shi, Xiao-Yan Song, Xiu-Lan Chen, and Yu-Zhong Zhang. 2012. "Cultivable Alginate Lyase-Excreting Bacteria Associated with the Arctic Brown Alga Laminaria" Marine Drugs 10, no. 11: 2481-2491. https://doi.org/10.3390/md10112481
APA StyleDong, S., Yang, J., Zhang, X.-Y., Shi, M., Song, X.-Y., Chen, X.-L., & Zhang, Y.-Z. (2012). Cultivable Alginate Lyase-Excreting Bacteria Associated with the Arctic Brown Alga Laminaria. Marine Drugs, 10(11), 2481-2491. https://doi.org/10.3390/md10112481




