Abstract
Monitoring and characterizing species biodiversity is essential for germplasm preservation, academic studies, and various practical applications. Duckweeds represent a group of tiny aquatic plants that include 36 species divided into 5 genera within the Lemnaceae family. They are an important part of aquatic ecosystems worldwide, often covering large portions of the water reservoirs they inhabit, and have many potential applications, including in bioremediation, biofuels, and biomanufacturing. Here, we evaluated the biodiversity of duckweeds in Ukraine and Eastern China by characterizing specimens using the two-barcode protocol with the chloroplast atpH–atpF and psbK–psbI spacer sequences. In total, 69 Chinese and Ukrainian duckweed specimens were sequenced. The sequences were compared against sequences in the NCBI database using BLAST. We identified six species from China (Spirodela polyrhiza, Landoltia punctata, Lemna aequinoctialis, Lemna minor, Lemna turionifera, and Wolffia globosa) and six from Ukraine (S. polyrhiza, Lemna gibba, Lemna minor, Lemna trisulca, Lemna turionifera, and Wolffia arrhiza). The most common duckweed species in the samples from Ukraine were Le. minor and S. polyrhiza, accounting for 17 and 15 out of 40 specimens, respectively. The most common duckweed species in the samples from China was S. polyrhiza, accounting for 15 out of 29 specimens. La. punctata and Le. aequinoctialis were also common in China, accounting for five and four specimens, respectively. According to both atpH–atpF and psbK–psbI barcode analyses, the species identified as Le. aequinoctialis does not form a uniform taxon similar to other duckweed species, and therefore the phylogenetic status of this species requires further clarification. By monitoring duckweeds using chloroplast DNA sequencing, we not only precisely identified local species and ecotypes, but also provided background for further exploration of native varieties with diverse genetic backgrounds. These data could be useful for future conservation, breeding, and biotechnological applications.
1. Introduction
Monitoring and characterizing species biodiversity is essential for germplasm preservation, academic studies, and various practical applications [1]. Duckweed is an important element in aquatic ecosystems worldwide, often covering large portions of the still water surface they inhabit. This group of tiny aquatic plants is composed of 36 species divided into five genera in the Lemnaceae family [2,3], an early diverging family of monocotyledonous plants [4].
Duckweeds are a diverse group and provide many opportunities for genetic, physiological, biochemical, and practical research [5,6]. After being important model plants in the 1950s–1970s, duckweeds became popular again in the 2010s, primarily due to their potential as a biofuel feedstock because of their high biomass growth rate, low lignin content, and high starch content [7,8]. In addition to starch, duckweed biomass is rich in proteins, carbohydrates, crude fiber, minerals, and lipids. This biomass composition makes duckweed a potential food source for animals, fish, and humans [9]. Duckweeds have also been studied for their use in wastewater treatment [10,11], biosensing [12,13], and phytoremediation of water reservoirs contaminated with various toxic chemicals [14,15]. Several duckweed species have been genetically engineered with the eventual aim of producing pharmaceutical proteins such as antigens, peptide hormones, and antibodies [16,17,18].
The multiple potential applications of duckweed have led to an increasing interest in duckweed genetics, molecular evolution, and diversity. The genome size of duckweeds varies by 14-fold, from 160 Mb in great duckweed (Spirodela polyrhiza (L.) Schleid) to ≈2.2 Gb in Wolffia arrhiza (L.) Horkel ex Wimm [19]. Researchers have sequenced the whole genomes of two representative ecotypes of Spirodela polyrhiza [20,21] and Spirodela intermedia W. Koch [22], as well as genomes of Lemna minor L. [23] and Wolffia australiana (Benth.) Hartog and Plas [24]. Moreover, there are ongoing whole-genome sequencing projects for Landoltia punctata (G.Mey.) Les and D.J.Crawford and Lemna gibba L. [25]. Four biannual international conferences specifically on duckweed have taken place since 2012, and there has been a tremendous surge in diverse academic and applied studies of various aspects of duckweed biology [5,26,27,28,29].
Duckweeds include the smallest known flowering plants and often have reduced morphology, making some species difficult to identify using traditional botanical approaches, not even mentioning ecotypes [30]. Recently, molecular methods have been developed to aid in identifying duckweed species and distinguishing ecotypes [31]. The Consortium for the Barcode of Life (CBOL) [32] recommends seven chloroplast DNA (cpDNA) barcodes to identify land plants simply and reliably [33]. The recommended barcodes have been adapted for identification of duckweeds supported by the constantly growing number of reference sequences deposited in DNA sequence databases. Most of these sequences came from studies of the samples deposited at the world’s largest live duckweed depository at the Rutgers University’s Duckweed Stock Cooperative (RDSC) in New Brunswick, NJ, USA (www.ruduckweed.org, accessed on 31 March 2022), with the rest coming from smaller collections or random samplings. Additionally, 12 chloroplast genome sequences, representing 7 duckweed species [34,35,36,37], have been sequenced and deposited in the NCBI database.
In many parts of the world, including Ukraine and China, farmers practicing intensive agriculture use substantial amounts of fertilizers. Fertilizer that is not fully utilized by crops eventually ends up in water reservoirs surrounding agricultural fields. Due to its ability to quickly assimilate nitrogen, phosphorous, and other nutrients, duckweed can rapidly grow, producing on average of 13–38 dry tons of biomass/hectare/year [38], converting agricultural and municipal wastewater into clean water and a high-value biomass ideal for animal/fish feed and numerous other applications [5]. In both Ukraine and China, duckweed is the dominant vegetation in ponds and lakes. In contrast to China, where different aspects of duckweed research are relatively well developed (for example, the RDSC collection hosts more than 200 duckweed ecotypes originated from China), there is rather scarce information on duckweed in Ukraine and Eastern Europe in general.
In this work, we evaluated the biodiversity of duckweed in different regions of Eastern China and Ukraine on the basis of the two-barcode protocol for sequencing the chloroplast atpH–atpF (ATP) and psbK–psbI (PSB) spacers in the collected duckweed specimens. With this approach, we precisely identified local species and ecotypes. Our results provide a foundation for further exploring native varieties with diverse genetic backgrounds and for duckweed breeding and biotechnological applications.
2. Results
2.1. Genotyping of the Duckweed Specimens
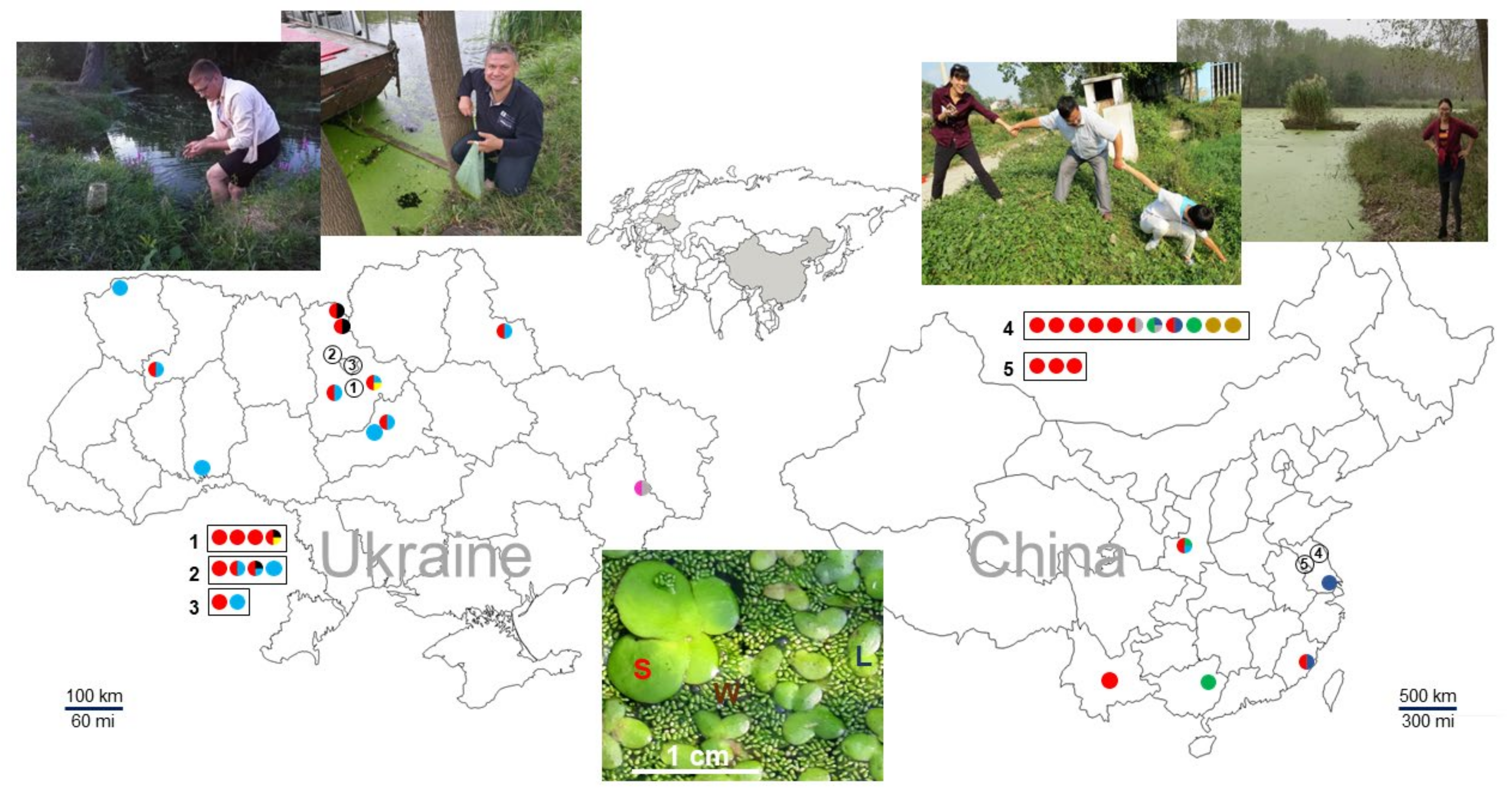
We collected 69 duckweed specimens from across Ukraine and southeastern China. Several locations contained more than one species, as illustrated in Figure 1. From these specimens, plus RDSC clone 8656, we obtained 140 representative sequences, which we deposited in GenBank (Table S1). Because the PCR primers for the atpH–atpF spacer are located further into the corresponding atpH and atpF gene sequences compared to psbK–psbI spacer, the ATP barcodes contain longer portions of the coding sequences compared to PSB. The high reliability of the represented barcodes is based on sequences generated using both forward and reverse primers following careful nucleotide validation.
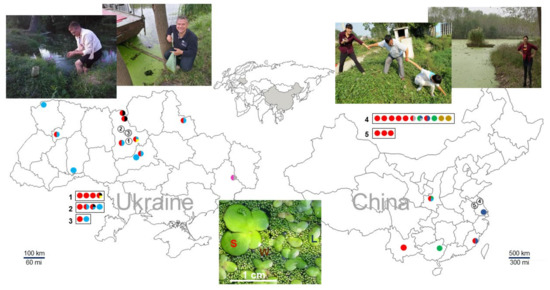
Figure 1.
Location of duckweed sampling sites in Ukraine and China.Dot colors correspond to different species: red, S. polyrhiza; green, La. punctata; light blue, Le. minor; dark blue, Le. aequinoctialis Welw.; pink, Le. gibba; black, Le. trisulca; grey, Le. turionifera; yellow, W. arrhiza; and brown, W. globosa. The image at the bottom was taken at a pond in Huai’an, China. It illustrates a community of three different species, S. polyrhiza (S), Le. aequinoctialis (L), and W. globosa (W), growing together. The exact GPS coordinates of the sites are listed in Table S2. Geographic maps were taken from the websites located at https://www.d-maps.com/m/asia/china/chine/chine58.gif (accessed on 11 February 2022) and https://www.d-maps.com/m/europa/ukraine/ukraine50.gif (accessed on 11 February 2022).
2.1.1. Great Duckweed, Spirodela polyrhiza
Barcoding showed that 15 of the 39 specimens collected in Ukraine and 15 of the 30 specimens collected in China were S. polyrhiza. The S. polyrhiza ATP sequences from our study and the reference sequences from the whole chloroplast genome of U.S. S. polyrhiza ecotype 7498 [35,37] had high sequence conservation. The main detected sequence variations were T↔C transitions at defined positions along the sequence and a couple of T↔A transversions, with no biases related to the specimen’s geographic origins (Figure S1A). The PSB sequences showed similar low sequence diversity but with different sequence polymorphisms, including single-nucleotide polymorphisms (SNPs), insertion/deletions (InDels), and more random nucleotide transitions/transversions (Figure S1B) compared to the ATP sequences. Our PSB sequences also contained single-nucleotide insertions of additional A nucleotides at positions 25 and 354 and an additional T at position 402, compared to the reference sequence of S. polyrhiza ecotype 7498.
2.1.2. Dotted Duckweed, Landoltia punctata
By chloroplast DNA barcoding, we identified five La. punctata ecotypes. Two ecotypes were collected near the Hongze lake (Jiangsu province, China) and kept in our in vitro collections (NB0014 and NB0022). Ecotype Ya3 was collected from Yanling and Gu1 from Guilin; the ecotype RDSC EL019 collected earlier in Kuhming was obtained from the RDSC (New Brunswick, USA). As La. punctata inhabits tropical areas [29], we did not find it in Ukraine. Sequence alignments showed a high stability of both the ATP and PSB sequences in La. punctata. The ATP sequences of the six ecotypes only shared two A↔G transitions, both in Gu1, and a single nucleotide deletion (Figure S2A); the six PSB sequences differed by a single G→T transversion in Ya3 (Figure S2B).
2.1.3. Common Duckweed, Lemna minor
Lemna minor was the most represented duckweed species in the Ukraine specimens. We identified 17 of the 39 specimens collected in Ukraine as Le. minor. We also identified one specimen from China, Ya2 collected in Yangling, as Le. minor. The ATP and PSB sequences of the Le. minor ecotypes had very low sequence divergence, with near 100% similarity to the corresponding 29 and 31 GenBank ATP and PSB sequences representing Le. minor ecotypes, respectively. We compared the sequences of our specimens with the corresponding sequences of a Russian ecotype for which the chloroplast genome was sequenced [34] and found only three nucleotide substitutions in the PSB sequences (Figure S3A) and five G→A transition and a single T→G transversion in the ATP sequences (Figure S3B).
2.1.4. Star Duckweed, Lemna trisulca
We identified four duckweed specimens from Ukraine as Le. trisulca. They had a 100% similarity (Figure S4A) to the ATP sequences previously reported for ecotypes from the USA and Canada [39]. However, alignment of PSB sequences clearly distinguished Ukrainian ecotypes from the North American ones on the basis of the duplication of an AT-rich 23-bp long DNA sequence in the North American ecotypes (Figure S4B). Moreover, alignment of a few Le. trisulca ATP and PSB sequences [39] revealed distinct variants in strain UTCC 399 of unknown origin, characterized by short 4–6-nucleotide insertions/deletions as compared to the Ukrainian and North American ecotypes.
2.1.5. Turion Duckweed, Lemna turionifera
We identified one specimen from Ukraine (from the southeast) and two collected from China (from near Hongze lake) as Le. turionifera. The ATP and PSB sequences of these specimens showed no sequence variation when aligned with Le. turionifera sequences from Canada, the Czech Republic, and Lake Tai in China [40]. The only variation we found was a single nucleotide deletion in the ATP sequence of the accession from the Czech Republic (Figure S5).
2.1.6. Swollen or Fat Duckweed, Lemna gibba
We identified one specimen from Ukraine (DW102) as Le. gibba. The PSB sequence showed homology with the corresponding sequences of four Le. gibba strains from the USA, Italy, Ethiopia, and Japan [39], as well as strain RDSC 5504, which originated from Shanghai, China. The ATP sequence of DW102 differed in two positions, a single insertion of A (which was also in the sequence of the Shanghai strain) and a unique C→T transition (Figure S6).
2.1.7. Lesser Duckweed, Lemna aequinoctialis
Lemna aequinoctialis had the highest variation in ATP and PSB sequences among the species analyzed in this study. We collected four Le. aequinoctialis strains: two from Huai’an city (I2 and NB0017), one from Shanghai (NB0007), and one from Fuzhou (Fu94). We divided the strains into two groups on the basis of their barcode sequences (Figure S7). Strains NB0017 and NB0007 differed from I2 and Fu94 by two tandem duplications of 21 and 5 bp, three specific SNPs in their PSB sequences (positions 71, 117, and 162), and five SNPs (positions 12, 189, 359, 364, and 395) in their ATP sequences. These two groups aligned with three American Le. aequinoctialis strains [39], with NB0017 and NB0007 having similar sequences to those of strains 6612 (Centerville, CA, USA) and 8656 (Argentina), whereas I2 and Fu94 aligned with strains 6746 (Plainsburg, CA, USA) and 7126 (Texas, USA) (Figure S7). All four Chinese strains had three specific SNPs in their PSB sequences (positions 357, 401, 444) compared to the three American strains.
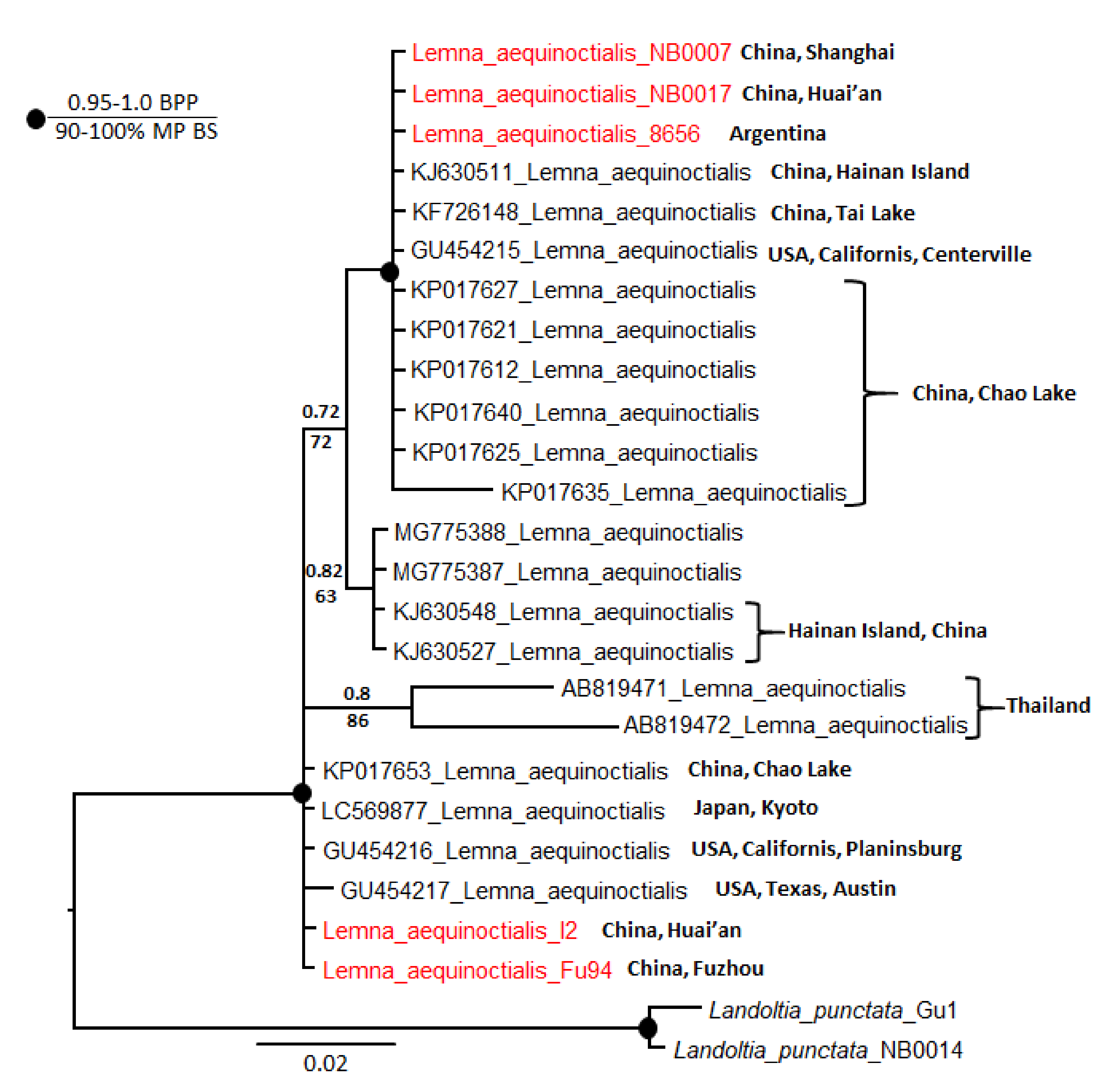
To examine polymorphism in Le. aequinoctialis barcodes in more detail, we analyzed the phylogeny of the 21 ATP sequences available in NCBI GenBank together with our five sequences. We included two La. punctata accessions as an outgroup. In total, there were 513 characters, of which 453 were constant. Of the variable characters, 18 were parsimony uninformative and 42 were parsimony informative. Parsimony and Bayesian analyses yielded the same topology but with lower bootstrap percentages than posterior probabilities. A heuristic search found most-parsimonious trees that were 70 steps long (consistency index 0.9143, retention index 0.9259). The resultant dendrogram from this analysis is shown in Figure 2. All sequences were divided into four subclades: two with very strong support and two with little support. Our accessions are subordinate to the two strongly supported clades.
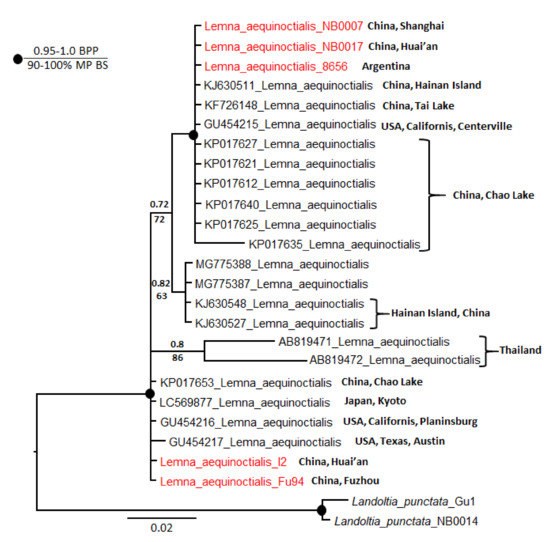
Figure 2.
Bayesian consensus tree based on analysis of atpH–atpF intergenic spacer sequences of Lemna aequinoctialis and Landoltia punctata as an outgroup. Bayesian posterior probabilities (BPP) and maximum parsimony bootstrap values (MP BS) are shown above and below the branches, respectively. Strongly supported clades (MP BS > 90% and BPP > 0.95) are indicated with black circles at the branchpoints. For the origin of specimens, see Table S2.
2.1.8. Least Duckweed, Wolffia arrhiza, and Watermeal Duckweed, Wolffia globosa
We identified two Ukrainian specimens as W. arrhiza and two Chinese specimens as W. globosa. Both specimens from Ukraine, DW32 and DW35, had high ATP sequence similarity with the homologous sequence of African and Italian specimens, but a high level of nucleotide mismatches with the sequence from a W. arrhiza specimens from Brazil (Figure S8A). There was 100% similarity between the PSB sequences of the two Ukrainian specimens, with the sequence blasting revealing a single hit in GenBank (Figure S8B).
There were more hits for W. globosa compared to W. arrhiza; 31 for ATP and 14 for PSB. The Chinese strains C2 and NB0015 (characterized in this study), together with strains DW2101-4 (Acc. KJ630544.1; Hainan) and LC49 (Lake Chao) [41], were more closely grouped with a specimen from the USA [39] than with those from other Asian countries India, Japan, and Thailand (Figure S9). This grouping was based on nucleotide substitutions at positions 248, 253, and 383 in the ATP sequence (Figure S9A) and, even more profoundly, by multiple SNPs and three deletions/insertions of short nucleotide sequences in the PSB sequence (Figure S9B).
2.2. Phylogenetic Analysis
Phylogenetic analysis of our 69 duckweed specimens using ATP and PSB sequences separately showed no conclusive results. Therefore, we performed a combined ATP and PSB analysis of 70 taxa, including Pistia stratiotes [42] as an outgroup species. The combined data matrix included 1197 characters divided in two partitions: 1–560 for ATP and 561–1197 for PSB, of which 751 were constant, 103 were parsimony uninformative, and 343 were parsimony informative.
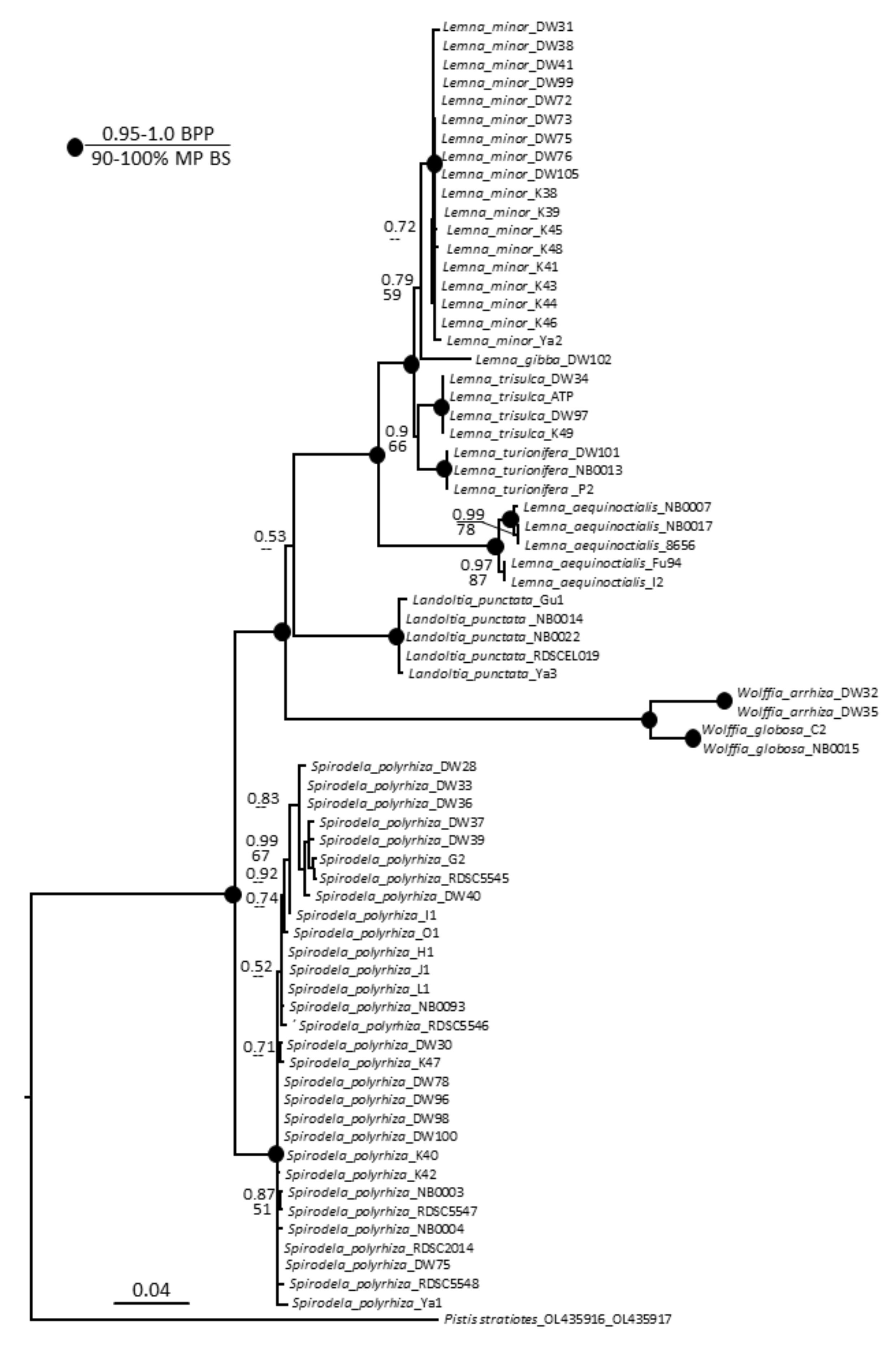
Parsimony and Bayesian analyses yielded the same topology but with lower bootstrap percentages than posterior probabilities. The heuristic search found most-parsimonious trees that were 635 steps long (consistency index 0.8551, retention index 0.9691). The resultant dendrogram from this analysis is shown in Figure 3. All species studied built monophyletic and mostly not polymorphic clades, with few exceptions. The S. polyrhiza clade had several small subclades with weak support, and the clade with L. aequinoctialis accessions was divided into two subclades with strong support. Overall, this phylogeny of Lemnaceae is congruent with previous studies [3,43,44,45].
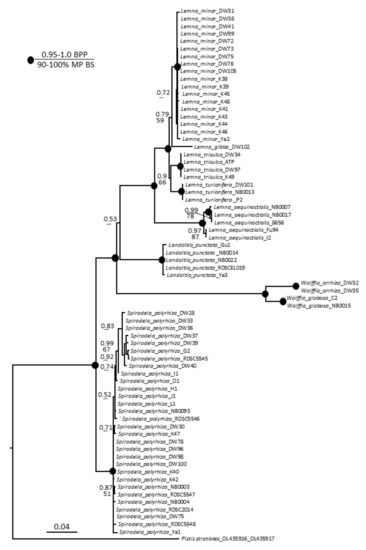
Figure 3.
Bayesian consensus tree based on analysis of the combined chloroplast DNA dataset (atpH–atpF and psbK–psbI intergenic spacers) of Lemnaceae taxa and Pistia stratiotes as an outgroup. Bayesian posterior probabilities (BPP, top value) and maximum parsimony bootstrap values (MP BS, bottom value) are shown at the branches. Strongly supported clades (MP BS > 90% and BPP > 0.95) are indicated with black circles at the branchpoints.
3. Discussion
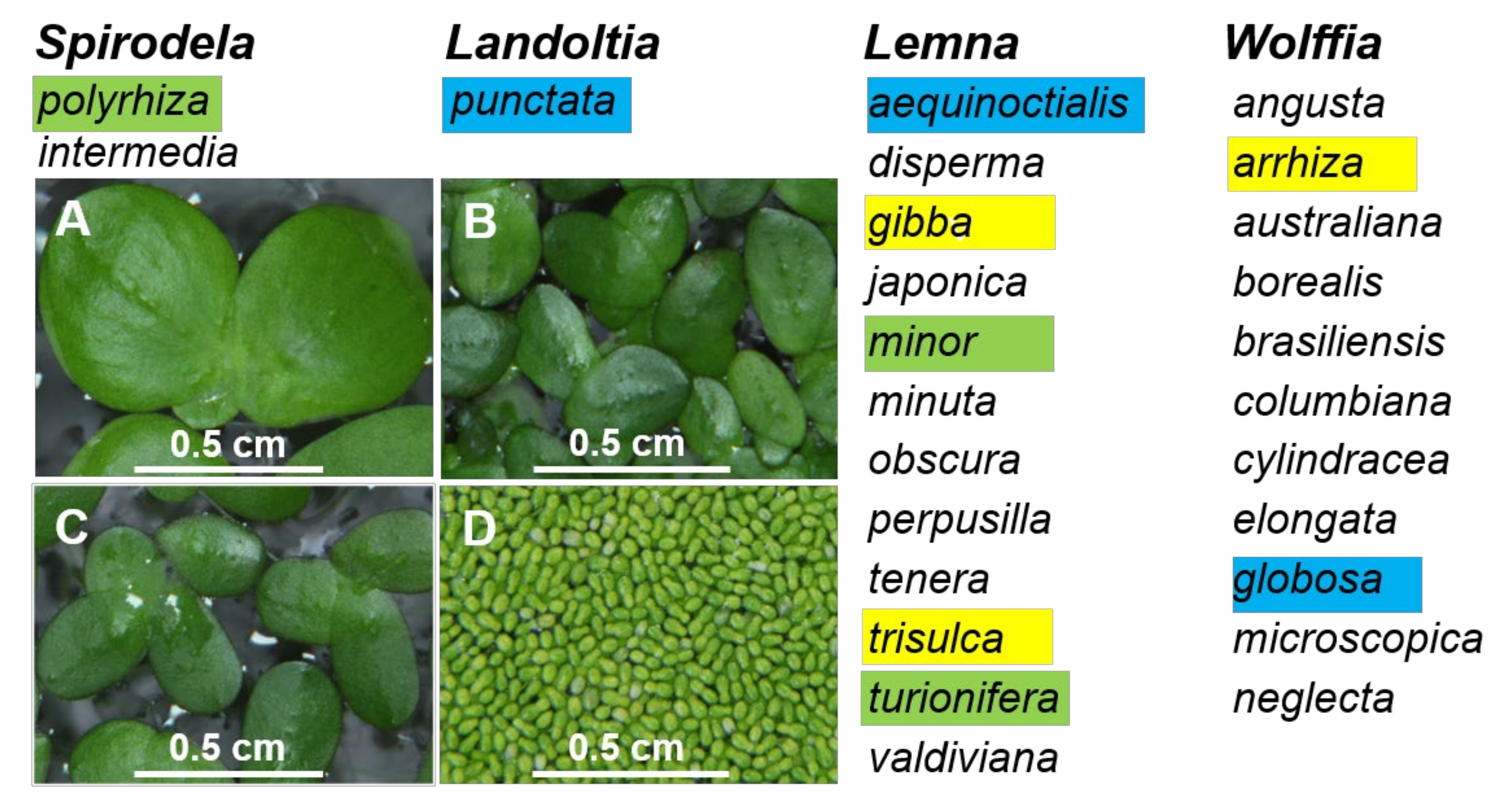
As the smallest known flowering plants, duckweeds have a reduced morphology, which makes them difficult to identify using traditional botanical approaches [30]. Therefore, molecular approaches [2] offer valuable alternatives for species monitoring of this ancient group of plants. Here, to provide additional data supporting molecular approaches for duckweed identification, as well as to examine duckweed diversity, we collected 39 duckweed specimens from Ukraine and 30 from China. Using DNA barcoding, we identified six duckweed species among the Ukrainian specimens (S. polyrhiza, Le. gibba, Le. minor, Le. trisulca, Le. turionifera, and W. arrhiza) and six species from China (S. polyrhiza, La. punctata, Le. aequinoctialis, Le. minor, Le. turionifera, and W. globosa). These species represent four out of the five genera in the Lemnaceae family (Figure 4). The only genus not represented was Wolffiella, which only occurs in the Americas and Africa [29].
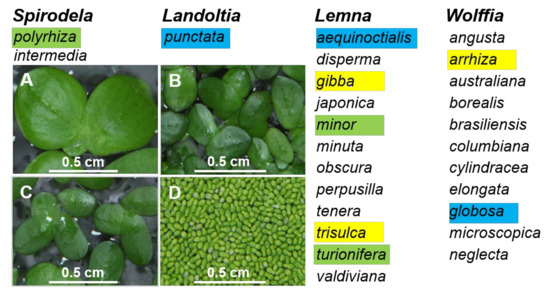
Figure 4.
Four genera of the Lemnaceae plant family were represented by duckweed species in Ukraine and China. Species that were found in Ukraine and China are highlighted green, those found only in Ukraine are highlighted yellow, and those found only in China are highlighted blue. A, B, C, and D are representative images of Spirodela polyrhiza, Landoltia punctata, Lemna minor, and Wolffia globose, respectively, from the in vitro collection of Huaiyin Normal University.
The distribution of duckweed species in this study generally matched previously identified duckweed distributions, with S. polyrhiza, Le. Minor, and W. arrhiza being the most common species in Europe and S. polyrhiza, La. Punctate, Le. aequinoctialis, and W. globoza the usual species in China [29,46]. However, there is little information on duckweed biodiversity in Eastern Europe in general and in Ukraine in particular [47,48]. To the best of our knowledge, this study is the first chloroplast-barcoding-based record of duckweed biodiversity in an Eastern European country. Compared to Ukraine, duckweed biodiversity in China is relatively well investigated [31,40,41], and there are numerous ecotypes from China deposited in the RDSC world collection in the USA (New Brunswick, NJ) and in different institutions in China [49,50].
Our molecular identification of randomly sampled specimens agrees that the great duckweed, S. polyrhiza, is the most cosmopolitan of the 36 duckweed species recognized worldwide [29]. It was the most dominant species in East China and in Ukraine. Although the phylogenetic analysis demonstrated a certain degree of clustering of the S. polyrhiza specimens on the basis of the limited barcode sequence variations (Figure 3), it did not show any clear links to the geographic origin of the specimens. Generally, the ATP and PSB sequences of our specimens had almost 100% similarity with the corresponding sequences of strain 7498 [37], with a low sequence variability between the specimens. Similarly, S. polyrhiza nuclear genomes sequenced from 63 specimens collected worldwide had high sequence conservation [51].
La. punctata is the only representative of the genus Landoltia. It is considered to be closely related to Spirodela [36] but is not as widely distributed as S. polyrhiza. It mostly inhabits tropical and subtropical areas [29]. Therefore, it is not surprising that we collected La. punctata specimens in China but none in Ukraine. Genetic analysis of the six Chinese La. punctata specimens revealed few nucleotide substitutions. This stability of ATP and PSB sequences was also observed among specimens from different geographic origins, including India, Africa, America, and Australia (Figure S2).
We identified five Lemna species among the specimens collected from Ukraine and China. The common duckweed, Le. minor, was the most predominant duckweed species in Ukraine, represented by 17 out of 40 specimens, closely followed by S. polyrhiza (15 specimens) (Figure 1). However, we only identified one specimen from China as Le. minor, a specimen that was collected in north-central China. Lemna turionifera was a minor species in both China and Ukraine [40,45]. We identified Le. gibba and Le. trisulca only in Ukraine, and Le. aequinoctialis only in China (Figure 1).
Three Lemna species, Le. minor, Le. gibba, and Le. turionifera, have stable species-specific ATP and PSB sequences as reflected in the phylogenetic dendrogram (Figure 3), with very few variations compared to their counterparts from other parts of the world available in GenBank. The three Le. trisulca specimens from Ukraine had perfect ATP sequence similarity; however, there were clear differences in PSB sequences compared (Figure S4) with specimens from the USA and Canada [39]. The differences were due to a 23 bp duplication.
The most intriguing results from this study were on the phylogeny of Le. aequinoctialis. On the basis of the ATP and PSB sequences, we grouped the four Le. aequinoctialis specimens (collected in Fuzhou, Shanghai, and Huai’an) into two clades (Figure S7). Sequences of these clades were aligned with the ATP sequences of Le. aequinoctialis strain DW0101-3 (Hainan, China; Acc. KJ630511.1) and strain LC42 (Lake Chao, China) [41]. We constructed a phylogeny based on all Le. aequinoctialis ATP sequences in GenBank, and the resulting tree was complex (Figure 2). On the basis of these results, we suggest that the status of this species needs further careful examination.
We identified two Wolffia species in our study: W. arrhiza, a common species in Europe and Africa, and W. globosa, which inhabits Southeast Asia [29], were identified in Ukraine and China, respectively. The W. arrhiza sequences showed high sequence similarity with other W. arrhiza sequences of specimens from Europe and Africa (Figure S8). The W. globosa sequences had several characteristic SNPs, both in the ATP and PSB sequences, compared with sequences of specimens from India, Japan, and Thailand (Figure S9); however, they had a higher similarity to sequences of specimens from the USA [39]. The USA specimen is likely a recent invasion in addition to the native Wolffia columbiana [29].
4. Materials and Methods
4.1. Plant Material
Duckweed specimens were collected from various still water reservoirs, lakes, and ponds across eastern China and Ukraine during 2016–2019. Prior to genotyping, most specimens from China were sterilized and kept under aseptic conditions on agar medium according to previously described methods [43]. These specimens were kept at the duckweed in vitro collection recently organized at Huaiyin Normal University, Hui’an, China [47], barcoded and used in our previous studies [11,18,52,53]. The Ukrainian duckweed specimens were collected from water reservoirs, sorted according to their morphological characteristics, and directly subjected to DNA extraction for further chloroplast DNA barcoding. All analyzed duckweed specimens and locations are listed in Table S2.
4.2. DNA Extraction, Fragment Amplification, Sequencing, and Alignment
Total DNA was extracted from plant tissue using a modified CTAB method [54]. The PCR amplifications were carried out as recommended by the CBOL Plant Working Group [32], described in [39], using primers 5′-TTAGCATTTGTTTGGCAAG and 5′-AAAGTTTGAGAGTAAGCAT for the psbK–psbI intergenic spacer and 5′-ACTCGCACACACTCCCTTTCC and 5′-GCTTTTATGGAAGCTTTAACAAT for the atpH–atpF intergenic spacer. Following amplification, the DNA fragments were sent to Sangon Biotech (Shanghai, China) for purification and sequencing. The raw sequences were preliminarily optimized using the ‘Online Analysis Tools’ package (http://molbiol-tools.ca, accessed on 25 March 2022), in particular, the program MAFFT, version 7 (https://mafft.cbrc.jp/alignment/server/ accessed on 25 March 2022). Multiple DNA sequence alignments were generated with ClustalW software [55], and the alignments were subsequently corrected manually in MEGA 5 [56].
For BLAST alignment analyses, a duckweed reference barcode set was compiled from ATP and PSB sequences that were generated in this study and those available from the NCBI database as of January 2022. Queried sequences were trimmed to include only intergenic regions and used in BLASTN (version 2.2.26+) searches to identify homologies to other barcode sequences in the set. The number of top hits for each query are presented in Table S1.
4.3. Phylogenetic Analysis
Phylogenetic analysis was carried out on individual and combined ATP and PSB sequences using parsimony and Bayesian methods. Pistia stratiotes, from the Araceae family, was used as an outgroup for the Lemnaceae family. Parsimony analysis was performed with PAUP* 4.0b10 [57] using heuristic searches with tree bisection–reconnection and 100 random additional sequence replicates. Bootstrap support (BS) [58] was estimated with 100 bootstrap replicates, each with 100 random addition sequence searches. Bayesian analyses were implemented with MrBayes 3.1.23 [59]. Sequence evolution models were evaluated using the Akaike information criterion (AIC) with the aid of jModelTest2 v2.1.6 [60]. Two independent runs each of eight chains and 10 million generations, sampling every 1000 generations, were executed, and 25% of initial trees were discarded as burn-in. The remaining 15,000 trees were combined into a 50% majority-rule consensus tree.
5. Conclusions
Our survey of the duckweed species in Ukraine and China makes a solid contribution to monitoring the biodiversity of aquatic flora in these countries. In addition to precisely identifying six major species and their geographic distribution in each these countries by double chloroplast DNA barcoding, our data highlighted the need to re-examine the phylogenetic status of one of those species, Lemna aequinoctialis. The study added 138 new chloroplast ATP and PSB barcodes to the 1754 corresponding barcodes for these species available in the NCBI database as of March 2022 (Table S1). These new resources might fuel future research on plant molecular evolution, biodiversity conservation, breeding, and various biotechnological applications.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11111468/s1, Figure S1: Nucleotide alignment of atpH–atpF and psbK–psbI spacers sequences of Spirodela polyrhiza collected in Ukraine and China (for ecotype details see supplemental Table S2). Matching residues are shown as dots. A. Nucleotide alignment of atpH–atpF barcodes including representative sequences of S. polyrhiza specimens available in GenBank: MN419335, USA; GU454201, Hong Kong; GU454202, India; GU454203, Malaysia; GU454204, USA; GU454205, Mexico; GU454206, Canada; GU454208, Europe. B. Nucleotide alignment of psbK-psbI barcodes including representative sequences of S. polyrhiza specimens available in GenBank: MN419335, USA; GU454297, Hong Kong; GU454298, India; GU454299, Malaysia; GU454300, USA; GU454301, Mexico; GU454302, Canada; GU454304, Europe. Figure S2: Nucleotide alignment of atpH-atpF and psbK-psbI spacers of Landoltia punctata (for ecotypes details see supplemental Table S2). Matching residues are shown as dots. A. Nucleotide alignment of atpH-atpF barcodes including the representative sequences of La. punctata from different parts of the world available in the GenBank: GU454209, South Africa; GU454211, India; GU454212, USA; GU454213, Australia. B. Nucleotide alignment of psbK-psbI barcodes including the representative sequences of La. punctata from different parts of the world available in the GenBank: GU454305, South Africa; GU454307, India; GU454308, USA; GU454309, Australia. Figure S3: Nucleotide alignment of atpH-atpF and psbK-psbI spacers of Lemna minor (for ecotypes details see supplemental Table S2). Matching residues are shown as dots. A. Nucleotide alignment of atpH-atpF barcodes including the representative sequences of Le. minor from different parts of the world available in the GenBank: DQ400350, Russia; GU454226, Turkey; GU454227, USA; GU454228, South Africa; GU454229, Japan; GU454230, Finland; GU454231, Germany. B. Nucleotide alignment of psbK-psbI barcodes including the representative sequences of Le. minor from different parts of the world available in the GenBank: DQ400350, Russia; GU454322, Turkey; GU454323, USA; GU454324, South Africa; GU454325, Japan; GU454326, Finland; GU454327, Germany. Figure S4: Nucleotide alignment of atpH-atpF and psbK-psbI spacers of Lemna trisulca (for ecotypes details see supplemental Table S2). Matching residues are shown as dots. A. Nucleotide alignment of atpH-atpF barcodes including the representative sequences of Le. trisulca from different parts of the world available in the GenBank: GU454236, Canada; GU454237, USA; GU454237, UTCC 399. B. Nucleotide alignment of psbK-psbI barcodes including the representative sequences of L. trisulca from different parts of the world available in the GenBank: GU454332, Canada; GU454333, USA; GU454334, UTCC 399. Figure S5: Nucleotide alignment of atpH-atpF and psbK-psbI spacers of Lemna turionifera (for ecotypes details see supplemental Table S2). Matching residues are shown as dots. A. Nucleotide alignment of atpH-atpF barcodes including the representative sequences of Le. turionifera from different parts of the world available in the GenBank: KF726146, China; MG000422, Canada; GU454240, Czech. B. Nucleotide alignment of psbK-psbI barcodes including the representative sequences of Le. turionifera from different parts of the world available in the GenBank: MG000496, Canada; GU454335, China; GU454336, Czech. Figure S6: Nucleotide alignment of atpH-atpF and psbK-psbI spacers of Lemna gibba, ecotype DW102 (Supplemental Table S2). Matching residues are shown as dots. A. Nucleotide alignment of the DW102 atpH-atpF barcode with the representative sequences of Le. gibba from different parts of the world available in the GenBank: KX212889, China; GU454219, USA; GU454220, Italy; GU454221, Ethiopia; GU454222, Japan. B. Nucleotide alignment of the DW102 psbK-psbI barcode with the representative sequences of Le. gibba from different parts of the world available in the GenBank: GU454315, USA; GU454316, Italy; GU454317, Ethiopia; GU454318, Japan. Figure S7: Nucleotide alignment of atpH-atpF and psbK-psbI spacers of Lemna aequinoctialis (for ecotypes details see supplemental Table S2). Matching residues are shown as dots. A. Nucleotide alignment of atpH-atpF barcodes including the representative sequences of Le. aequinoctialis from different parts of the world available in the GenBank: GU454215, USA, California, Centerville; GU454216, USA, California, Plainsburg; GU454217, USA, Texas, Austin. B. Nucleotide alignment of psbK-psbI barcodes including the representative sequences of Le. aequinoctialis from different parts of the world available in the GenBank: GU454311, USA, California, Centerville; GU454312, USA, California, Plainsburg; GU454313, USA, Texas, Austin. Figure S8: Nucleotide alignment of atpH-atpF and psbK-psbI spacers of Wolffia arrhiza (for ecotypes details see supplemental Table S2). Matching residues are shown as dots. A. Nucleotide alignment of atpH-atpF barcodes including the representative sequences of W. arrhiza from different parts of the world available in the GenBank: MG812317, Kenya; MG775412, Italy; MG775413, Brazil. B. Nucleotide alignment of psbK-psbI barcodes including the representative sequence of W. arrhiza available in the GenBank: GU454364, Hungary. Figure S9: Nucleotide alignment of atpH-atpF and psbK-psbI spacers of Wolffia globosa (for ecotypes details see supplemental Table S2). Matching residues are shown as dots. A. Nucleotide alignment of atpH-atpF barcodes including the representative sequences of W. globosa from different parts of the world available in the GenBank: GU454281, USA; KJ630544, China; KP017660, China; MG812325, India; MN881100, Japan. B. Nucleotide alignment of psbK-psbI barcodes including the representative sequences of W. globosa from different parts of the world available in the GenBank: GU454378, USA; GU454379, Thailand; MG812330, India; MN881100, Japan. References [61,62,63,64,65,66,67,68,69,70,71] are cited in the supplementary materials.
Author Contributions
Conceptualization, B.M., Y.Z. and M.B.; methodology, G.C., A.S., N.F. and M.B.; software and validation, A.S., N.F. and M.B.; formal analysis, G.C., A.S., N.F. and M.B.; investigation, G.C., A.S., O.L., O.K., A.P., D.C. and H.Z.; resources, O.K., A.P., Y.Z., B.M., D.G. and J.X.; data curation, A.S., N.F. and M.B.; writing—original draft preparation, A.S., N.F. and M.B.; writing—review and editing, M.B.; visualization, G.C., A.S., A.P. and M.B.; supervision, Y.Z., B.M. and M.B.; project administration, J.X., B.M., D.G. and M.B.; funding acquisition, M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by an individual grant provided by Huaiyin Normal University (Huai’an, China) to MB.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Acknowledgments
We thank the following people for their help collecting duckweed samples: Roman Volkov and Irina Panchuk (Yuriy Fedkovych University of Chernivtsi), Volodymyr Borysyuk (Kyiv, Ukraine), Raisa Stepanenko (Vysochne, Ukraine), Antonina Melnyk (Pasiky, Ukraine), Oleksandr and Laura Chekanov (Sumy, Ukraine), and Vasyl Stepanenko (Korsun-Shevchenkivskyi, Ukraine), as well as many students from the Huaiyin Normal University and owners of fish and crab farms around Huai’an city (China).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Langridge, P.; Waugh, R. Harnessing the Potential of Germplasm Collections. Nat. Genet. 2019, 51, 200–201. [Google Scholar] [CrossRef] [PubMed]
- Appenroth, K.J.; Borisjuk, N.; Lam, E. Telling Duckweed Apart: Genotyping Technologies for the Lemnaceae. Chin. J. Appl. Environ. Biol. 2013, 19, 1–10. [Google Scholar] [CrossRef]
- Acosta, K.; Appenroth, K.J.; Borisjuk, L.; Edelman, M.; Heinig, U.; Jansen, M.A.K.; Oyama, T.; Pasaribu, B.; Schubert, I.; Sorrels, S.; et al. Return of the Lemnaceae: Duckweed as a Model Plant System in the Genomics and Postgenomics Era. Plant Cell 2021, 33, 3207–3234. [Google Scholar] [CrossRef] [PubMed]
- Nauheimer, L.; Metzler, D.; Renner, S.S. Global History of the Ancient Monocot Family Araceae Inferred with Models Accounting for Past Continental Positions and Previous Ranges Based on Fossils. New Phytol. 2012, 195, 938–950. [Google Scholar] [CrossRef]
- Zhou, Y.; Borisjuk, N. Small Aquatic Duckweed Plants with Big Potential for Production of Valuable Biomass and Wastewater Remediation. Int. J. Environ. Sci. Nat. Resour. 2019, 16, 555942. [Google Scholar] [CrossRef]
- Baek, G.; Saeed, M.; Choi, H.-K. Duckweeds: Their Utilization, Metabolites and Cultivation. Appl. Biol. Chem. 2021, 64, 73. [Google Scholar] [CrossRef]
- Xu, J.; Cui, W.; Cheng, J.J.; Stomp, A.-M. Production of High-Starch Duckweed and Its Conversion to Bioethanol. Biosyst. Eng. 2011, 110, 67–72. [Google Scholar] [CrossRef]
- Cui, W.; Cheng, J.J. Growing Duckweed for Biofuel Production: A Review. Plant Biol. 2015, 17 (Suppl. 1), 16–23. [Google Scholar] [CrossRef]
- Appenroth, K.-J.; Sree, K.S.; Böhm, V.; Hammann, S.; Vetter, W.; Leiterer, M.; Jahreis, G. Nutritional Value of Duckweeds (Lemnaceae) as Human Food. Food Chem. 2017, 217, 266–273. [Google Scholar] [CrossRef]
- Ge, X.; Zhang, N.; Phillips, G.C.; Xu, J. Growing Lemna Minor in Agricultural Wastewater and Converting the Duckweed Biomass to Ethanol. Bioresour. Technol. 2012, 124, 485–488. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, G.; Peterson, A.; Zha, X.; Cheng, J.; Li, S.; Cui, D.; Zhu, H.; Kishchenko, O.; Borisjuk, N. Biodiversity of Duckweeds in Eastern China and Their Potential for Bioremediation of Municipal and Industrial Wastewater. J. Geosci. Environ. Protect. 2018, 6, 108–116. [Google Scholar] [CrossRef][Green Version]
- Cayuela, M.L.; Millner, P.; Slovin, J.; Roig, A. Duckweed (Lemna gibba) Growth Inhibition Bioassay for Evaluating the Toxicity of Olive Mill Wastes before and during Composting. Chemosphere 2007, 68, 1985–1991. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, P.D.; Sree, K.S.; Appenroth, K.J. Duckweeds for Water Remediation and Toxicity Testing. Toxicol. Environ. Chem. 2016, 98, 1127–1154. [Google Scholar] [CrossRef]
- Bokhari, S.H.; Ahmad, I.; Mahmood-Ul-Hassan, M.; Mohammad, A. Phytoremediation Potential of Lemna minor L. for Heavy Metals. Int. J. Phytoremediat. 2016, 18, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, P.; Sree, K.S.; Appenroth, K.-J. Duckweed Biomarkers for Identifying Toxic Water Contaminants? Environ. Sci. Pollut. Res. Int. 2019, 26, 14797–14822. [Google Scholar] [CrossRef] [PubMed]
- Stomp, A.-M. The Duckweeds: A Valuable Plant for Biomanufacturing. Biotechnol. Annu. Rev. 2005, 11, 69–99. [Google Scholar] [CrossRef] [PubMed]
- Rival, S.; Wisniewski, J.-P.; Langlais, A.; Kaplan, H.; Freyssinet, G.; Vancanneyt, G.; Vunsh, R.; Perl, A.; Edelman, M. Spirodela (Duckweed) as an Alternative Production System for Pharmaceuticals: A Case Study, Aprotinin. Transgenic Res. 2008, 17, 503–513. [Google Scholar] [CrossRef]
- Peterson, A.; Kishchenko, O.; Zhou, Y.; Vasylenko, M.; Giritch, A.; Sun, J.; Borisjuk, N.; Kuchuk, M. Robust Agrobacterium-Mediated Transient Expression in Two Duckweed Species (Lemnaceae) Directed by Non-Replicating, Replicating, and Cell-to-Cell Spreading Vectors. Front. Bioeng. Biotechnol. 2021, 9, 5. [Google Scholar] [CrossRef]
- Hoang, P.T.N.; Schubert, V.; Meister, A.; Fuchs, J.; Schubert, I. Variation in Genome Size, Cell and Nucleus Volume, Chromosome Number and RDNA Loci among Duckweeds. Sci. Rep. 2019, 9, 3234. [Google Scholar] [CrossRef]
- Wang, W.; Haberer, G.; Gundlach, H.; Gläßer, C.; Nussbaumer, T.; Luo, M.C.; Lomsadze, A.; Borodovsky, M.; Kerstetter, R.A.; Shanklin, J.; et al. The Spirodela polyrhiza Genome Reveals Insights into Its Neotenous Reduction Fast Growth and Aquatic Lifestyle. Nat. Commun. 2014, 5, 3311. [Google Scholar] [CrossRef]
- Michael, T.P.; Bryant, D.; Gutierrez, R.; Borisjuk, N.; Chu, P.; Zhang, H.; Xia, J.; Zhou, J.; Peng, H.; El Baidouri, M.; et al. Comprehensive Definition of Genome Features in Spirodela polyrhiza by High-Depth Physical Mapping and Short-Read DNA Sequencing Strategies. Plant J. 2017, 89, 617–635. [Google Scholar] [CrossRef] [PubMed]
- Hoang, P.T.N.; Fiebig, A.; Novák, P.; Macas, J.; Cao, H.X.; Stepanenko, A.; Chen, G.; Borisjuk, N.; Scholz, U.; Schubert, I. Chromosome-Scale Genome Assembly for the Duckweed Spirodela intermedia, Integrating Cytogenetic Maps, PacBio and Oxford Nanopore Libraries. Sci. Rep. 2020, 10, 19230. [Google Scholar] [CrossRef] [PubMed]
- Van Hoeck, A.; Horemans, N.; Monsieurs, P.; Cao, H.X.; Vandenhove, H.; Blust, R. The First Draft Genome of the Aquatic Model Plant Lemna minor Opens the Route for Future Stress Physiology Research and Biotechnological Applications. Biotechnol. Biofuels 2015, 8, 188. [Google Scholar] [CrossRef]
- Michael, T.P.; Ernst, E.; Hartwick, N.; Chu, P.; Bryant, D.; Gilbert, S.; Ortleb, S.; Baggs, E.L.; Sree, K.S.; Appenroth, K.J.; et al. Genome and Time-of-Day Transcriptome of Wolffia australiana Link Morphological Minimization with Gene Loss and Less Growth Control. Genome Res. 2021, 31, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Vu, G.T.H.; Fourounjian, P.; Wang, W.; Cao, X.H. Future Prospects of Duckweed Research and Applications. In The Duckweed Genomes; Cao, X.H., Fourounjian, P., Wang, W., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 179–185. [Google Scholar]
- Appenroth, K.-J.; Sree, K.S.; Fakhoorian, T.; Lam, E. Resurgence of Duckweed Research and Applications: Report from the 3rd International Duckweed Conference. Plant Mol. Biol. 2015, 89, 647–654. [Google Scholar] [CrossRef]
- Cao, H.X.; Fourounjian, P.; Wang, W. The Importance and Potential of Duckweeds as a Model and Crop Plant for Biomass-Based Applications and Beyond. In Handbook of Environmental Materials Management; Hussain, C.M., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–16. [Google Scholar]
- An, D.; Zhou, Y.; Li, C.; Xiao, Q.; Wang, T.; Zhang, Y.; Wu, Y.; Li, Y.; Chao, D.-Y.; Messing, J.; et al. Plant Evolution and Environmental Adaptation Unveiled by Long-Read Whole-Genome Sequencing of Spirodela. Proc. Natl. Acad. Sci. USA 2019, 116, 18893–18899. [Google Scholar] [CrossRef]
- Tippery, N.P.; Les, D.H. Tiny Plants with Enormous Potential: Phylogeny and Evolution of Duckweeds. In The Duckweed Genomes; Cao, X.H., Fourounjian, P., Wang, W., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 19–38. [Google Scholar]
- Les, D.H.; Crawford, D.J.; Landolt, E.; Gabel, J.D.; Kimball, R.T. Phylogeny and Systematics of Lemnaceae, the Duckweed Family. Syst. Bot. 2002, 27, 221–240. [Google Scholar] [CrossRef]
- Zhang, J.; Azizullah, A. Genetic Diversity and DNA Barcoding in the Duckweed Family. In The Duckweed Genomes; Cao, X.H., Fourounjian, P., Wang, W., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 59–65. [Google Scholar]
- CBOL Plant Working Group. A DNA Barcode for Land Plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Hollingsworth, P.M. Refining the DNA Barcode for Land Plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19451–19452. [Google Scholar] [CrossRef]
- Mardanov, A.V.; Ravin, N.V.; Kuznetsov, B.B.; Samigullin, T.H.; Antonov, A.S.; Kolganova, T.V.; Skyabin, K.G. Complete Sequence of the Duckweed (Lemna minor) Chloroplast Genome: Structural Organization and Phylogenetic Relationships to Other Angiosperms. J. Mol. Evol. 2008, 66, 555–564. [Google Scholar] [CrossRef]
- Wang, W.; Messing, J. High-Throughput Sequencing of Three Lemnoideae (Duckweeds) Chloroplast Genomes from Total DNA. PLoS ONE 2011, 6, e24670. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Fang, Y.; Guo, L.; Li, Z.; He, K.; Zhao, Y.; Zhao, H. Phylogenic Study of Lemnoideae (Duckweeds) through Complete Chloroplast Genomes for Eight Accessions. PeerJ 2017, 5, e4186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; An, D.; Li, C.; Zhao, Z.; Wang, W. The Complete Chloroplast Genome of Greater Duckweed (Spirodela polyrhiza 7498) Using PacBio Long Reads: Insights into the Chloroplast Evolution and Transcription Regulation. BMC Genom. 2020, 21, 76. [Google Scholar] [CrossRef] [PubMed]
- Fourounjian, P.; Fakhoorian, T.; Cao, X.H. Importance of Duckweeds in Basic Research and Their Industrial Applications. In The Duckweed Genomes; Cao, X.H., Fourounjian, P., Wang, W., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 1–17. [Google Scholar]
- Wang, W.; Wu, Y.; Yan, Y.; Ermakova, M.; Kerstetter, R.; Messing, J. DNA Barcoding of the Lemnaceae, a Family of Aquatic Monocots. BMC Plant Biol. 2010, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zhang, F.; Cui, W.; Ma, J. Genetic Structure of Duckweed Population of Spirodela, Landoltia and Lemna from Lake Tai, China. Planta 2014, 239, 1299–1307. [Google Scholar] [CrossRef]
- Tang, J.; Li, Y.; Ma, J.; Cheng, J.J. Survey of Duckweed Diversity in Lake Chao and Total Fatty Acid, Triacylglycerol, Profiles of Representative Strains. Plant Biol. 2015, 17, 1066–1072. [Google Scholar] [CrossRef]
- Stepanenko, A.; Chen, G.; Hoang, P.T.N.; Fuchs, J.; Schubert, I.; Borisjuk, N. The Ribosomal DNA Loci of the Ancient Monocot Pistia Stratiotes L. (Araceae) Contain Different Variants of the 35S and 5S Ribosomal RNA Gene Units. Front. Plant Sci. 2022, 13, 819750. [Google Scholar] [CrossRef]
- Borisjuk, N.; Chu, P.; Gutierrez, R.; Zhang, H.; Acosta, K.; Friesen, N.; Sree, K.S.; Garcia, C.; Appenroth, K.J.; Lam, E. Assessment, Validation and Deployment Strategy of a Two-Barcode Protocol for Facile Genotyping of Duckweed Species. Plant Biol. 2015, 17 (Suppl. 1), 42–49. [Google Scholar] [CrossRef]
- Bog, M.; Baumbach, H.; Schween, U.; Hellwig, F.; Landolt, E.; Appenroth, K.-J. Genetic Structure of the Genus Lemna L. (Lemnaceae) as Revealed by Amplified Fragment Length Polymorphism. Planta 2010, 232, 609–619. [Google Scholar] [CrossRef]
- Bog, M.; Schneider, P.; Hellwig, F.; Sachse, S.; Kochieva, E.Z.; Martyrosian, E.; Landolt, E.; Appenroth, K.-J. Genetic Characterization and Barcoding of Taxa in the Genus Wolffia Horkel Ex Schleid. (Lemnaceae) as Revealed by Two Plastidic Markers and Amplified Fragment Length Polymorphism (AFLP). Planta 2013, 237, 1–13. [Google Scholar] [CrossRef]
- Landolt, E. The family of Lemnaceae—A monographic study. Veröff. Geobot. Inst. Rübel 1986, 71, 1–563. [Google Scholar]
- Orlov, O.O.; Iakushenko, D.M. Lemna turionifera Landolt (Araceae) a new species for the flora of Ukraine. Ukr. Bot. J. 2013, 70, 224–231. [Google Scholar] [CrossRef]
- Sabliy, L.; Konontsev, S.; Grokhovska, J.; Widomski, M.K.; Łagód, G. Nitrogen Removal from Fish Farms Water by Lemna minor and Wolffia arrhiza. Proc. ECOpole 2016, 10, 499–504. [Google Scholar] [CrossRef]
- Lam, E.; Appenroth, K.J.; Ma, Y.; Shoham, T.; Sree, K.S. Registration of duckweed clones/strains-future approach. Duckweed Forum 2020, 8, 35–37. Available online: http://www.ruduckweed.org (accessed on 31 March 2022).
- Sree, K.S.; Appenroth, K.-J. Worldwide Genetic Resources of Duckweed: Stock Collections. In The Duckweed Genomes; Cao, X.H., Fourounjian, P., Wang, W., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 39–46. [Google Scholar]
- Xu, S.; Stapley, J.; Gablenz, S.; Boyer, J.; Appenroth, K.J.; Sree, K.S.; Gershenzon, J.; Widmer, A.; Huber, M. Low Genetic Variation Is Associated with Low Mutation Rate in the Giant Duckweed. Nat. Commun. 2019, 10, 1243. [Google Scholar] [CrossRef] [PubMed]
- Borisjuk, N.; Peterson, A.; Lv, J.; Qu, G.; Luo, Q.; Shi, L.; Chen, Q.; Kishchenko, O.; Zhou, Y.; Shi, J. Structural and biochemical properties of duckweed surface cuticle. Front. Chem. 2018, 6, 317. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Stepanenko, A.; Borisjuk, N. Mosaic Arrangement of the 5S rDNA in the Aquatic Plant Landoltia punctata (Lemnaceae). Front. Plant Sci. 2021, 12, 678689. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid Isolation of High Molecular Weight Plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Wilgenbusch, J.C.; Swofford, D. Inferring evolutionary trees with PAUP*. Curr. Protoc. Bioinform. 2003, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ma, S.; Huang, M.; Peng, M.; Bog, M.; Sree, K.S.; Appenroth, K.-J.; Zhang, J. Species Distribution, Genetic Diversity and Barcoding in the Duckweed Family (Lemnaceae). Hydrobiologia 2015, 743, 75–87. [Google Scholar] [CrossRef]
- Zhou, Y.; Kishchenko, O.; Stepanenko, A.; Chen, G.; Wang, W.; Zhou, J.; Pan, C.; Borisjuk, N. The Dynamics of NO3− and NH4+ Uptake in Duckweed Are Coordinated with the Expression of Major Nitrogen Assimilation Genes. Plants 2021, 11, 11. [Google Scholar] [CrossRef]
- Bog, M.; Lautenschlager, U.; Landrock, M.; Landolt, E.; Fuchs, J.; Sree, K.S.; Oberprieler, C.; Appenroth, K.-J. Genetic Characterization and Barcoding of Taxa in the Genera Landoltia and Spirodela (Lemnaceae) by Three Plastidic Markers and Amplified Fragment Length Polymorphism (AFLP). Hydrobiologia 2015, 749, 169–182. [Google Scholar] [CrossRef]
- Kittiwongwattana, C.; Thawai, C. Rhizobium Lemnae Sp. Nov., a Bacterial Endophyte of Lemna aequinoctialis. Int. J. Syst. Evol. Microbiol. 2014, 64, 2455–2460. [Google Scholar] [CrossRef]
- Lee, Y.; Choi, H.J.; Shiga, T. Taxonomic Identity of Landoltia punctata (Araceae, Lemnoideae) in Korea. J. Asia-Pac. Biodivers. 2020, 13, 494–498. [Google Scholar] [CrossRef]
- Barks, P.; Dempsey, Z.; Burg, T.; Laird, R. Among-Strain Consistency in the Pace and Shape of Senescence in Duckweed. J. Ecol. 2018, 106, 2132–2145. [Google Scholar] [CrossRef]
- Fu, L.; Huang, M.; Han, B.; Sun, X.; Sree, K.S.; Appenroth, K.-J.; Zhang, J. Flower Induction, Microscope-Aided Cross-Pollination, and Seed Production in the Duckweed Lemna gibba with Discovery of a Male-Sterile Clone. Sci. Rep. 2017, 7, 3047. [Google Scholar] [CrossRef] [PubMed]
- Burgess, K.; Fazekas, A.; Kesanakurti, P.; Graham, S.; Husband, B.; Newmaster, S.; Percy, D.; Hajibabaei, M.; Barrett, S. Discriminating Plant Species in a Local Temperate Flora Using the RbcL+matK DNA Barcode. Methods Ecol. Evol. 2011, 2, 333–340. [Google Scholar] [CrossRef]
- Bog, M.; Appenroth, K.-J.; Sree, K.S. Duckweed (Lemnaceae): Its Molecular Taxonomy. Front. Sustain. Food Syst. 2019, 3, 117. [Google Scholar] [CrossRef]
- Appenroth, K.-J.; Sree, K.S.; Bog, M.; Ecker, J.; Seeliger, C.; Böhm, V.; Lorkowski, S.; Sommer, K.; Vetter, W.; Tolzin-Banasch, K.; et al. Nutritional Value of the Duckweed Species of the Genus Wolffia (Lemnaceae) as Human Food. Front. Chem. 2018, 6, 483. [Google Scholar] [CrossRef]
- Park, H.; Park, J.H.; Jeon, H.H.; Woo, D.U.; Lee, Y.; Kang, Y.J. Characterization of the Complete Chloroplast Genome Sequence of Wolffia globosa (Lemnoideae) and Its Phylogenetic Relationships to Other Araceae Family. Mitochondrial DNA Part B 2020, 5, 1905–1907. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).