Abstract
The demosponge Suberites domuncula has been described to contain high levels of a proteinaceous toxin, Suberitine, that displays haemolytic activityIn the present study this 7–8 kDa polypeptide has been isolated and was shown to exhibit also cytotoxic effects on cells of the same species. Addition of retinal, a recently identified metabolite of β-carotene that is abundantly present in S. domuncula was found to reduce both the haemolytic and the cell toxic activity of Suberitine at a molar ratio of 1:1. Spectroscopic analyses revealed that the interaction between β-carotene and Suberitine can be ascribed to a reversible energy transfer reaction. The enzyme that synthesises retinal in the sponge system is the β,β-carotene-15,15′-dioxygenase [carotene dioxygenase]. In order to clarify if this enzyme is the only β-carotene-metabolizing enzyme a further oxygenase had been identified and cloned, the (related) carotenoid oxygenase. In contrast to the dioxygenase, the carotenoid oxygenase could not degrade β-carotene or lycopene in Escherichia coli strains that produced these two carotenoids; therefore it had been termed related-carotenoid oxygenase. Exposure of primmorphs to light of different wavelengths from the visible spectrum resulted after 3 days in a strong upregulation of the dioxygenase in those 3D-cell aggregates that had been incubated with β-carotene. The strongest effect is seen with blue light at a maximum around 490 nm. It is concluded that the toxin Suberitine is non-covalently modified by retinal, the cleavage product from β-carotene via the enzyme carotene dioxygenase, a light inducible oxygenase. Hence, this study highlights that in S. domuncula the bioactive metabolite, retinal, has the property to detoxify its homologous toxin.
1. Introduction
Since the discovery of the first clinically relevant secondary metabolite isolated from the sponge Cryptotethya crypta (phylum Porifera) [1], 1-β-D-arabinofuranosylthymine [ara-T], a cornucopia of unique chemical compounds has been identified. They all have the potential for industrial development as pharmaceuticals, cosmetics, nutritional supplements, molecular probes, enzymes, fine chemicals, and agrichemicals [2,3,4]. From ara-T two derivatives have been chemically synthesized that proved to be potent anti-leukemic (1-β-D-arabinofuranosylcytosine [ara-C] [5]) or anti-viral (9-β-D-arabinofuranosyladenine [ara-A] [6]) drugs. In contrast to the secondary metabolites, proteinaceous bioactive substances were given less attention despite their presumed higher biological and biotechnological importance [7]. This changed gradually with the first cloning of such a bioactive polypeptide, the hemagglutinin from Geodia cydonium [8]. Subsequently, further marine bioactive proteins were identified [9] with the ASABF-type antimicrobial peptide from Suberites domuncula being the most recent one [10]. The proteinaceous compounds have, if their genes are known, the advantage over the secondary metabolites that they can be modified straightforwardly by molecular biological techniques [11].
Alike the secondary metabolites also the bioactive proteins are used by sponges as defense molecules against attacking pro- and eukaryotic organisms [12]. Until their use for defense, the secondary metabolites remain either compartmented in cellular vesicles, as e.g., avarol [13], or are stored as pro-toxins, as e.g., aeroplysinin [14], and by that remain functionally inactive for the sponge producer. In the bioactive peptides, however, the molecules undergo activation during post-translational modification and extracellular transport [10]. In the present study a novel strategy has been detected in sponges by which a toxic peptide remains functionally inactive for the host producer, by the interaction of the peptide with a low molecular bioactive metabolite. Following our earlier finding that the demosponge S. domuncula is rich in retinoids [15], we elucidated recently the basic pathway of the retinoids in this species [16]. The retinoid β-carotene is produced by bacteria that are symbiotically associated with this sponge. This precursor molecule is enzymatically processed by cleavage through the β,β-carotene-15,15′-dioxygenase (carotene dioxygenase) to retinal, followed by oxidation via the retinal dehydrogenase to retinoic acid. The end-product retinoic acid functions as morphogen in the sponge via interaction with the ligand-activated transcription factor, the retinoid X receptor [17]. In the present study we give evidence that retinal interacts with the toxic peptide in S. domuncula, with Suberitine.
The biological active Suberitine has been discovered in S. domuncula already in 1906 [18] and was then isolated and purified by Cariello and Zanetti [19]. It has been proposed that this peptide acts as a defense molecule against attacking invaders [20]. The peptide with a size of about 7 kDa is composed of four subunits [19], is neurotoxic and displays hemolytic activity [21,22]. The biological activity of Suberitine is abolished after modification of the cysteinyl and tryptophyl residues within the molecule [23]. Importantly, these authors described later a protein from the same sponge, the blue carotenoprotein, with a similar size like the native Suberitine that has the capacity to bind to carotenoids [24]. In the present study we describe that Suberitine in S. domuncula is inactivated after binding to retinal. In order to elucidate the tuned activation/inactivation pathway of Suberitine we studied if the carotenoids themselves control the expression of the gene(s) involved in the formation of retinal. Consequently we screened for additional genes that might encode enzymes that cleave carotenoids [25]. We identified two further (potential) carotenoid oxygenases that, however, after expression, did not cleave β-carotene. Therefore, these enzymes had been termed related-carotenoid oxygenases. The data presented here indicate that low molecular bioactive compound(s) that are produced by associated microorganisms and processed by sponge host-encoded enzymes control the biological activity of bioactive peptides in sponges.
2. Results
2.1. Suberitine: Purification
Suberitine was purified from freshly collected animals (Figure 1A). Using a purification procedure, basically proposed by [19], a 55.8-fold enrichment was obtained using the consecutively performed steps of ammonium sulfate precipitation, Sephadex G-100 and Sephadex G-50 gel chromatography, as outlined under “Materials and Methods”. During this procedure the specific hemagglutination activity increased from 469.6 units/mg to 26,219.1 units/mg (Table 1). The purity of the fractions was analyzed by SDS-PAGE. Likewise the samples were subjected to SDS treatment and heat. It was seen that both Fraction II (ammonium sulfate fractionation; Figure 2, lane a) and Fraction III (Sephadex G-100 filtration) contained many protein bands (Figure 2, lane b), while after fractionation by Sephadex G-50 the pooled fractions that contained the majority of the hemagglutination activity displayed only one band corresponding to an apparent size of 7000 to 8000 Da (Figure 2, lane c). This fraction (0.29 mg protein/mL) was used for the following experiments.
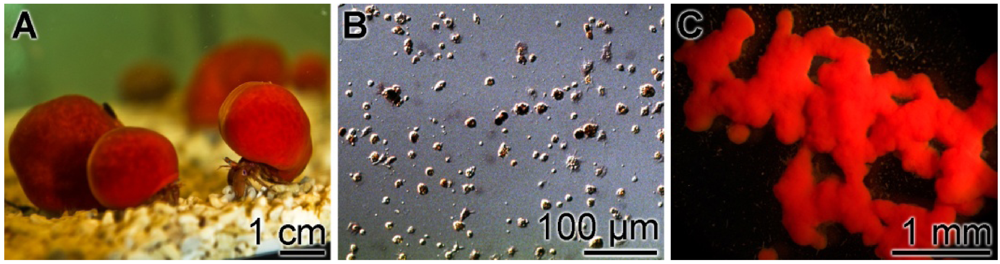
Figure 1.
The sponge S. domuncula and primmorphs formed from this species. (A) Specimens maintained in aquaria; (B) Single cells were prepared and (C) allowed to reaggregate to 3D-cell aggregates, the primmorphs, for three days in the incubation medium.

Table 1.
Purification of Suberitine. The crude extract started from 50 g fresh tissue.
| Fraction | Total Protein (mg) | Total Hemagglutination (units) | Purification (fold) | Recovery (%) | Specific Activity (units/mg) |
|---|---|---|---|---|---|
| I: Crude fraction | 218.4 | 102,370 | 1 | 100 | 469.6 |
| II: (NH4)2SO4 precipitation | 13.02 | 47,940 | 7.8 | 47 | 3687.7 |
| III: Sephadex G-100 | 3.24 | 32,640 | 21.4 | 32 | 10,074.1 |
| IV: Sephadex G-50 | 1.05 | 27,530 | 55.8 | 27 | 26,219.1 |
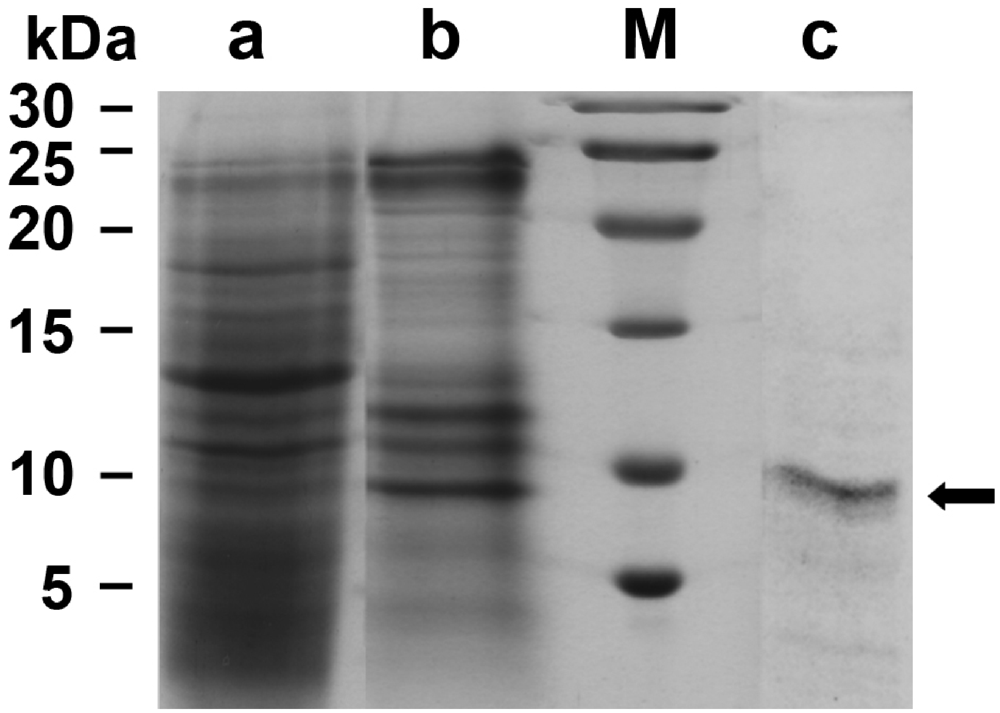
Figure 2.
Analysis of the fractions, containing Suberitine, obtained during the purification procedure by SDS-PAGE (15% acrylamide/0.1% SDS); 3 µg of protein were applied per slot. (lane a) Fraction II obtained after ammonium sulfate fractionation; (lane b) Fraction III after gel chromatography on Sephadex G-100; (lane c) Fraction IV after a final gel filtration chromatography step (Sephadex G-50). The arrow marks the single protein band of an apparent size of 7000 to 8000: (M) Size markers.
2.2. Suberitine: Biological Activity
As reported, the purified Suberitine displayed a pronounced biological hemagglutination activity [19,21].
In order to determine the interacting effect of metabolites of the β-carotene/retinoic acid pathway in sponges, a modified hemagglutination assay was applied, allowing an assessment on a percental basis. Among the metabolites tested (β-carotene, all-E-retinal, all-E-retinol and all-E-retinoic acid) all-E-retinal was the strongest interactor molecule, on the stoichiometric basis, reducing the hemagglutination activity of Suberitine at a [molar] stoichiometric basis (Suberitine/retinoid). At a molar ratio of 1.7 (Suberitine/retinal) the activity was lowered significantly by 40%, from 52 ± 8% down to 31 ± 6% (Figure 3); in comparison the reduction by β-carotene was 27%, by all-E-retinol 34% and by all-E-retinoic acid 19%, under the same concentration ratio conditions. Increasing the concentration of retinal and leaving the level of Suberitine constant in the assay, and by that decreasing the molar ratio (Suberitine/retinal) to 0.5, resulted in an about 40% reduction of the hemagglutination activity, down to 15 ± 4%. A further increase of retinal (ratio of 0.2) augmented the reducing activity of retinal to 10 ± 2%. In the absence of Suberitine or the presence of 3 µM retinoid, no significant hemolysis was seen under the conditions used (Figure 3 and not shown here).
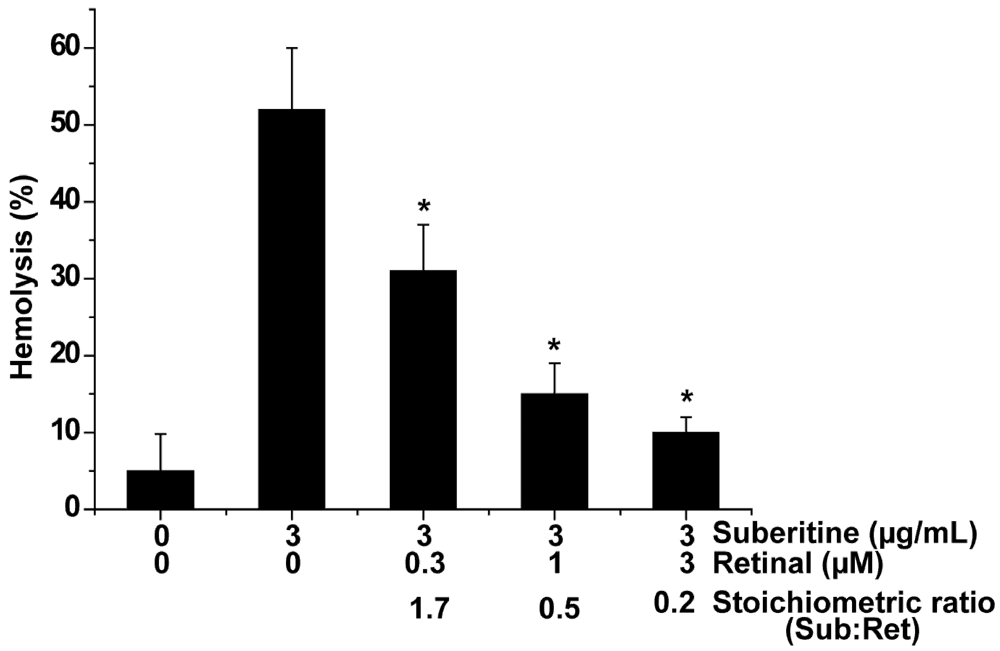
Figure 3.
Abolishment of the hemagglutination activity of Suberitine by retinal. Constant concentrations of Suberitine (3 µg/mL, equal to 0.4 µM of the purified peptide) were incubated with 0.3 to 3 µM of retinal and then added to the hemolytic test system. For the evaluation of the hemagglutination activity an assay system was used, allowing a percental assessment of the degree of hemolysis, as described under “Materials and Methods”. The values represent the means ± SEM of ten experiments each; *P < 0.05.
These data suggest that retinal interferes with Suberitine and reduces the hemagglutination activity of this peptide.
2.3. Interaction of Suberitine with Retinal
In order to assess the (potential) interfering effect of retinal with Suberitine, the optical absorption spectrum at 380 nm was followed after addition of the components. For the assay, the concentration of retinal remained unchanged, while the concentration of Suberitine was increased (Figure 4). Under the conditions used, 1 µM of retinal showed an absorbance of 0.02, a value that agrees with the extinction coefficient of 42,800 M−1 cm−1 that had been measured in ethanol [26]. Addition of Suberitine at a molar ratio (Suberitine:retinal) of 0.17 caused a reduction to 0.016; a further increase of the concentration of Suberitine in the assay caused a further reduction of the absorbance value of retinal, down to 0.01 at a 1.7 molar ratio. Suberitine alone showed no measurable absorbance at 380 nm.
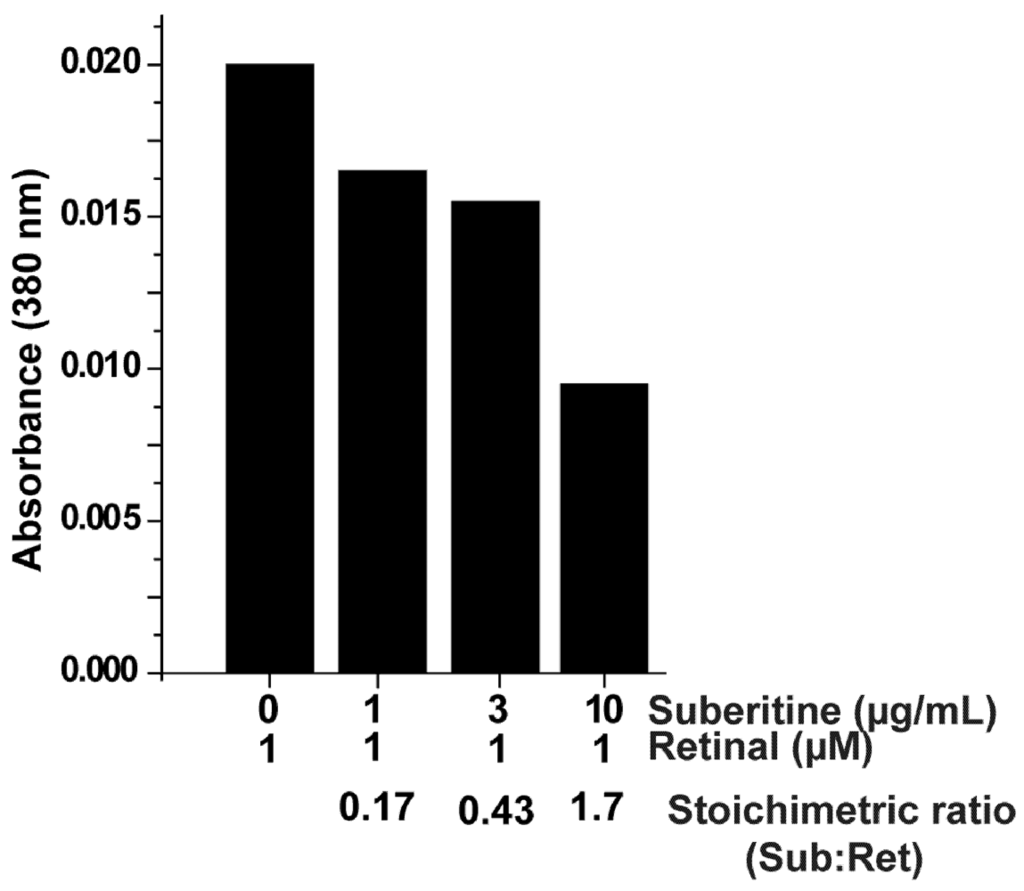
Figure 4.
Effect of Suberitine on the absorbance spectrum of retinal, here measured at the wavelength of 380 nm.
2.4. Effect of Suberitine on the Viability of Sponge Cells
The effect of Suberitine on the viability of sponge cells was determined after an incubation period of 24 h by applying the MTT assay system. The amount of formazan formation was used as an indicator for metabolically active mitochondria in viable cells. The cell viability in assays not subjected to either Suberitine or retinal was set to 100% (Figure 5). Addition of 3 µg/mL of Suberitine caused already after the 24 h incubation period a significant reduction of cell viability by 46%. This inhibitory effect was abolished by retinal at a stoichiometric ratio of 0.2 (Suberitine/retinal). Retinal alone displayed no effect on cell viability.
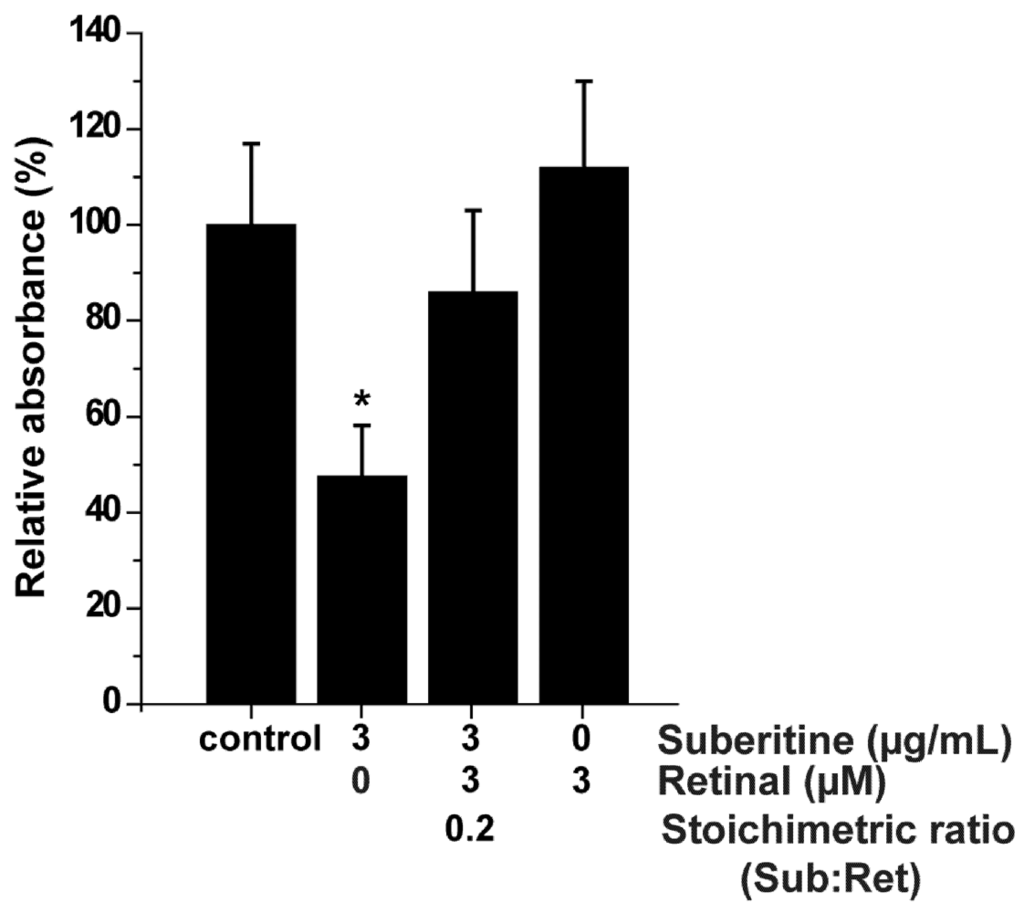
Figure 5.
MTT assay was used to determine viability of S. domuncula cells treated with Suberitine, in the absence or presence of retinal. Each data point represents the means ± SEM (n = 10); *P < 0.05 correlated to the controls.
2.5. Molecular Cloning of the Two (Related) Carotenoid Oxygenases
The complete cDNAs (SDrBCO1 and SDrBCO2), encoding the S. domuncula related β-carotenoid oxygenases, termed rBCO1_SUBDO and rBCO2_SUBDO, were isolated. The sizes of the nt sequences were 1763 bp for SDrBCO1 and 1772 bp for SDrBCO2. The completeness of the cDNAs was proven by Northern blot analysis (1.8 kb; data not shown). The two nt sequences comprise one ORF each, coding for putative 533 aa long polypeptides with a size of 60,519 Da and a pI [isoelectric point] of 8.53 for rBCO1_SUBDO; for rBCO2_SUBDO the calculated values are 60,258 Da and a pI of 8.60. The two new polypeptide sequences share a similarity/identity of 90%/89% to each other and 21%/37% between rBCO1_SUBDO and the recently identified sponge CDO1_SUBDO sequence (accession number FR848951) [20%/36% rBCO2_SUBDO to CDO1_SUBDO]. The polypeptides comprise the following signatures: the pyrokinins signature (aa449 to aa457 [with respect to rBCO1_SUBDO]), characteristic for insect neuropeptides; and the signature for the retinal pigment epithelial membrane protein superfamily (aa8 to aa517; expect value = 2.4e−29). Both motifs are also found in the sponge CDO1_SUBDO sequence (Figure 6A). In the following experiments, the sequence of the related carotenoid oxygenase rBCO2_SUBDO (SDrBCO2) was selected together with the carotene dioxygenase (CDO1_SUBDO/SDCDO1). (Database: The following sequences have been deposited in the EMBL/GenBank database; the Suberites domuncula mRNA for the putative β-carotenoid oxygenase (BCO1 gene) with the accession number HE603240, and the S. domuncula putative β-carotenoid oxygenase (BCO2 gene) with HE603239.)
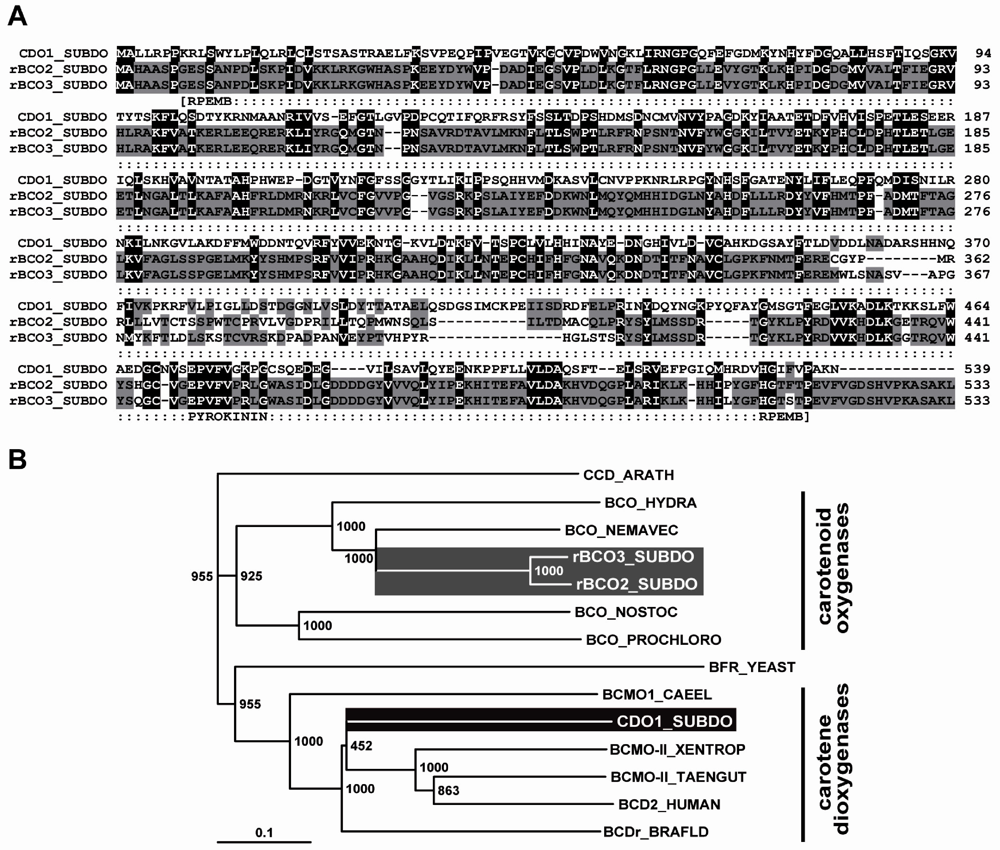
Figure 6.
S. domuncula related β-carotenoid oxygenases. (A) The two sponge putative carotenoid oxygenases (rBCO1_SUBDO; rBCO2_SUBDO) were aligned with the S. domuncula carotene dioxygenase (CDO1_SUBDO). Residues conserved (identical or similar with respect to their physico-chemical properties) in all three sequences are shown in white on black; those that share similarity only in two sequences are in black on gray. The characteristic signatures, the pyrokinin signature [pyrokinin] and the signature for the retinal pigment epithelial membrane protein superfamily [RPEMB], are marked; (B) For phylogenetic analyses, the three sponge oxygenases were aligned with the two cnidarian putative β-carotenoid oxygenases from N. vectensis (BCO_NEMAVEC) and from H. magnipapillata (BCO_HYDRA) together with the carotenoid oxygenase from the cyanobacteria, from N. punctiforme (BCO_NOSTOC) and from P. marinus (BCO_PROCHLORO). Additionally, selected carotene dioxygenases have been included; from human (BCD2_HUMAN; EAW67189.1), from Xenopus tropicalis (BCMO-II_XENTROP; XP_002942801.1), from Taeniopygia guttata (BCMO-II_TAENGUT; XP_002189781.1), from Branchiostoma floridae (BCDr_BRAFLD; XP_002605888.1) and from Caenorhabditis elegans (BCMO1_CAEEL; NP_496729.2), as well as the yeast Bfr2p from Saccharomyces cerevisiae (BFR_YEAST; EGA87437.1). As an outgroup sequence to root the tree, the 9-cis-epoxycarotenoid dioxygenase from Arabidopsis thaliana (CCD_ARATH; NP_191911.1) has been selected. The degree of support of internal branches was assessed by bootstrapping (1000 replicates) and the evolutionary distance was calculated (0.1 aa substitutions per position in the sequence).
The highest sequence similarity of the sponge related carotenoid oxygenase was identified with the two cnidarian putative β-carotenoid oxygenases from Nematostella vectensis (sea anemone; XP_001635106.1) and from Hydra magnipapillata (fresh water polyp; XP_002163128.1) with E-values of <2e−125. No similar sequence could be identified among more evolved metazoan taxa, having an E-value of <e−20. However, significant similarity was found by searching the databases to carotenoid oxygenase from cyanobacteria, e.g., from Nostoc punctiforme (YP_001864026.1; E-value = 4e−96), or from Prochlorococcus marinus (NP_874706.1; E-value = 4e−53). A phylogenetic tree was constructed with the two sponge and the two cnidarian carotenoid oxygenases together with the sponge carotene dioxygenase and some selected related polypeptides. After alignment and application of the distantly related 9-cis-epoxycarotenoid dioxygenase from Arabidopsis thaliana (NP_191911.1) as an outgroup the rooted tree showed that the β-carotenoid oxygenases and the carotene dioxygenases fall into two branches (Figure 6B). Among the carotenoid oxygenases the eukaryotic oxygenases are separated from the prokaryotic oxygenases.
2.6. Determination of Oxygenase Activity in Bacterial System
The function of the S. domuncula SDrBCO was examined in E. coli, which had been transformed with the gene clusters maintaining the accumulation of either β-carotene [E. coli(β-car+)] or lycopene [E. coli(lyco+)] in the microorganisms. These strains were subsequently additionally transformed with the S. domuncula SDrBCO gene inserted into pBAD-TOPO. As a positive control, the carotinoid synthesizing bacteria were transformed with SDCDO [16]. The bacteria, lacking the sponge oxygenase genes, either SDrBCO or SDCDO, termed E. coli(−), turned to a yellowish (for E. coli(β-car+)) or reddish color (for E. coli(lyco+)) due to the accumulation of β-carotene or lycopene, respectively (Figure 7). The E. coli strain carrying the S. domuncula SDCDO gene remained after incubation almost white or gray, indicating that those microorganisms metabolized the blue color absorbing retinoid metabolites β-carotene or lycopene, respectively. This visual assessment was supported by a quantitative analysis of the amount of β-carotene (or lycopene) in the respective cultures. A quantitative analysis of the amount of β-carotene present in overnight cultures of E. coli(−), which did not carry the SDCDO gene, revealed a concentration of 42.7 pmol/mg (dry matter), while the bacteria that had been transformed with the SDCDO gene contained only 17.3 pmol/mg of β-carotene. The same effect is seen for the E. coli(lyco+) system; the SDCDO gene-lacking bacteria contained 57.4 pmol/mg lycopene, while the double-transformed bacteria had 23.6 pmol/mg (Figure 7).
In contrast, the transformed E. coli strains (E. coli(β-car+) or E. coli(lyco+)) that had been super-transfected with the new carotenoid oxygenase gene, SDrBCO2, showed both the same coloration and almost the same amount of β-carotene (E. coli(β-car+):E. coli(+) system) or lycopene (E. coli(lyco+):E. coli(+) system), like the microorganisms not super-transfected with SDrBCO2 (E. coli(−)). Based on these findings we conclude that the carotenoid oxygenase 2 gene (SDrBCO2) does not encode an enzyme that is able to metabolize either β-carotene or lycopene (Figure 7). Therefore, we termed these new oxygenases “related carotenoid oxygenases”.
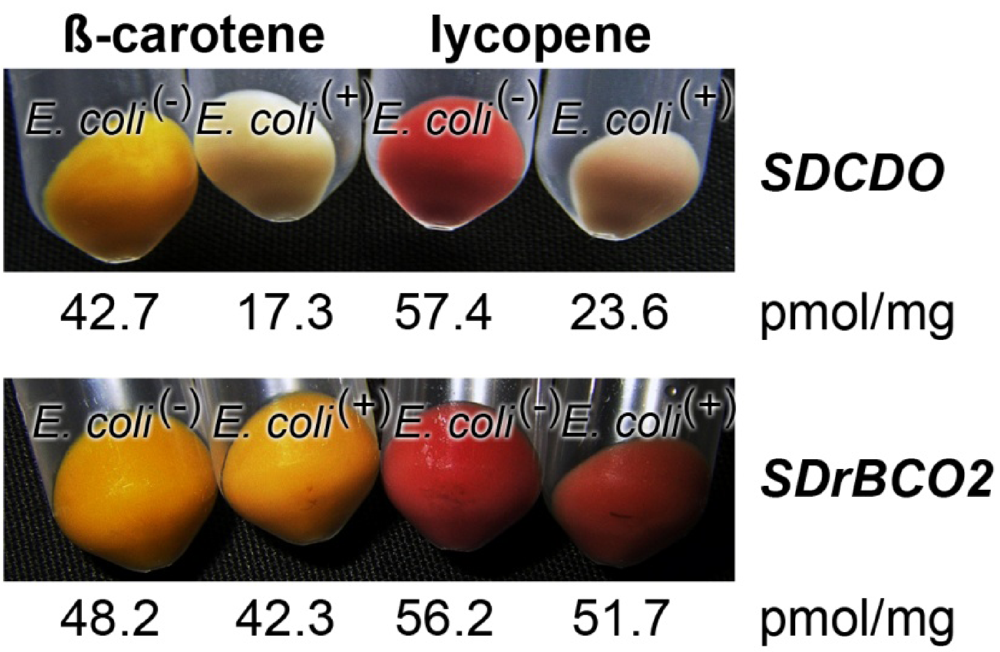
Figure 7.
Proof that the newly described oxygenase gene SDrBCO does encode an enzyme that can metabolize neither β-carotene nor the related carotenoid, lycopene, by application of the color shift E. coli system. In a positive control series, (Upper Panel) the β-carotene synthesizing E. coli(β-car+) or lycopene producing E. coli(lyco+) strains were additionally transformed with the sponge SDCDO gene (E. coli(+)), while the controls remained empty (E. coli(−)). After induction with L-(+)-arabinose the super- transformed strain (E. coli(β-car+) [E. coli(lyco+)]:E. coli(+) system) remained whitish, due to the metabolic cleavage of either β-carotene or lycopene, while the empty strain (E. coli(β-car+) [E. coli(lyco+)]:E. coli(−) system) became colored yellowish or reddish and contained high amounts of the respective retinoid β-carotene or lycopene. Testing this system (E. coli(β-car+) or lycopene producing E. coli(lyco+)) with the new oxygenase gene SDrBCO (Lower Panel) both the super-transformed strains, carrying the SDrBCO gene (E. coli(β-car+) [E. coli(lyco+)]:E. coli(+) system) or lacking it (E. coli(β-car+) [E. coli(lyco+)]:E. coli(−) system) have the same color and contain almost the same amount of retinoid, either β-carotene or lycopene, respectively.
2.7. Effect of Light on the Expression of the Carotene Dioxygenase and the Carotenoid Oxygenase 2
The steady-state expression levels of the two S. domuncula oxygenase genes, SDCDO1 and SDrBCO2 has been determined in primmorphs, a special form of cell aggregates (Figure 1C) that developed during reaggregation from single cells (Figure 1B), after exposure to light of different wavelengths. The primmorphs were incubated either in the absence or presence of 3 µM β-carotene. One day prior to the experiments the primmorphs were kept in complete darkness. Three days after starting the cultivation of the aggregates they were exposed to light of different wavelength spectra for blue, green and red light. After an exposure period of 3 days the expression levels were determined by the technique of qPCR and correlated to the expression of the house-keeping gene ß-tubulin.
In the dark the expression level of the carotene dioxygenase is low with approximately 0.2 × 10−5 transcripts with respect to that of the house-keeping gene tubulin. Addition of β-carotene has no effect on the expression level of the dioxygenase in the dark. Exposure of the primmorphs not incubated in the presence of β-carotene, to light of different wavelengths showed a non-significant increase in the level of transcripts. However, if the β-carotene-treated primmorphs were exposed to light a significant increase of the expression level was seen. The level of the dioxygenase expression increased by 10.5-fold if exposed to blue light, by 5.1-fold with green light and by 4.9-fold after exposure to red light (Figure 8).
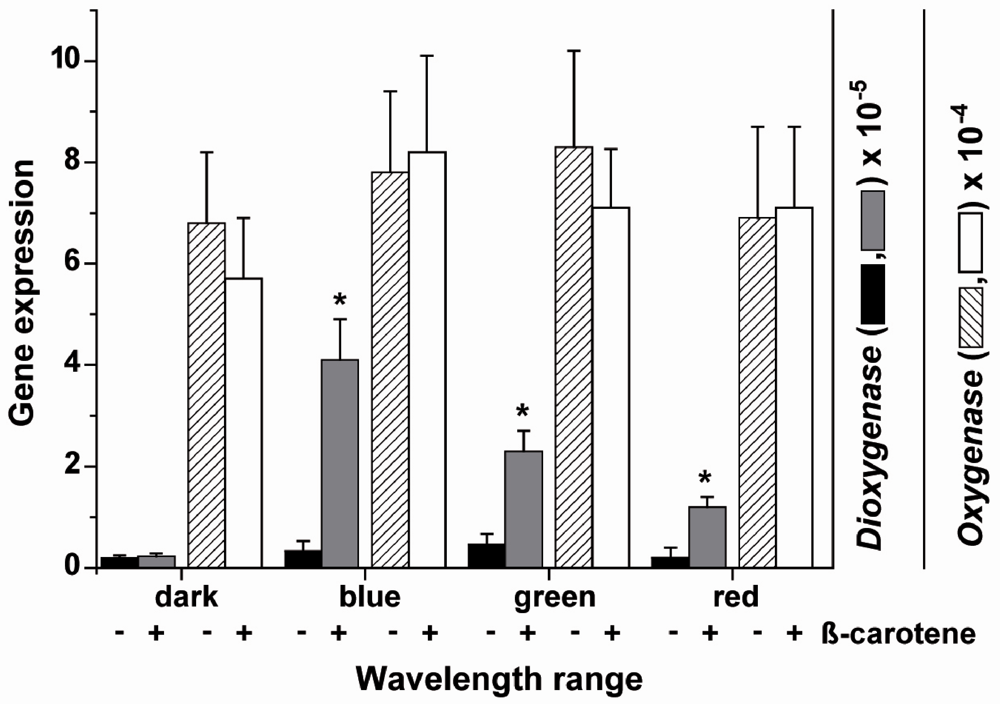
Figure 8.
Expression of the carotene dioxygenase and the carotenoid oxygenase 2 gene in primmorphs. The aggregates remained in the absence of or were incubated with 3 µM β-carotene. Three days after formation of the primmorphs the aggregates were exposed to blue, green or red light for an additional 3 days. Then the steady-state level of carotene dioxygenase and of carotenoid oxygenase 2 was determined by qPCR using tubulin as reference gene. The expression levels are shown of the two oxygenases of primmorphs kept either in the absence (−) or presence (+) of 3 µM β-carotene and exposed to light of different wavelengths. The increase of the carotene dioxygenase expression is significant (*P < 0.05 referred) in primmorphs if the light exposed cultures had been incubated also with β-carotene. The bars represent the means (±SD) from 10 separate experiments each.
The expression level of the carotenoid oxygenase 2 in primmorphs in the dark was higher (around 7 × 10−4, correlated to the level of tubulin), compared to that of the dioxygenase (Figure 8). This level did not change if the primmorphs were incubated with β-carotene or not (Figure 8; only the values for the expression in the dark are shown). However, in contrast to the carotene dioxygenase, the steady-state expression of the carotenoid oxygenases 2 in the primmorphs did not change significantly, if the aggregates were exposed to blue, green or red light (Figure 8), irrespective if they were β-carotene treated or not (data not shown).
3. Discussion
Suberitine is a neurotoxic (for crabs) and hemolytic (for mammalian erythrocytes) protein [22] which is even cytotoxic for the cells of the producer animal, S. domuncula, as shown in this study. Therefore, this sponge had to develop a mechanism that controls the potential self-directed bioactivity within the own body. Until now the genetic blueprint for Suberitine is not known. Therefore, it was not yet possible to elucidate if Suberitine is activated after translation, perhaps by formation of disulfide linkage or by proteolytic processing of a biologically inactive pro-toxin.
Due to the preliminary work by Cariello and Zanetti [24], describing a S. domuncula blue carotenoprotein, it was suggestive to postulate a connection between retinoids and Suberitine, an association that might modulate the bioactivity. In a recent study it had been shown that S. domuncula cells and tissue are especially rich in β-carotene and retinal [16]. While β-carotene is a product of the sponge-associated microorganisms, retinal is the enzymatically cleaved metabolite formed by the host-encoded carotene dioxygenase. The concentration of β-carotene in freshly collected specimens from the field is high with 137 ± 12 nmol/g dry tissue (carrots in comparison contain about 300 nmol/g [27]), and drops in animals kept in aquaria to 11 ± 4 nmol/g in parallel with the decrease of the load with bacteria [16]. In view of the findings that (i) the tryptophyl residues within the Suberitine molecule are crucial for its activity [23] and (ii) that retinal binds to tryptophan residues in some proteins, e.g., rhodopsin [28], by a reversible energy transfer reaction, we studied the quenching effect of Suberitine on retinal on a molar stoichiometric basis. The data revealed that a distinct reduction of the absorbance of retinal occurred at 380 nm at a stoichiometric Suberitine/retinal ratio of ≥1, suggesting a non-covalent interaction between these two components. This assumption is supported by gel chromatographic studies, which revealed that Suberitine, incubated with retinal and subsequently chromatographed with a 1 M NaCl containing PBS buffer through the Sephadex G-50 matrix, had the same elution behavior like untreated Suberitine (data not shown). This spectroscopic finding was confirmed by functional studies as well. Both the hemolytic activity and the cell toxicity of Suberitine were significantly reduced by retinal at a stoichiometric Suberitine/retinal ratio of around 1. The interplay of retinal with especially light-sensitive retinylidene proteins is well studied [29]. This interaction can be attributed to the retinal-mediated protein conformation changes caused by tryptophan-retinal coupling and subsequent quenching [30]. From these in vitro data we conclude that retinal controls the activity of Suberitine at a stoichiometric ratio of around 1. Moreover, we propose that also in vivo retinal modulates the biological activity of Suberitine. This view would imply that the β-carotene level, and in turn also the retinal level, affects the self-directed toxicity of the toxin (Figure 9). If the level of the retinoid is high the toxin displays no cytotoxic effect on the host producer cells, while at lower levels of these metabolites Suberitine also causes self-directed cell toxicity. This proposed mechanism may not be as far-fetched as it sounds at first glance. From previous studies it is well established that in S. domuncula self-destruction of tissue occurs after (i) tissue impairment in response to low aeration or after the adverse influence of xenobiotics [10] or (ii) as allograft reaction [31]. These two processes are the results of (self-directed) apoptotic responses. The self-directed cell toxicity caused by Suberitine is likely caused by necrosis, since no apoptotic DNA fragmentation could be indentified in sponge cells, exposed to the homologous toxin.
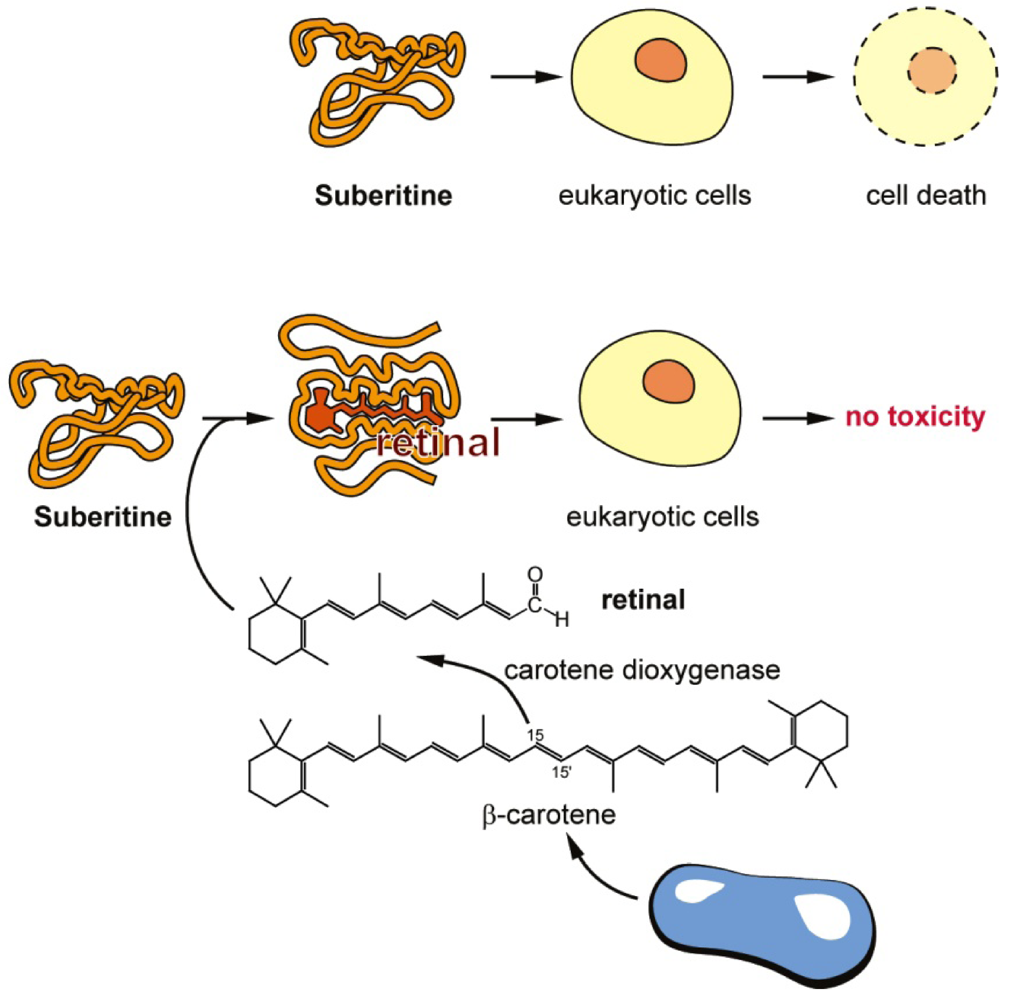
Figure 9.
Proposed interaction of retinal with Suberitine (scheme). (Upper panel) In the absence of retinal Suberitine interacts with eukaryotic cells by killing them either by hemolysis or by (likely) necrotic cell death; (Lower panel) Retinal interacts with Suberitine by non-covalent, reversible energy transfer reaction under formation of a complex that is not toxic for eukaryotic cells; (Bottom) The route of formation of retinal from β-carotene that is produced in bacteria is outlined.
From previous studies it is known that the retinoids (existing in sponges) in the species S. domuncula are derived from β-carotene through sponge-associated/symbiotic microorganisms [15,16]. In more evolved metazoans, e.g., in vertebrates, this pro-vitamin A carotenoid is either symmetrically cleaved (via the β,β-carotene-15,15′-dioxygenase) to retinal [25] or asymmetrically metabolized (via the asymmetrically cleaving by β-carotene dioxygenases) to β-apo-10′-carotenal and β-ionone [32]. Since in sponges, until now only the β,β-carotene-15,15′-dioxygenase has been identified and cloned [16], a screening for potential additional genes that might be involved in the β-carotene metabolism was needed. Searching the S. domuncula database that comprises over 40,000 sequences [33], two clusters of ESTs (Expressed Sequence Tag) sequences were found whose deduced proteins were closely related to carotenoid oxygenases. The cDNA sequences of both groups were completed. A phylogenetic analysis of the resulting two deduced polypeptides revealed that these new sponge polypeptides share highest sequence similarity to the hydrozoan hypothetical oxygenases from N. vectensis and from H. magnipapillata. Surprisingly, no further related polypeptides could be identified in higher metazoans; however, distinct similarities exist to cyanobacterial carotenoid oxygenases. Since the two new sponge oxygenases are highly similar in their sequences (90% on the protein level), only one of them, the rBCO3_SUBDO (SDrBCO3) was selected for the further analyses. The fact that these mentioned sponge genes/cDNAs, encoding for proteins involved in retinoid metabolism, have been (initially) identified in an EST (Expressed Sequence Tag) database from S. domuncula (see [10]) implies that the cDNAs were obtained from poly(A)+ mRNAs, sequences that exist only in multicellular eukaryotic organisms. Hence, already this piece of evidence supports that these genes are described from sponges derived from the S. domuncula host and not from associated microorganism. Furthermore, the phylogenetic analyses of the deduced proteins, given here and previously [16], clearly identify the sequences as sponge-derived.
In order to verify a potential β-carotene-degrading tomato carotenoid, lycopene [34], metabolizing activity that might be caused by the new sponge oxygenase, bacterial β-carotene- and lycopene-synthesizing and accumulating E. coli strains were used [32,35]. The data revealed that E. coli that had been transformed with the new sponge oxygenase did not degrade β-carotene or lycopene. In turn, the new sponge oxygenases were termed “related carotenoid oxygenases”. Based on these findings we suggest that the new related carotenoid oxygenase from S. domuncula might hydrolyze other retinoids in sponges. From studies in this animal it is known that a series of retinoids, retinal, retinol or retinoic acid, exists that is distinctly different from β-carotene [15,16]. Furthermore, the high abundance of carotenoids in sponges is well described [36].
Published studies described that in S. domuncula retinoic acid acts as a morphogen during the development of aquiferous canals [17] and as an inducer of the bone morphogenetic protein-1, required for the processing of silintaphin-2 during spicule formation [16]. Since S. domuncula comprises a light-reacting protein network with the luciferase as light-producing [37] and the cryptochrome as a (potential) photon-responding molecule [38], our group intensively screened for light sensory photopigments. β-Carotene, retinol and retinal are surely candidates that might act as putative cofactor(s) of the suspected photopigment protein. Therefore, we incubated primmorphs with β-carotene, retinol or retinal, and determined their effect on the expression of the carotene dioxygenase as well as on the new described, related carotenoid oxygenase in dependence on light of different wavelengths. From such a study we wanted to obtain a first indication if the enzymatic processing pathway of β-carotene, either β-carotene itself or the retinal/retinol metabolites, is linked to distinct light wavelengths. The data revealed that indeed light causes a differential effect on gene expression of the dioxygenases. At first, it was established that only the expression of the carotene dioxygenase is influenced by light, while the steady-state level of the carotenoid oxygenase gene remained unchanged. This effect is seen only in those primmorphs that had been incubated with β-carotene. Primmorphs that were treated with retinol or retinal showed a much lower response to light (data not shown). In the dark the level of the carotene dioxygenase transcripts was relatively low. Exposure to light significantly increased the expression of the carotene dioxygenase; especially the response to blue light is high (10.5-fold) compared to the expression in the dark. Red or green light was less effective. This finding can be explained on the basis of the habitat where S. domuncula lives. In the sea these animals are found in a depth of about 20 m, an environment that is predominantly penetrated by the shorter wavelength spectrum of visible light, of around 380 to 500 nm (blue) [39]. Hence, the carotene dioxygenase gene is, after the cryptochrome [38], the second sponge gene whose expression is light-controlled. To the best of our knowledge no report has been published showing that the expression of the carotene dioxygenase in metazoans is light sensitive. In contrast, in plants the gene encoding the related 9,10/9′,10′ carotenoid cleavage dioxygenase is light-inducible [40]. Since the blue light-dependent increase of the sponge gene expression is only seen after incubation of the primmorphs with β-carotene we take this finding as a first evidence that β-carotene, or a deriving metabolite of it, is implicated in the light-reacting protein network in sponges.
4. Experimental Section
4.1. Sponge and Primmorphs
Specimens of S. domuncula (Porifera, Demospongiae, Hadromerida) were collected in the Northern Adriatic Sea near Rovinj (Croatia) at depths between 18 and 25 m, and then kept in aquaria in artificial seawater (Tropic Marine, Tropic Marine Centre Ltd., Rickmansworth; UK) in Mainz (Germany) at 17 °C [41]. For the experiments animals that had been kept in the aquarium for over three months were used. From those specimens primmorphs, 3D cell aggregates were prepared as described [16,42] and cultivated in natural seawater (Sigma-Aldrich, Taufkirchen; Germany) supplemented with 1% RPMI 1640 medium (Sigma). Where indicated, the primmorphs were incubated with 1 µM β-carotene (Sigma; C4582).
4.2. Exposure of Primmorphs to Light
The experimental design was given previously [43]. In short, primmorphs were prepared and kept for one day in complete darkness; at day 3 the aggregates were used for the experiments. Then the primmorphs were exposed to light of different wavelength ranges from a distance of 5–6 cm by using optical filters (Schott-Advanced Optics, Mainz, Germany) that were placed in front of a white light bulb [50 m2 kg/s3 (General Electric, Fairfield, CT, USA) 29208 projector white light bulb]. The following filters were used: for blue light a DMZ 12 filter (cutoff below 400 nm; maximum at 490 nm; cutoff above 599 nm), for green light VG9 (400 nm maximum at 520–650 nm), and red light KMZ 50 OG590 (700 nm maximum at 780–900 nm). The controls remained in the dark. Where indicated, the primmorphs were incubated from the beginning of the re-aggregation from single cells with 3 µM β-carotene.
4.3. Purification of Suberitine
For the isolation and purification of Suberitine the procedure of Cariello and Zanetti [19,24] has been used with modifications adopted from Müller et al. [44]. Sponge cubes were extracted in Ca2+- and Mg2+-free seawater; the suspension was centrifuged (5 °C; 80,000 × g; 30 min). Starting from 50 g of tissue a fraction (Fraction I) of 78 mL (2.8 mg protein/mL) was obtained. This fraction was subjected to ammonium sulfate precipitation (final concentration 80%). The precipitate formed was collected by centrifugation (5 °C; 80,000 × g; 30 min) and dissolved in 3 mL of PBS (pH = 7.4; 137 mM NaCl, 10 mM Na-phosphate, 2.7 mM KCl). The resulting Fraction II (3.1 mL; 4.2 mg protein/mL) was dialyzed exhaustively against PBS. Subsequently Fraction II was fractionated (in aliquots of 1 mL) by gel chromatography on Sephadex G-100 (Sigma-Aldrich; column size: diameter 1 cm, height 40 cm; fraction volume 1.2 mL). The fractions containing the hemagglutination activity, which eluted at a ratio Ve/Vo (elution volume/void volume) [45] of 2.2 to 2.4 (3.6 mL), were collected (Fraction III; 0.9 mg protein/mL) and finally purified by gel filtration using Sephadex G-50 superfine (Sigma-Aldrich; column size: diameter 1 cm, height 50 cm; fraction volume 1.2 mL), as described [19]. The fractions with the highest hemagglutination activity (3.6 mL) were determined at a Ve/Vo ratio of 1.60 to 1.72 (Fraction IV; 0.29 mg protein/mL).
4.4. SDS-PAGE Analysis
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described [16]. Samples of 3 μg of protein were mixed with loading buffer (Roti-Load; Carl Roth, Karlsruhe; Germany), boiled for 8 min, and subjected to SDS-PAGE (15% acrylamide and 0.1% SDS). The gels were stained with Coomassie brilliant blue. The protein size standards (“Low Range Protein Ladder”) from Thermo Fisher Scientific (Rockford, IL USA) were used to estimate protein sizes.
4.5. Testing for Hemolytic Activity
The hemolytic action of Suberitine on red blood cells was tested with human type A erythrocytes as recently described [10]. In brief, a 10% (v/v) suspension of PBS-washed erythrocytes was exposed to serial dilutions of the respective Suberitine fraction in microtiter plates (Nunc/Thermo Electron, Langenselbold; Germany). For comparison and calibration of total hemolysis the samples were subjected to 1% (v/v) Triton X-100. After incubation (1 h, 37 °C) the plates were centrifuged (20 °C; 2000 × g; 10 min) and a 150 µL aliquot from the supernatant was taken and used for absorbance measurements at 450 nm. The reciprocal of the greatest dilution at which agglutination occurred was taken and given as titer (units) [46]. The values are related to the protein concentration in the sample.
For the competition experiments of Suberitine with retinal, we applied a different hemagglutination assay in order to assess the change of the activity on a percental basis [10]. The percentage of hemolysis was calculated as follows: (A450 of the Suberitine-treated sample − A450 of buffer-treated sample)/(A450 of Triton X-100-treated sample − A450 of buffer-treated sample) × 100%.
In the co-incubation series of Suberitine with retinoids the purified peptide sample (either 3 µg/mL [0.4 µM]; or 1 µg/mL [0.14 µM]) was incubated with those compounds for 15 min at 37 °C and then used for the experiments.
4.6. Interaction of Suberitine with Retinal
In an earlier study it was clarified that besides cysteinyl residues, tryptophyl residues are also crucial for full activity of Suberitine [23]. Furthermore, it is known that retinal interacts with tryptophan residues in proteins [47], such as in the human retinol-binding protein, most likely by quenching the absorbance of the retinoid [48]. In order to assess the (potential) interfering effect of retinal with Suberitine the optical absorption spectrum at 380 nm was followed.
4.7. Quenching of the Optical Absorption Spectrum of Retinal by Suberitine
The effect of retinal on the absorbance spectrum by Suberitine was measured optically using a Zeiss Spectrophotometer PMQ II at 380 nm, the maximum of the absorbance spectrum of retinal. The components were dissolved in PBS (pH 7.4).
4.8. MTT Sponge Cell Viability Assay
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay system, optimized for the determination of sponge cells, was applied [49]. Sponge single cells were obtained by dissociation in Ca2+- and Mg2+-free seawater; 200 µL each of a cell suspension (106 cells/mL, adjusted with a hemocytometer) were added to wells of microtiter plates and subsequently incubated for 24 h in natural seawater, supplemented with 1% RPMI 1640 medium. The incubation was terminated by addition of MTT (0.5 mg/mL) and the optical densities were measured at 495 nm (595 nm as reference wavelength) in an ELISA Reader (model 450; Bio-Rad).
4.9. Molecular Cloning of S. domuncula (Related) Carotenoid Oxygenases
By searching the S. domuncula EST (Expressed Sequence Tag) database [33] several fragments were identified that encoded parts of a putative carotenoid oxygenase polypeptide. Their ORFs (open reading frame) were completed by the 3'- and 5'-racing technique using the CapFishing Full-length cDNA Premix Kit (Seegene; Rockville, MD, USA) as described [50]. Two different sequences were identified that were termed putative β-carotene oxygenases [BCO]-1 and -2. Since (as will be described later) these oxygenases do not cleave β-carotene, we termed these putative molecules “related to β-carotene oxygenases”. The complete nucleotide (nt) clones of the sponge related β-carotene oxygenases, SDrBCO1 and SDBrCO2, comprise 1763 base pairs (bp) and 1772 bp, respectively, excluding the poly(A) streches; the ORFs rang from nt45–47 to nt1644–1646(stop) and from nt45–47 to nt1644–1646.
4.10. Sequence Analyses
The sponge sequences were analyzed with computer programs BLAST [51] and FASTA [52]. Multiple alignments were performed with CLUSTAL-W Ver. 1.6 [53] and the degree of support for internal branches was further assessed by bootstrapping [53]. Motif scan was performed using the internet database [54]. The graphic presentations were performed by using the Genedoc program [55].
4.11. Determination of the Potential Carotenoid Oxygenase Activity in the Bacterial System
Like in the recent study to prove that the sponge SDCDO cDNA encodes a carotene dioxygenase enzyme that cleaves β-carotene to retinal [16] by using the β-carotene- and lycopene-synthesizing and accumulating E. coli strains (E. coli JM109 [32,35]), this bacterial system was also applied to clarify if the carotenoid oxygenases (SDrBCO), cloned here, metabolize this retinoid. These indicator E. coli strains were termed E. coli(β-car+) and E. coli(lyco+); the transformed strains become yellow/reddish since they form blue light absorbing β-carotene or lycopene, respectively. In the present study the bacteria were transformed with the S. domuncula gene SDrBCO2 using the vector pBAD-TOPO (Invitrogen/Life Technologies, Darmstadt; Germany) following the described procedure [35]. Overnight cultures were induced with 0.5% (w/v) of l-(+)-arabinose for 3 h [32]. Subsequently the cells were collected, broken, and the amounts of β-carotene and lycopene were determined by reversed phase HPLC. In a parallel series of experiments the bacteria were transformed with the SDCDO cDNA [16] and used as a positive control. As one control, the strain lacking the genes for the oxygenase enzymes, named E. coli(−), were used.
4.12. Quantitative Analysis of β-carotene and Lycopene
Defined amounts (wet weight) of broken bacterial cells were extracted with acetone. After evaporation identical aliquots (10 µL) were separated by reversed phase HPLC [35]. The quantification was achieved by calibration with peaks from defined amounts of β-carotene reference compound.
4.13. Quantitative Real-Time RT-PCR (qRT-PCR)
The determination of the steady-state level of the oxygenase transcripts was performed by quantitative real-time RT-PCR (qRT-PCR) as described [16,44]. In brief, RNA was extracted and reverse transcribed and then subjected to qPCR in an iCycler (Bio-Rad; Hercules, CA). The following primer pairs were applied; for amplification of the carotene dioxygenase (accession number FR848951) Fwd: 5'-ATGCACGTTCTCATCACAATC-3' (nt1088 to nt1108) and Rev: 5'-GCACATAATACTCCCGTCACTC-3' (nt1239 to nt1218; size of the fragment, 152 bp) and for the amplification of the S. domuncula (related) carotenoid oxygenase 2 (SDrBCO2) Fwd: 5'-TGGATACAAACTACCCTACAGAG-3' (nt1,257 to nt1,279) and Rev: 5'-ACATAGCCATCATCATCATCAC-3' (nt1412 to nt1391; 156 bp). The level of transcripts the house-keeping molecule ß-tubulin (AJ550806) was quantified with the primer pair Fwd: 5'-AACCGCTGTTTGCGACATCC-3' (nt1126 to nt1145) and Rev: 5'-CAATGCAAGAAAGCCTTTCGCC-3' (nt1266 to nt1245; 141 bp). The threshold position was set to 50.0 RFU above PCR subtracted baseline for all runs. Mean Ct values and efficiencies were calculated by the iCycler software. The estimated PCR efficiencies were in a range of 92%–103%. Expression levels of the respective transcripts for the oxygenases were correlated with the one for ß-tubulin to assess the relative expression levels (EtubulinCt oxygenase/EoxygenaseCt oxygenase), whereby “E” describes the PCR efficiency and “Ct” represents the threshold cycle [56].
4.14. Additional Analytical Method
Protein concentrations were determined by the Bradford method [57] using Roti-Quant solution (Roth, Karlsruhe; Germany).
4.15. Statistical Analysis
The results were statistically evaluated using the paired Student’s t-test [58].
4. Conclusion
The data in the present study show that retinal controls the activity of the toxin Suberitine in its homologous system as well as in the vertebrate erythrocyte system. At a molar, stoichiometric ratio of 1:1 the retinoid abolishes the function of this toxin. The experiments suggest that the change of the biological activity is caused by a non-covalent interaction which, very likely, results in a change of the conformation of the protein. In turn, this is the first report which highlights that in sponges a low molecular bioactive metabolite can interact with a bioactive peptide and thus alters its function. This result also indicates that bioactive peptides/secondary metabolites in sponges might not only function as toxic agents but can also have the capacity to modulate the bioactivity of another toxic compound produced in the homologous system.
Acknowledgements
W.E.G. Müller is a holder of an ERC Advanced Investigator Grant (no. 268476-–BIOSILICA). This work was supported by grants from the German Bundesministerium für Bildung und Forschung, the Deutsche Forschungsgemeinschaft, the European Commission/EUREKA (EUROSTARS, No. 4289–SILIBACTS), the International Human Frontier Science Program, the European Commission (project No. 244967-Mem-S), the Public Welfare Project of Ministry of Land and Resources of the People’s Republic of China (Grant No. 201011005-06) and the International S & T Cooperation Program of China (Grant No. 2008DFA00980).
- Samples Availability: Available from the authors.
References
- Bergmann, W.; Feeney, R.J. The isolation of a new thymine pentoside from sponges. J. Am. Chem. Soc. 1950, 72, 2809–2810. [Google Scholar]
- Sarma, A.S.; Daum, T.; Müller, W.E.G. Secondary metabolites from marine sponges: Part I: Origin and chemistry of new metabolites, and synthetic studies; Part II: Biological properties of new metabolites and physiological activities of avarol and related compounds isolated from Dysidea sp. In Akademie gemeinnütziger Wissenschaften zu Erfurt; Ullstein-Mosby Verlag: Berlin, Germany, 1993; pp. 1–168. [Google Scholar]
- Pomponi, S.A. The potential for the marine biotechnology industry. Trends Future Chall. US Natl. Ocean Coast. Policy 1998, 101–104. [Google Scholar]
- Dictionary of Marine Natural Products with CD-ROM; Blunt, J.W.; Munro, M.H.G. (Eds.) Chapman and Hall/CRC: Boca Raton, FL, USA, 2008; p. 2534.
- Müller, W.; Yamazaki, Z.; Sögtrop, H.H.; Zahn, R.K. Action of 1-β-D-arabinofuranosylcytosine on mammalian tumor cells. 2. Inhibition of mammalian and oncogenic viral polymerases. Eur. J. Cancer 1972, 8, 421–428. [Google Scholar] [PubMed]
- Müller, W.E.G.; Zahn, R.K.; Beyer, R.; Falke, D. 9-β-D-arabinofuranosyladenine as a tool to study Herpes simplex virus DNA replication in vitro. Virology 1977, 76, 787–796. [Google Scholar]
- Mebs, D.; Weiler, I.; Heinke, H.F. Bioactive proteins from marine sponges: Screening of sponge extracts for hemagglutinating, hemolytic, ichthyotoxic and lethal properties and isolation and characterization of hemagglutinins. Toxicon 1985, 23, 955–962. [Google Scholar]
- Pfeifer, K.; Haasemann, M.; Gamulin, V.; Bretting, H.; Fahrenholz, F.; Müller, W.E.G. S-type lectins occur also in invertebrates: Unusual subunit composition and high conservation of the carbohydrate recognition domain in the lectin genes from the marine sponge Geodia cydonium. Glycobiology 1993, 3, 179–184. [Google Scholar]
- Ghosh, D.; Saha, M.; Sana, B.; Mukherjee, J. Marine enzymes. Adv. Biochem. Engin/Biotechnol. 2005, 96, 189–218. [Google Scholar] [CrossRef]
- Wiens, M.; Schröder, H.C.; Korzhev, M.; Wang, X.H.; Batel, R.; Müller, W.E.G. Inducible ASABF-type antimicrobial peptide from the sponge Suberites domuncula: microbicidal and hemolytic activity in vitro and toxic effect on molluscs in vivo. Mar. Drugs 2011, 9, 1969–1994. [Google Scholar]
- Wang, G. Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies; CABI: Oxfordshire, UK, 2010. [Google Scholar]
- Müller, W.E.G. Sponges (Porifera); Springer Press: Berlin, Germany, 2003. [Google Scholar]
- Müller, W.E.G.; Diehl-Seifert, B.; Sobel, C.; Bechtold, A.; Kljajic, Z.; Dorn, A. Sponge secondary metabolites: biochemical and ultrastructural localization of the antimitotic agent avarol in Dysidea avara. J. Histochem. Cytochem. 1986, 34, 1687–1690. [Google Scholar]
- Ebel, R.; Brenzinger, M.; Kunze, A.; Gross, H.J.; Proksch, P. Wound activation of protoxins in marine sponge Aplysina aerophoba. J. Chem. Ecol. 1997, 23, 1451–1462. [Google Scholar]
- Biesalski, H.K.; Doepner, G.; Tzimas, G.; Gamulin, V.; Schröder, H.C.; Batel, R.; Nau, H.; Müller, W.E.G. Modulation of myb gene expression in sponges by retinoic acid. Oncogene 1992, 7, 1765–1774. [Google Scholar]
- Müller, W.E.G.; Binder, M.; von Lintig, J.; Guo, Y.W.; Wang, X.H.; Kaandorp, J.A.; Wiens, M.; Schröder, H.C. Interaction of the retinoic acid signaling pathway with spicule formation in the marine sponge Suberites domuncula through activation of bone morphogenetic protein-1. Biochim. Biophys. Acta 2011, 1810, 1178–1194. [Google Scholar]
- Wiens, M.; Batel, R.; Korzhev, M.; Müller, W.E.G. Retinoid X receptor and retinoic acid response in the marine sponge Suberites domuncula. J. Exp. Biol. 2003, 206, 3261–3271. [Google Scholar]
- Richet, C. De l'action toxique de la subéritine (extrait aqueux de Suberites domuncula). C. R. Séanc. Soc. Biol. 1906, 61, 598–600. [Google Scholar]
- Cariello, L.; Zanetti, L. Suberitine, the toxic protein from the marine sponge, Suberites domuncula. Comp. Biochem. Physiol. 1979, 64C, 15–19. [Google Scholar]
- Arndt, W. Die Spongien als kryptotoxische Tiere. Zool. Jb. Abt. Allg. Zool. 1928, 45, 343–360. [Google Scholar]
- Cariello, L.; Salvato, B.; Jori, G. Partial characterization of suberitine, the neurotoxic protein purified from Suberites domuncula. Comp. Biochem. Physiol. 1980, 67B, 337–344. [Google Scholar]
- Cariello, L.; Tosti, E.; Zanetti, L. The hemolytic activity of suberitine. Comp. Biochem. Physiol. 1982, 73C, 91–93. [Google Scholar]
- Cariello, L.; Salvato, B.; Jori, G. The role of the cysteinyl and one of the tryptophyl residues in the neurotoxic action of suberitine. Experientia 1981, 37, 801–803. [Google Scholar]
- Cariello, L.; Zanetti, L. A blue carotenoprotein from the marine sponge Suberites domuncula: purification and properties. Mar. Biol. 1981, 62, 151–155. [Google Scholar]
- Giuliano, G.; Al-Babili, S.; von Lintig, J. Carotenoid oxygenases: Cleave it or leave it. Trends Plant Sci. 2003, 8, 145–149. [Google Scholar]
- Rehorek, M.; Heyn, M.P. Binding of all-trans-retinal to the purple membrane. Evidence for cooperativity and determination of the extinction coefficient. Biochemistry 1979, 18, 4977–4983. [Google Scholar]
- Edwards, A.J.; Nguyen, C.H.; You, C.S.; Swanson, J.E.; Emenhiser, C.; Parker, R.S. α- and β-Carotene from a commercial carrot puree are more bioavailable to humans than from boiled-mashed carrots, as determined using an extrinsic stable isotope reference method. J. Nutr. 2002, 132, 159–167. [Google Scholar] [PubMed]
- Farrens, D.L.; Khorana, H.G. Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J. Biol. Chem. 1995, 270, 5073–5076. [Google Scholar]
- Ottolenghi, M.; Sheves, M. The photophysics and photochemistry of retinal proteins. Isr. J. Chem. 1995, 35, 193–513. [Google Scholar]
- Jang, D.J.; el-Sayed, M.A. Tryptophan fluorescence quenching as a monitor for the protein conformation changes occurring during the photocycle of bacteriorhodopsin under different perturbations. Proc. Natl. Acad. Sci. USA 1989, 86, 5815–5819. [Google Scholar]
- Wiens, M.; Perović-Ottstadt, S.; Müller, I.M.; Müller, W.E.G. Allograft rejection in the mixed cell reaction system of the demosponge Suberites domuncula is controlled by differential expression of apoptotic genes. Immunogenetics 2004, 56, 597–610. [Google Scholar]
- Kiefer, C.; Hessel, S.; Lampert, J.M.; Vogt, K.; Lederer, M.O.; Breithaupt, D.E.; von Lintig, J. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J. Biol. Chem. 2001, 276, 14110–14116. [Google Scholar]
- Sponge sequence database found under SpongeBase. Available online: https://octavia.vk.medizin.uni-mainz.de/login.cgi (accessed on 15 August 2010).
- Amir, H.; Karas, M.; Giat, J.; Danilenko, M.; Levy, R.; Yermiahu, T.; Levy, J.; Sharoni, Y. Lycopene and 1,25-dihydroxyvitamin D3 cooperate in the inhibition of cell cycle progression and induction of differentiation in HL-60 leukemic cells. Nutr. Cancer 1999, 33, 105–112. [Google Scholar]
- von Lintig, J.; Vogt, K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving β-carotene to retinal. J. Biol. Chem. 2000, 275, 11915–11920. [Google Scholar]
- Liaaen-Jensen, S.; Renstrøm, B.; Ramdahl, T.; Hallenstvet, M.; Bergquist, P. Carotenoids of marine sponges. Biochem. Syst. Ecol. 1982, 10, 167–174. [Google Scholar]
- Müller, W.E.G.; Kasueske, M.; Wang, X.H.; Schröder, H.C.; Wang, Y.; Pisignano, D.; Wiens, M. Luciferase a light source for the silica-based optical waveguides (spicules) in the demosponge Suberites domuncula. Cell Mol. Life. Sci. 2009, 66, 537–552. [Google Scholar]
- Müller, W.E.G.; Wang, X.H.; Schröder, H.C.; Korzhev, M.; Grebenjuk, V.A.; Markl, J.S.; Jochum, K.P.; Pisignano, D.; Wiens, M. A cryptochrome-based photosensory system in the siliceous sponge Suberites domuncula (Demospongiae). FEBS J. 2010, 277, 1182–1201. [Google Scholar]
- Rangarajan, K. Light penetration in the inshore waters of Porto Novo. Proc. Indian Acad. Sci 1959, 49B, 271–279. [Google Scholar]
- Simkin, A.J.; Underwood, B.A.; Auldridge, M.; Loucas, H.M.; Shibuya, K.; Schmelz, E.; Clark, D.G.; Klee, H.J. Circadian regulation of the PhCCD1 carotenoid cleavage dioxygenase controls emission of β-ionone, a fragrance volatile of Petunia flowers. Plant Physiol. 2004, 136, 3504–3514. [Google Scholar]
- LePennec, G.; Perović, S.; Ammar, M.S.A.; Grebenjuk, V.A.; Steffen, R.; Müller, W.E.G. Cultivation of primmorphs from the marine sponge Suberites domuncula: morphogenetic potential of silicon and iron. J. Biotechnol. 2003, 100, 93–108. [Google Scholar]
- Müller, W.E.G.; Wiens, M.; Batel, R.; Steffen, R.; Schröder, H.C.; Borojevic, R.; Custodio, M.R. Establishment of a primary cell culture from a sponge: primmorphs from Suberites domuncula. Mar. Ecol. Progr. Ser. 1999, 178, 205–219. [Google Scholar]
- Wiens, M.; Wang, X.; Unger, A.; Schröder, H.C.; Grebenjuk, V.A.; Pisignano, D.; Jochum, K.P.; Müller, W.E.G. Flashing light signaling circuit in sponges: Endogenous light generation after tissue ablation in Suberites domuncula. J. Cell Biochem. 2010, 111, 1377–1389. [Google Scholar]
- Müller, W.E.G.; Müller, I.; Pondeljak, V.; Kurelec, B.; Zahn, R.K. Species-specific aggregation factor in sponges. Isolation, purification and characterization of the aggregation factor from Suberites domuncula. Differentiation 1978, 10, 45–53. [Google Scholar] [CrossRef]
- Determann, H. Gel Chromatography-Gel Filtration, 2nd ed; Springer Press: Berlin, Germany, 1969. [Google Scholar]
- Kljajic, Z.; Schröder, H.C.; Rottmann, M.; Cuperlovic, M.; Movesian, M.; Uhlenbruck, G.; Gasic, M.; Zahn, R.K.; Müller, W.E.G. A D-mannose specific lectin from Gerardia savaglia that inhibits nucleocytoplasmic transport of mRNA. Eur. J. Biochem. 1987, 169, 97–104. [Google Scholar]
- Horwitz, J.; Heller, J. Modification of tryptophan residues in retinol-binding protein and prealbumin with 2-hydroxy-5-nitrobenzyl bromide. Effects of the modification of the protein-retinol and protein-protein interaction. J. Biol. Chem. 1974, 249, 7181–7185. [Google Scholar] [PubMed]
- Gonzalez-Fernandez, F.; Bevilacqua, T.; Lee, K.I.; Chandrashekar, R.; Hsu, L.; Garlipp, M.A.; Griswold, J.B.; Crouch, R.K.; Ghosh, D. Retinol-binding site in interphotoreceptor retinoid-binding protein (IRBP): A novel hydrophobic cavity. Invest. Ophthalmol. Vis. Sci. 2009, 50, 5577–5586. [Google Scholar]
- Zhang, X.; Le Pennec, G.; Steffen, R.; Müller, W.E.G.; Zhang, W. Application of a MTT assay for screening nutritional factors in growth media of primary sponge cell culture. Biotechnol. Progr. 2004, 20, 151–155. [Google Scholar]
- Eckert, C.; Schröder, H.C.; Brandt, D.; Perovic-Ottstadt, S.; Müller, W.E.G. A histochemical and electron microscopic analysis of the spiculogenesis in the demosponge Suberites domuncula. J. Histochem. Cytochem. 2006, 54, 1031–1040. [Google Scholar]
- BLAST Assembled RefSeq Genomes. Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 31 January 2011).
- FASTA Assembled RefSeq Genomes. Available online: http://www.ebi.ac.uk/fasta33/ (accessed on 31 January 2011).
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar]
- Motif scan analysis using the internet database. Available online: http://myhits.isbsib.ch/cgi-bin/motif_scan (accessed on 31 January 2011).
- Nicholas, K.B.; Nicholas, H.B.; Deerfield, D.W. GeneDoc: Analysis and visualization of genetic variation. EMBnet News 1997, 4, 1–4. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar]
- Compton, S.; Jones, C.G. Mechanism of dye response and interference in the Bradford protein assay. Anal. Biochem. 1985, 151, 369–374. [Google Scholar]
- Sachs, L. Angewandte Statistik; Springer: Berlin, Germany, 1984; p. 242. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).