Bioengineered Efficacy Models of Skin Disease: Advances in the Last 10 Years
Abstract
1. Introduction
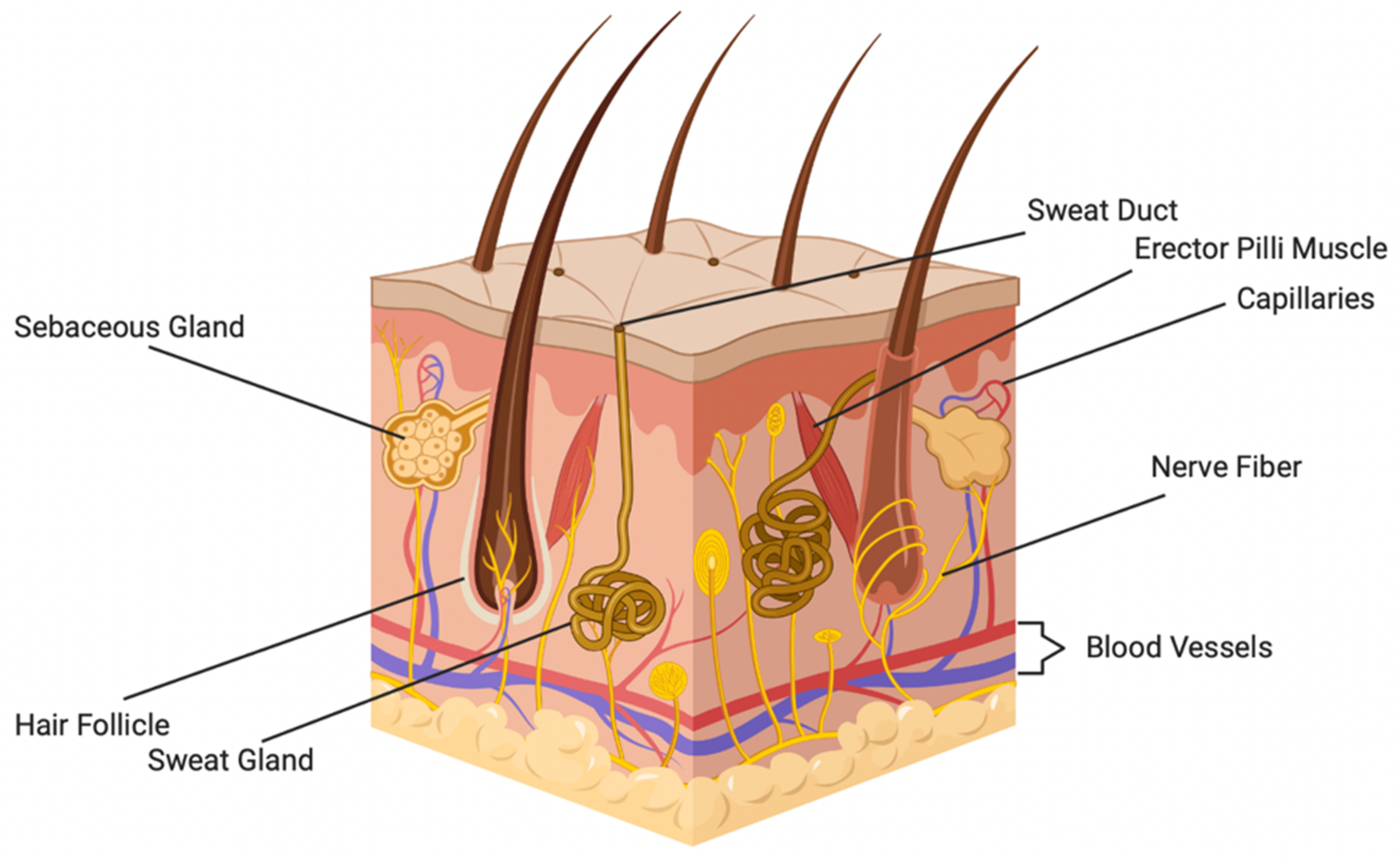
2. Overview of Skin Structure and Function
3. Skin Diseases
4. Current 3D Bioengineered Models of Skin Disease
4.1. Epidermolysis Bullosa Models
4.2. Ichthyosis Models
4.3. Atopic Dermatitis Models
4.4. Psoriasis Models
4.5. Scleroderma Models
4.6. Melanoma Models
5. Methods for Generating 3D Human Skin Equivalents
6. Challenges in Engineering 3D Skin Models
7. Current Advances in the Field of Skin Engineering
| Method | Adnexal Structure(s) | Cells Used | Reference(s) |
|---|---|---|---|
| 3D Bioprinting, Extrusion | Vasculature | IL-4-Treated NHKs, iPSCs, NHFs, pericytes | [71] |
| Manual Deposition | Immune System | MUTZ-LC, NHFs, NHKs | [128,129,130] |
| Manual Deposition | Immune System | NHFs, NHKs, LCs, DCs | [131] |
| Manual Deposition | Immune System | NHFs, NHKs, MUTZ-3-LCs | [132] |
| Manual Deposition | Immune System | NHFs, NHKs, DCs | [133] |
| Manual Deposition | Immune System | NHFs, NHKs, Macrophages | [134] |
| Manual Deposition | Immune System | NHKs, NHFs, Peripheral Blood Mononuclear Cells, CD4+ T cells | [135] |
| Skin-On-A-Chip | Vasculature | HaCaT Cells, HS27 Fibroblasts, HUVECs | [94] |
| 3D Bioprinting, Extrusion | Nervous System | hNSCs | [115] |
| Manual Deposition | Nervous System | hNSCs | [116] |
| 3D Bioprinting, Extrusion | Nervous System | Schwann Cells | [117] |
| Manual Deposition | Immune System, Nervous System | NHKs, NHFs, hiNSCs | [118] |
| Manual Deposition | Hair Follicle | SKPs, Epi-SCs | [119] |
| 3D Bioprinting, Extrusion | Hair Follicle, Sweat Gland | NHKs, NHFs, MSCs | [120] |
| Manual Deposition | Hair Follicle | Dermal Progenitor Cells, Epi-SCs | [121] |
| Manual Deposition | Vasculature, Hair Follicle | DPCs, NHKs, NHFs, HUVECs | [107] |
| 3D Bioprinting, Extrusion | Vasculature | NHKs, NHFs, Pericytes, Endothelial Cells | [124] |
| 3D Bioprinting, Extrusion | Sweat Gland | Epithelial Progenitor Cells | [122] |
| Manual Deposition | Sebaceous Gland | hiPSCs | [123] |
| 3D Bioprinting, Extrusion | Vasculature | NHKs, NHFs, HMVECs | [125] |
| 3D Bioprinting, Extrusion | Vasculature | Adipose-Derived Stem Cells, Endothelial Progenitor Cells | [126] |
| Skin-On-A-Chip | Vasculature | NHKs, NHFs, HUVECs | [101] |
| Manual Deposition | Lymphatic System, Vasculature | NHFs, HUVECs, NHKs, NHDLMECs | [127] |
| Manual Deposition | Lymphatic System, Vasculature | LECs, NHFs | [127] |
8. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karimkhani, C.; Dellavalle, R.P.; Coffeng, L.E.; Flohr, C.; Hay, R.J.; Langan, S.M.; Nsoesie, E.O.; Ferrari, A.J.; Erskine, H.E.; Silverberg, J.I.; et al. Global Skin Disease Morbidity and Mortality. JAMA Dermatol. 2017, 153, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.W.; Collins, S.A.B.; Resneck, J.S.; Bolognia, J.L.; Hodge, J.A.; Rohrer, T.A.; Van Beek, M.J.; Margolis, D.J.; Sober, A.J.; Weinstock, M.A.; et al. The burden of skin disease in the United States. J. Am. Acad. Dermatol. 2017, 76, 973–974. [Google Scholar] [CrossRef] [PubMed]
- Van Norman, G.A. Drugs, Devices, and the FDA: Part 1. JACC Basic Transl. Sci. 2016, 1, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Wouters, O.J.; Mckee, M.; Luyten, J. Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009–2018. JAMA 2020, 323, 844–853. [Google Scholar] [CrossRef]
- Fisher, J.A.; Cottingham, M.D.; Kalbaugh, C.A. Peering into the pharmaceutical “pipeline”: Investigational drugs, clinical trials, and industry priorities. Soc. Sci. Med. 2015, 131, 322–330. [Google Scholar] [CrossRef]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Baker, B.M.; Chen, C.S. Deconstructing the third dimension—How 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012, 125, 3015–3024. [Google Scholar] [CrossRef]
- Beauchamp, P.; Moritz, W.; Kelm, J.M.; Ullrich, N.D.; Agarkova, I.; Anson, B.D.; Suter, T.M.; Zuppinger, C. Development and Characterization of a Scaffold-Free 3D Spheroid Model of Induced Pluripotent Stem Cell-Derived Human Cardiomyocytes. Tissue Eng. Part C Methods 2015, 21, 852–861. [Google Scholar] [CrossRef]
- Gauvin, R.; Chen, Y.-C.; Lee, J.W.; Soman, P.; Zorlutuna, P.; Nichol, J.W.; Bae, H.; Chen, S.; Khademhosseini, A. Microfabrication of complex porous tissue engineering scaffolds using 3D projection stereolithography. Biomaterials 2012, 33, 3824–3834. [Google Scholar] [CrossRef]
- Shuster, E. Fifty Years Later: The Significance of the Nuremberg Code. N. Engl. J. Med. 1997, 337, 1436–1440. [Google Scholar] [CrossRef]
- Wall, R.J.; Shani, M. Are animal models as good as we think? Theriogenology 2008, 69, 2–9. [Google Scholar] [CrossRef]
- Akasaka, E.; Kleiser, S.; Sengle, G.; Bruckner-Tuderman, L.; Nyström, A. Diversity of Mechanisms Underlying Latent TGF-β Activation in Recessive Dystrophic Epidermolysis Bullosa. J. Investig. Dermatol. 2021, 141, 1450–1460.E9. [Google Scholar] [CrossRef]
- Clarysse, K.; Pfaff, C.M.; Marquardt, Y.; Huth, L.; Kortekaas Krohn, I.; Kluwig, D.; Lüscher, B.; Gutermuth, J.; Baron, J. JAK1/3 inhibition preserves epidermal morphology in full-thickness 3D skin models of atopic dermatitis and psoriasis. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 367–375. [Google Scholar] [CrossRef]
- Lincoln, V.; Cogan, J.; Hou, Y.; Hirsch, M.; Hao, M.; Alexeev, V.; De Luca, M.; De Rosa, L.; Bauer, J.W.; Woodley, D.T.; et al. Gentamicin induces LAMB3 nonsense mutation readthrough and restores functional laminin 332 in junctional epidermolysis bullosa. Proc. Natl. Acad. Sci. 2018, 115, E6536–E6545. [Google Scholar] [CrossRef]
- Lee, S.H.; Bae, I.-H.; Choi, H.; Choi, H.W.; Oh, S.; Marinho, P.A.; Min, D.J.; Kim, D.-Y.; Lee, T.R.; Lee, C.S.; et al. Ameliorating effect of dipotassium glycyrrhizinate on an IL-4- and IL-13-induced atopic dermatitis-like skin-equivalent model. Arch. Dermatol. Res. 2019, 311, 131–140. [Google Scholar] [CrossRef]
- Li, S.; Teegarden, A.; Bauer, E.M.; Choi, J.; Messaddeq, N.; Hendrix, D.A.; Ganguli-Indra, G.; Leid, M.; Indra, A.K. Transcription Factor CTIP1/BCL11A Regulates Epidermal Differentiation and Lipid Metabolism During Skin Development. Sci. Rep. 2017, 7, 13427. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, L.J.; Guha, G.; Li, S.; Kyrylkova, K.; Kioussi, C.; Leid, M.; Ganguli-Indra, G.; Indra, A.K. Selective ablation of Ctip2/Bcl11b in epidermal keratinocytes triggers atopic dermatitis-like skin inflammatory responses in adult mice. PLoS ONE 2012, 7, e51262. [Google Scholar] [CrossRef]
- Yin, J.; Xie, X.; Ye, Y.; Wang, L.; Che, F. BCL11A: A potential diagnostic biomarker and therapeutic target in human diseases. Biosci. Rep. 2019, 39, BSR20190604. [Google Scholar] [CrossRef]
- Luc, S.; Huang, J.; Mceldoon, J.L.; Somuncular, E.; Li, D.; Rhodes, C.; Mamoor, S.; Hou, S.; Xu, J.; Orkin, S.H. Bcl11a Deficiency Leads to Hematopoietic Stem Cell Defects with an Aging-like Phenotype. Cell Rep. 2016, 16, 3181–3194. [Google Scholar] [CrossRef]
- Niehues, H.; Bouwstra, J.A.; El Ghalbzouri, A.; Brandner, J.M.; Zeeuwen, P.L.J.M.; van den Bogaard, E.H. 3D skin models for 3R research: The potential of 3D reconstructed skin models to study skin barrier function. Exp. Dermatol. 2018, 27, 501–511. [Google Scholar] [CrossRef]
- Hargis, A.M.; Myers, S. The Integument. In Pathologic Basis of Veterinary Disease; Mosby: St. Louis, MO, USA, 2017; pp. 1009–1146.e1. [Google Scholar] [CrossRef]
- Venus, M.; Waterman, J.; McNab, I. Basic physiology of the skin. Surgery 2010, 28, 469–472. [Google Scholar] [CrossRef]
- Meglio, P.D.; Conrad, C. Psoriasis, Cutaneous Lupus Erithematosus and Immunobiology of the Skin. In Encyclopedia of Immunobiology; Ratcliffe, M.J.H., Ed.; Academic Press: Oxford, UK, 2016; pp. 192–203. [Google Scholar]
- Menon, G.K.; Cleary, G.W.; Lane, M.E. The structure and function of the stratum corneum. Int. J. Pharm. 2012, 435, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Falanga, V.; Faria, K.; Bollenbach, T. Chapter 77—Bioengineered Skin Constructs. In Principles of Tissue Engineering, 4th ed.; Lanza, R., Langer, R., Vacanti, J., Eds.; Academic Press: Boston, MA, USA, 2014; pp. 1619–1643. [Google Scholar]
- Braverman, I.M. The Cutaneous Microcirculation. J. Investig. Dermatol. Symp. Proc. 2000, 5, 3–9. [Google Scholar] [CrossRef]
- Schneider, M.R.; Schmidt-Ullrich, R.; Paus, R. The Hair Follicle as a Dynamic Miniorgan. Curr. Biol. 2009, 19, R132–R142. [Google Scholar] [CrossRef]
- Makrantonaki, E.; Ganceviciene, R.; Zouboulis, C.C. An update on the role of the sebaceous gland in the pathogenesis of acne. Derm. Endocrinol. 2011, 3, 41–49. [Google Scholar] [CrossRef]
- Kobielak, K.; Kandyba, E.; Leung, Y. Chapter 22—Skin and Skin Appendage Regeneration. In Translational Regenerative Medicine; Atala, A., Allickson, J.G., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 269–292. [Google Scholar]
- López-Bigas, N.; Ouzounis, C.A. Genome-wide identification of genes likely to be involved in human genetic disease. Nucleic Acids Res. 2004, 32, 3108–3114. [Google Scholar] [CrossRef]
- Dudbridge, F. Polygenic Epidemiology. Genet. Epidemiol. 2016, 40, 268–272. [Google Scholar] [CrossRef]
- Peate, W.E. Occupational skin disease. Am. Fam. Physician 2002, 66, 1025–1032. [Google Scholar]
- Satolli, F.; Rovesti, M.; Feliciani, C. Autoimmune Skin Disorders. In Advances in Integrative Dermatology; Wiley Online Library: Hoboken, NJ, USA, 2019; pp. 89–102. [Google Scholar] [CrossRef]
- Fine, J.-D. Inherited epidermolysis bullosa. Orphanet J. Rare Dis. 2010, 5, 12. [Google Scholar] [CrossRef]
- McGrath, J.A. Recently Identified Forms of Epidermolysis Bullosa. Ann. Dermatol. 2015, 27, 658–666. [Google Scholar] [CrossRef]
- Koga, H.; Prost-Squarcioni, C.; Iwata, H.; Jonkman, M.F.; Ludwig, R.J.; Bieber, K. Epidermolysis Bullosa Acquisita: The 2019 Update. Front. Med. 2019, 5, 362. [Google Scholar] [CrossRef]
- Kasperkiewicz, M.; Sadik, C.D.; Bieber, K.; Ibrahim, S.M.; Manz, R.A.; Schmidt, E.; Zillikens, D.; Ludwig, R.J. Epidermolysis Bullosa Acquisita: From Pathophysiology to Novel Therapeutic Options. J. Investig. Dermatol. 2016, 136, 24–33. [Google Scholar] [CrossRef]
- Oji, V.; Traupe, H. Ichthyosis. Am. J. Clin. Dermatol. 2009, 10, 351–364. [Google Scholar] [CrossRef]
- Vahlquist, A.; Fischer, J.; Törmä, H. Inherited Nonsyndromic Ichthyoses: An Update on Pathophysiology, Diagnosis and Treatment. Am. J. Clin. Dermatol. 2018, 19, 51–66. [Google Scholar] [CrossRef]
- Wells, R.S. Ichthyosis. Br. Med. J. 1966, 2, 1504. [Google Scholar] [CrossRef]
- Patel, N.; Spencer, L.A.; English, J.C.; Zirwas, M.J. Acquired ichthyosis. J. Am. Acad. Dermatol. 2006, 55, 647–656. [Google Scholar] [CrossRef]
- De Benedetto, A.; Agnihothri, R.; McGirt, L.Y.; Bankova, L.G.; Beck, L.A. Atopic Dermatitis: A Disease Caused by Innate Immune Defects? J. Investig. Dermatol. 2009, 129, 14–30. [Google Scholar] [CrossRef]
- Bieber, T. Atopic dermatitis. N. Engl. J. Med. 2008, 358, 1483–1494. [Google Scholar] [CrossRef]
- Langley, R.G.B. Psoriasis: Epidemiology, clinical features, and quality of life. Ann. Rheum. Dis. 2005, 64, ii18–ii23. [Google Scholar] [CrossRef]
- Capon, F. The Genetic Basis of Psoriasis. Int. J. Mol. Sci. 2017, 18, 2526. [Google Scholar] [CrossRef]
- Abraham, D.J.; Varga, J. Scleroderma: From cell and molecular mechanisms to disease models. Trends Immunol. 2005, 26, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Parihar, A.K.S.; Patel, S.; Srivastava, S.; Diwan, P.; Singh, M.R. Scleroderma: An insight into causes, pathogenesis and treatment strategies. Pathophysiology 2019, 26, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Kasperkiewicz, M.; Ellebrecht, C.T.; Takahashi, H.; Yamagami, J.; Zillikens, D.; Payne, A.S.; Amagai, M. Pemphigus. Nat. Rev. Dis. Prim. 2017, 3, 17026. [Google Scholar] [CrossRef] [PubMed]
- Bystryn, J.-C.; Rudolph, J.L. Pemphigus. Lancet 2005, 366, 61–73. [Google Scholar] [CrossRef]
- Vodo, D.; Sarig, O.; Sprecher, E. The Genetics of Pemphigus Vulgaris. Front. Med. 2018, 5, 226. [Google Scholar] [CrossRef]
- Schadendorf, D.; Van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef]
- Tang, M.-S. Ultraviolet A light: Potential underlying causes of melanoma. Future Med. 2010, 6, 1523–1526. [Google Scholar] [CrossRef]
- Ribero, S.; Glass, D.; Bataille, V. Genetic epidemiology of melanoma. Eur. J. Dermatol. 2016, 26, 335–339. [Google Scholar] [CrossRef]
- Roberson, E.D.O.; Bowcock, A.M. Psoriasis genetics: Breaking the barrier. Trends Genet. 2010, 26, 415–423. [Google Scholar] [CrossRef]
- Borodzicz, S.; Rudnicka, L.; Mirowska-Guzel, D.; Cudnoch-Jedrzejewska, A. The role of epidermal sphingolipids in dermatologic diseases. Lipids Health Dis. 2016, 15, 13. [Google Scholar] [CrossRef]
- Li, S.; Villarreal, M.; Stewart, S.; Choi, J.; Ganguli-Indra, G.; Babineau, D.C.; Philpot, C.; David, G.; Yoshida, T.; Boguniewicz, M.; et al. Altered composition of epidermal lipids correlates with Staphylococcus aureus colonization status in atopic dermatitis. Br. J. Dermatol. 2017, 177, e125–e127. [Google Scholar] [CrossRef]
- Leung, D.Y. New insights into atopic dermatitis: Role of skin barrier and immune dysregulation. Allergol. Int. 2013, 62, 151–161. [Google Scholar] [CrossRef]
- Ganguli-Indra, G.; Liang, X.; Hyter, S.; Leid, M.; Hanifin, J.; Indra, A.K. Expression of COUP-TF-interacting protein 2 (CTIP2) in human atopic dermatitis and allergic contact dermatitis skin. Exp. Dermatol. 2009, 18, 994–996. [Google Scholar] [CrossRef]
- Golonzhka, O.; Liang, X.; Messaddeq, N.; Bornert, J.M.; Campbell, A.L.; Metzger, D.; Chambon, P.; Ganguli-Indra, G.; Leid, M.; Indra, A.K. Dual role of COUP-TF-interacting protein 2 in epidermal homeostasis and permeability barrier formation. J. Investig. Dermatol. 2009, 129, 1459–1470. [Google Scholar] [CrossRef]
- Liang, X.; Bhattacharya, S.; Bajaj, G.; Guha, G.; Wang, Z.; Jang, H.S.; Leid, M.; Indra, A.K.; Ganguli-Indra, G. Delayed cutaneous wound healing and aberrant expression of hair follicle stem cell markers in mice selectively lacking Ctip2 in epidermis. PLoS ONE 2012, 7, e29999. [Google Scholar] [CrossRef]
- Itoh, M.; Umegaki-Arao, N.; Guo, Z.; Liu, L.; Higgins, C.A.; Christiano, A.M. Generation of 3D Skin Equivalents Fully Reconstituted from Human Induced Pluripotent Stem Cells (iPSCs). PLoS ONE 2013, 8, e77673. [Google Scholar] [CrossRef]
- Itoh, M.; Kiuru, M.; Cairo, M.S.; Christiano, A.M. Generation of keratinocytes from normal and recessive dystrophic epidermolysis bullosa-induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2011, 108, 8797–8802. [Google Scholar] [CrossRef]
- Desmet, E.; Van Gele, M.; Grine, L.; Remaut, K.; Lambert, J. Towards the development of a RNAi-based topical treatment for psoriasis: Proof-of-concept in a 3D psoriasis skin model. Exp. Dermatol. 2018, 27, 463–469. [Google Scholar] [CrossRef]
- Bocheńska, K.; Smolińska, E.; Moskot, M.; Jakóbkiewicz-Banecka, J.; Gabig-Cimińska, M. Models in the Research Process of Psoriasis. Int. J. Mol. Sci. 2017, 18, 2514. [Google Scholar] [CrossRef]
- Kandarova, H.; Hayden, P.J. Standardised Reconstructed Skin Models in Toxicology and Pharmacology: State of the Art and Future Development. In Organotypic Models in Drug Development; Schäfer-Korting, M., Stuchi Maria-Engler, S., Landsiedel, R., Eds.; Springer International Publishing: Cham, Germany, 2021; pp. 57–71. [Google Scholar]
- Shin, J.U.; Abaci, H.E.; Herron, L.; Guo, Z.; Sallee, B.; Pappalardo, A.; Jackow, J.; Wang, E.H.C.; Doucet, Y.; Christiano, A.M. Recapitulating T cell infiltration in 3D psoriatic skin models for patient-specific drug testing. Sci. Rep. 2020, 10, 4123. [Google Scholar] [CrossRef]
- Mildner, M.; Jin, J.; Eckhart, L.; Kezic, S.; Gruber, F.; Barresi, C.; Stremnitzer, C.; Buchberger, M.; Mlitz, V.; Ballaun, C.; et al. Knockdown of Filaggrin Impairs Diffusion Barrier Function and Increases UV Sensitivity in a Human Skin Model. J. Investig. Dermatol. 2010, 130, 2286–2294. [Google Scholar] [CrossRef]
- Mildner, M.; Ballaun, C.; Stichenwirth, M.; Bauer, R.; Gmeiner, R.; Buchberger, M.; Mlitz, V.; Tschachler, E. Gene silencing in a human organotypic skin model. Biochem. Biophys. Res. Commun. 2006, 348, 76–82. [Google Scholar] [CrossRef]
- Enjalbert, F.; Dewan, P.; Caley, M.P.; Jones, E.M.; Morse, M.A.; Kelsell, D.P.; Enright, A.J.; O’Toole, E.A. 3D model of harlequin ichthyosis reveals inflammatory therapeutic targets. J. Clin. Investig. 2020, 130, 4798–4810. [Google Scholar] [CrossRef] [PubMed]
- Sriram, G.; Bigliardi, P.L.; Bigliardi-Qi, M. Full-Thickness Human Skin Equivalent Models of Atopic Dermatitis. In Skin Stem Cells; Springer: New York, NY, USA, 2018; pp. 367–383. [Google Scholar]
- Liu, X.; Michael, S.; Bharti, K.; Ferrer, M.; Song, M.J. A biofabricated vascularized skin model of atopic dermatitis for preclinical studies. Biofabrication 2020, 12, 035002. [Google Scholar] [CrossRef] [PubMed]
- Luchetti, M.M.; Moroncini, G.; Jose Escamez, M.; Svegliati Baroni, S.; Spadoni, T.; Grieco, A.; Paolini, C.; Funaro, A.; Avvedimento, E.V.; Larcher, F.; et al. Induction of Scleroderma Fibrosis in Skin-Humanized Mice by Administration of Anti−Platelet-Derived Growth Factor Receptor Agonistic Autoantibodies. Arthritis Rheumatol. 2016, 68, 2263–2273. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.L.; Corinaldesi, C.; Migneco, G.; Carr, I.; Antanaviciute, A.; Carriero, A.; Wasson, C.; Georgiou, I.; Distler, J.H.W.; Holmes, S.; et al. Targeting human Plasmacytoid dendritic cells through BDCA2 prevents inflammation and fibrosis in xenotransplant mouse model of Scleroderma. Ann. Rheum. Dis. 2021, 80, 920–929. [Google Scholar] [CrossRef] [PubMed]
- Müller, I.; Kulms, D. A 3D Organotypic Melanoma Spheroid Skin Model. J. Vis. Exp. 2018, 135, 57500. [Google Scholar] [CrossRef]
- Haridas, P.; McGovern, J.A.; McElwain, S.D.L.; Simpson, M.J. Quantitative comparison of the spreading and invasion of radial growth phase and metastatic melanoma cells in a three-dimensional human skin equivalent model. Peer J. 2017, 5, e3754. [Google Scholar] [CrossRef]
- Rossi, A.; Appelt-Menzel, A.; Kurdyn, S.; Walles, H.; Groeber, F. Generation of a Three-dimensional Full Thickness Skin Equivalent and Automated Wounding. J. Vis. Exp. 2015, 96, e52576. [Google Scholar] [CrossRef]
- Bauer, J.W.; Schaeppi, H.; Kaserer, C.; Hantich, B.; Hintner, H. Large melanocytic nevi in hereditary epidermolysis bullosa. J. Am. Acad. Dermatol. 2001, 44, 577–584. [Google Scholar] [CrossRef]
- Gupta, R.; Woodley, D.T.; Chen, M. Epidermolysis bullosa acquisita. Clin. Dermatol. 2012, 30, 60–69. [Google Scholar] [CrossRef]
- Leman, G.; Moosbrugger-Martinz, V.; Blunder, S.; Pavel, P.; Dubrac, S. 3D-Organotypic Cultures to Unravel Molecular and Cellular Abnormalities in Atopic Dermatitis and Ichthyosis Vulgaris. Cells 2019, 8, 489. [Google Scholar] [CrossRef]
- Niehues, H.; Schalkwijk, J.; Van Vlijmen-Willems, I.M.J.J.; Rodijk-Olthuis, D.; Van Rossum, M.M.; Wladykowski, E.; Brandner, J.M.; Van Den Bogaard, E.H.J.; Zeeuwen, P.L.J.M. Epidermal equivalents of filaggrin null keratinocytes do not show impaired skin barrier function. J. Allergy Clin. Immunol. 2017, 139, 1979–1981.E13. [Google Scholar] [CrossRef]
- Blunder, S.; Rühl, R.; Moosbrugger-Martinz, V.; Krimmel, C.; Geisler, A.; Zhu, H.; Crumrine, D.; Elias, P.M.; Gruber, R.; Schmuth, M.; et al. Alterations in Epidermal Eicosanoid Metabolism Contribute to Inflammation and Impaired Late Differentiation in FLG-Mutated Atopic Dermatitis. J. Investig. Dermatol. 2017, 137, 706–715. [Google Scholar] [CrossRef]
- Gruber, R.; Elias, P.M.; Crumrine, D.; Lin, T.K.; Brandner, J.M.; Hachem, J.P.; Presland, R.B.; Fleckman, P.; Janecke, A.R.; Sandilands, A.; et al. Filaggrin genotype in ichthyosis vulgaris predicts abnormalities in epidermal structure and function. Am. J. Pathol 2011, 178, 2252–2263. [Google Scholar] [CrossRef]
- Lowes, M.A.; Bowcock, A.M.; Krueger, J.G. Pathogenesis and therapy of psoriasis. Nature 2007, 445, 866–873. [Google Scholar] [CrossRef]
- Ali, G.; Elsayed, A.K.; Nandakumar, M.; Bashir, M.; Younis, I.; Abu Aqel, Y.; Memon, B.; Temanni, R.; Abubaker, F.; Taheri, S.; et al. Keratinocytes Derived from Patient-Specific Induced Pluripotent Stem Cells Recapitulate the Genetic Signature of Psoriasis Disease. Stem Cells Dev. 2020, 29, 383–400. [Google Scholar] [CrossRef]
- Hunzelmann, N.; Krieg, T. Scleroderma: From pathophysiology to novel therapeutic approaches. Exp. Dermatol. 2010, 19, 393–400. [Google Scholar] [CrossRef]
- Bourland, J.; Fradette, J.; Auger, F.A. Tissue-engineered 3D melanoma model with blood and lymphatic capillaries for drug development. Sci. Rep. 2018, 8, 13191. [Google Scholar] [CrossRef]
- Vandyck, H.H.; Hillen, L.M.; Bosisio, F.M.; Van Den Oord, J.; Zur Hausen, A.; Winnepenninckx, V. Rethinking the biology of metastatic melanoma: A holistic approach. Cancer Metastasis Rev. 2021, 40, 603–624. [Google Scholar] [CrossRef]
- Passarelli, A.; Mannavola, F.; Stucci, L.S.; Tucci, M.; Silvestris, F. Immune system and melanoma biology: A balance between immunosurveillance and immune escape. Oncotarget 2017, 8, 106132–106142. [Google Scholar] [CrossRef] [PubMed]
- Bedogni, B.; Paus, R. Hair(y) Matters in Melanoma Biology. Trends Mol. Med. 2020, 26, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Dika, E.; Veronesi, G.; Misciali, C.; Corti, B.; Dika, I.; Riefolo, M.; Scarfì, F.; Lambertini, M.; Patrizi, A. Malignant Melanoma Cells and Hair Follicles: Focusing on Head and Neck Melanomas. Am. J. Clin. Pathol. 2019, 152, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Gomez Garcia, A.M.; Mclaren, C.E.; Meyskens, F.L. Melanoma: Is hair the root of the problem? Pigment. Cell Melanoma Res. 2011, 24, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.; Donahue, L.R.; Choi, E.; Scumpia, P.O.; Lowry, W.E.; Grenier, J.K.; Zhu, J.; White, A.C. Melanocyte Stem Cell Activation and Translocation Initiate Cutaneous Melanoma in Response to UV Exposure. Cell Stem Cell 2017, 21, 665–678.e6. [Google Scholar] [CrossRef]
- Mathes, S.H.; Ruffner, H.; Graf-Hausner, U. The use of skin models in drug development. Adv. Drug Deliv. Rev. 2014, 69–70, 81–102. [Google Scholar] [CrossRef]
- Wufuer, M.; Lee, G.; Hur, W.; Jeon, B.; Kim, B.J.; Choi, T.H.; Lee, S. Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci. Rep. 2016, 6, 37471. [Google Scholar] [CrossRef]
- Hafezi, F.; Shorter, S.; Tabriz, A.G.; Hurt, A.; Elmes, V.; Boateng, J.; Douroumis, D. Bioprinting and Preliminary Testing of Highly Reproducible Novel Bioink for Potential Skin Regeneration. Pharmaceutics 2020, 12, 550. [Google Scholar] [CrossRef]
- Murphy, S.V.; Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef]
- Cui, X.; Boland, T.; D’Lima, D.D.; Lotz, M.K. Thermal Inkjet Printing in Tissue Engineering and Regenerative Medicine. Recent Pat. Drug Deliv. Formul. 2012, 6, 149–155. [Google Scholar] [CrossRef]
- Varkey, M.; Visscher, D.O.; Van Zuijlen, P.P.M.; Atala, A.; Yoo, J.J. Skin bioprinting: The future of burn wound reconstruction? Burn. Trauma 2019, 7, 4. [Google Scholar] [CrossRef]
- Moncal, K.K.; Ozbolat, V.; Datta, P.; Heo, D.N.; Ozbolat, I.T. Thermally-controlled extrusion-based bioprinting of collagen. J. Mater. Sci. Mater. Med. 2019, 30, 55. [Google Scholar] [CrossRef]
- Risueño, I.; Valencia, L.; Jorcano, J.L.; Velasco, D. Skin-on-a-chip models: General overview and future perspectives. APL Bioeng. 2021, 5, 030901. [Google Scholar] [CrossRef]
- Mori, N.; Morimoto, Y.; Takeuchi, S. Skin integrated with perfusable vascular channels on a chip. Biomaterials 2017, 116, 48–56. [Google Scholar] [CrossRef]
- Ataç, B.; Wagner, I.; Horland, R.; Lauster, R.; Marx, U.; Tonevitsky, A.G.; Azar, R.P.; Lindner, G. Skin and hair on-a-chip: In vitro skin models versus ex vivo tissue maintenance with dynamic perfusion. Lab Chip 2013, 13, 3555–3561. [Google Scholar] [CrossRef]
- Kim, B.S.; Ahn, M.; Cho, W.W.; Gao, G.; Jang, J.; Cho, D.W. Engineering of diseased human skin equivalent using 3D cell printing for representing pathophysiological hallmarks of type 2 diabetes in vitro. Biomaterials 2021, 272, 120776. [Google Scholar] [CrossRef]
- Tomasina, C.; Bodet, T.; Mota, C.; Moroni, L.; Camarero-Espinosa, S. Bioprinting Vasculature: Materials, Cells and Emergent Techniques. Materials 2019, 12, 2701. [Google Scholar] [CrossRef]
- Velasquillo, C.; Galue, E.A.; Rodriquez, L.; Ibarra, C.; Ibarra-Ibarra, L.G. Skin 3D Bioprinting. Applications in Cosmetology. J. Cosmet. Dermatol. Sci. Appl. 2013, 3, 85–89. [Google Scholar] [CrossRef]
- Abaci, H.E.; Coffman, A.; Doucet, Y.; Chen, J.; Jacków, J.; Wang, E.; Guo, Z.; Shin, J.U.; Jahoda, C.A.; Christiano, A.M. Tissue engineering of human hair follicles using a biomimetic developmental approach. Nat. Commun. 2018, 9, 5301. [Google Scholar] [CrossRef]
- Moysidou, C.-M.; Barberio, C.; Owens, R.M. Advances in Engineering Human Tissue Models. Front. Bioeng. Biotechnol. 2021, 8, 620962. [Google Scholar] [CrossRef]
- Richards, D.; Jia, J.; Yost, M.; Markwald, R.; Mei, Y. 3D Bioprinting for Vascularized Tissue Fabrication. Ann. Biomed. Eng. 2017, 45, 132–147. [Google Scholar] [CrossRef]
- Li, H.; Chen, L.; Zhang, M.; Tang, S.; Fu, X. Three-dimensional culture and identification of human eccrine sweat glands in matrigel basement membrane matrix. Cell Tissue Res. 2013, 354, 897–902. [Google Scholar] [CrossRef]
- Wilke, K.; Martin, A.; Terstegen, L.; Biel, S.S. A short history of sweat gland biology. Int. J. Cosmet. Sci. 2007, 29, 169–179. [Google Scholar] [CrossRef]
- Niemann, C.; Horsley, V. Development and homeostasis of the sebaceous gland. Semin. Cell Dev. Biol. 2012, 23, 928–936. [Google Scholar] [CrossRef]
- Pupovac, A.; Senturk, B.; Griffoni, C.; Maniura-Weber, K.; Rottmar, M.; Mcarthur, S.L. Toward Immunocompetent 3D Skin Models. Adv. Healthc. Mater. 2018, 7, 1701405. [Google Scholar] [CrossRef]
- Griffoni, C. Towards Advanced Immunocompetent Skin Wound Models for In Vitro Drug Evaluation. Doctoral Thesis, Universität Würzburg, Wurzburg, Germany, 2019. [Google Scholar]
- Kühbacher, A.; Sohn, K.; Burger-Kentischer, A.; Rupp, S. Immune Cell-Supplemented Human Skin Model for Studying Fungal Infections. In Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 439–449. [Google Scholar]
- Tomaskovic-Crook, E.; Crook, J.M. 3D Bioprinting Electrically Conductive Bioink with Human Neural Stem Cells for Human Neural Tissues. In Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 159–170. [Google Scholar]
- Tomaskovic-Crook, E.; Zhang, P.; Ahtiainen, A.; Kaisvuo, H.; Lee, C.Y.; Beirne, S.; Aqrawe, Z.; Svirskis, D.; Hyttinen, J.; Wallace, G.G.; et al. Human Neural Tissues from Neural Stem Cells Using Conductive Biogel and Printed Polymer Microelectrode Arrays for 3D Electrical Stimulation. Adv. Healthc. Mater. 2019, 8, 1900425. [Google Scholar] [CrossRef]
- Ning, L.; Sun, H.; Lelong, T.; Guilloteau, R.; Zhu, N.; Schreyer, D.J.; Chen, X. 3D bioprinting of scaffolds with living Schwann cells for potential nerve tissue engineering applications. Biofabrication 2018, 10, 035014. [Google Scholar] [CrossRef]
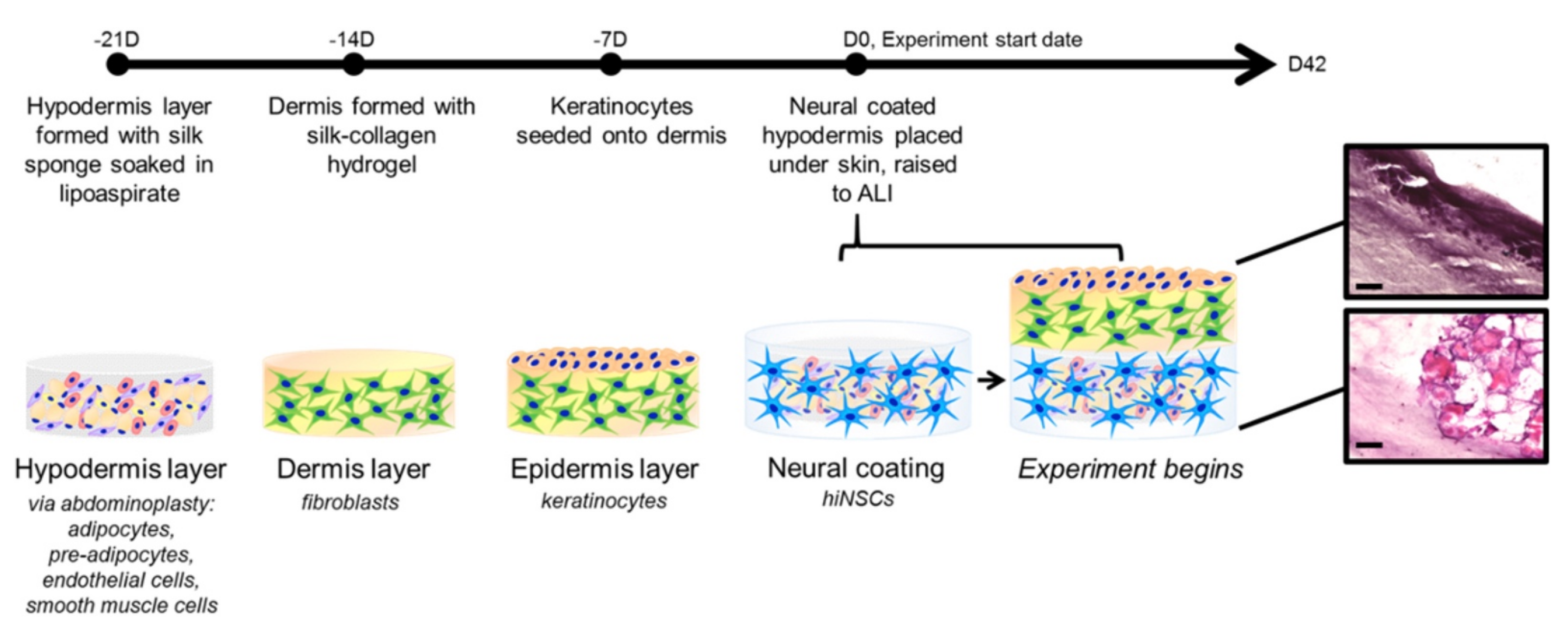
- Vidal, S.E.L.; Tamamoto, K.A.; Nguyen, H.; Abbott, R.D.; Cairns, D.M.; Kaplan, D.L. 3D biomaterial matrix to support long term, full thickness, immuno-competent human skin equivalents with nervous system components. Biomaterials 2019, 198, 194–203. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Xie, J.; Yao, B.; Mo, M.; Ma, D.; Huang, C.; Xu, R.; Fu, X.; Tredget, E.E.; et al. Engineered Skin Substitute Regenerates the Skin with Hair Follicle Formation. Biomedicines 2021, 9, 400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wen, J.; Liu, C.; Ma, C.; Bai, F.; Leng, X.; Chen, Z.; Xie, Z.; Mi, J.; Wu, X. Early-stage bilayer tissue-engineered skin substitute formed by adult skin progenitor cells produces an improved skin structure in vivo. Stem Cell Res. Ther. 2020, 11, 407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Enhejirigala; Yao, B.; Li, Z.; Song, W.; Li, J.; Zhu, D.; Wang, Y.; Duan, X.; Yuan, X.; et al. Using bioprinting and spheroid culture to create a skin model with sweat glands and hair follicles. Burn. Trauma 2021, 9, 407. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Yao, B.; Xie, J.; Fu, X. 3D bioprinted extracellular matrix mimics facilitate directed differentiation of epithelial progenitors for sweat gland regeneration. Acta Biomater. 2016, 32, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Albouy, M.; Tanguy, M.; Onteniente, B.; Thepot, A.; Maruotti, J.; Santos, M.D. LB1611 Development of a 3D full-thickness skin equivalent model containing sebocyte organoids derived from human iPS cells. J. Investig. Dermatol. 2018, 138, B24. [Google Scholar] [CrossRef]
- Baltazar, T.; Merola, J.; Catarino, C.; Xie, C.B.; Kirkiles-Smith, N.C.; Lee, V.; Hotta, S.; Dai, G.; Xu, X.; Ferreira, F.C.; et al. Three Dimensional Bioprinting of a Vascularized and Perfusable Skin Graft Using Human Keratinocytes, Fibroblasts, Pericytes, and Endothelial Cells. Tissue Eng. Part A 2020, 26, 227–238. [Google Scholar] [CrossRef]
- Huyan, Y.; Lian, Q.; Zhao, T.; Li, D.; He, J. Pilot Study of the Biological Properties and Vascularization of 3D Printed Bilayer Skin Grafts. Int. J. Bioprint. 2020, 6, 246. [Google Scholar] [CrossRef]
- Kim, B.S.; Kwon, Y.W.; Kong, J.-S.; Park, G.T.; Gao, G.; Han, W.; Kim, M.-B.; Lee, H.; Kim, J.H.; Cho, D.-W. 3D cell printing of in vitro stabilized skin model and in vivo pre-vascularized skin patch using tissue-specific extracellular matrix bioink: A step towards advanced skin tissue engineering. Biomaterials 2018, 168, 38–53. [Google Scholar] [CrossRef]
- Matsusaki, M.; Fujimoto, K.; Shirakata, Y.; Hirakawa, S.; Hashimoto, K.; Akashi, M. Development of full-thickness human skin equivalents with blood and lymph-like capillary networks by cell coating technology. J. Biomed. Mater. Res. Part A 2015, 103, 3386–3396. [Google Scholar] [CrossRef]
- Ouwehand, K.; Spiekstra, S.W.; Waaijman, T.; Scheper, R.J.; de Gruijl, T.D.; Gibbs, S. Technical advance: Langerhans cells derived from a human cell line in a full-thickness skin equivalent undergo allergen-induced maturation and migration. J. Leukoc. Biol. 2011, 90, 1027–1033. [Google Scholar] [CrossRef]
- Ouwehand, K.; Spiekstra, S.W.; Waaijman, T.; Breetveld, M.; Scheper, R.J.; de Gruijl, T.D.; Gibbs, S. CCL5 and CCL20 mediate immigration of Langerhans cells into the epidermis of full thickness human skin equivalents. Eur. J. Cell Biol. 2012, 91, 765–773. [Google Scholar] [CrossRef]
- Kosten, I.J.; Spiekstra, S.W.; de Gruijl, T.D.; Gibbs, S. MUTZ-3 derived Langerhans cells in human skin equivalents show differential migration and phenotypic plasticity after allergen or irritant exposure. Toxicol. Appl. Pharmacol. 2015, 287, 35–42. [Google Scholar] [CrossRef]
- Bechetoille, N.; Dezutter-Dambuyant, C.; Damour, O.; André, V.; Orly, I.; Perrier, E. Effects of solar ultraviolet radiation on engineered human skin equivalent containing both Langerhans cells and dermal dendritic cells. Tissue Eng. 2007, 13, 2667–2679. [Google Scholar] [CrossRef] [PubMed]
- Laubach, V.; Zöller, N.; Rossberg, M.; Görg, K.; Kippenberger, S.; Bereiter-Hahn, J.; Kaufmann, R.; Bernd, A. Integration of Langerhans-like cells into a human skin equivalent. Arch. Dermatol. Res. 2011, 303, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Chau, D.Y.; Johnson, C.; MacNeil, S.; Haycock, J.W.; Ghaemmaghami, A.M. The development of a 3D immunocompetent model of human skin. Biofabrication 2013, 5, 035011. [Google Scholar] [CrossRef]
- Linde, N.; Gutschalk, C.M.; Hoffmann, C.; Yilmaz, D.; Mueller, M.M. Integrating Macrophages into Organotypic Co-Cultures: A 3D In Vitro Model to Study Tumor-Associated Macrophages. PLoS ONE 2012, 7, e40058. [Google Scholar] [CrossRef]
- Kühbacher, A.; Henkel, H.; Stevens, P.; Grumaz, C.; Finkelmeier, D.; Burger-Kentischer, A.; Sohn, K.; Rupp, S. Central Role for Dermal Fibroblasts in Skin Model Protection against Candida albicans. J. Infect. Dis. 2017, 215, 1742–1752. [Google Scholar] [CrossRef]





| Skin Disease | Method | Cells Used in Model | Reference |
|---|---|---|---|
| Recessive Dystrophic Epidermolysis Bullosa | Manual Deposition | RDEBKs, RDEBFs, NHKs, RDEB PS-iPSC-Derived Keratinocytes, RDEB PS-iPSC-Derived Fibroblasts | [12,62] |
| Herlitz Junctional Epidermolysis Bullosa | Manual Deposition | H-JEBKs, H-JEBFs, NHKs, NHFs | [14] |
| Psoriasis | n/a | Psoriatic Fibroblasts, NHKs | [63,64,65] |
| Psoriasis | Manual Deposition | IL-17A-, IL-22-, and TNF-Treated NHKs and NHFs | [13] |
| Psoriasis | Manual Deposition | Polarized Th1/Th17 cells, CD4+ T cells, NHKs, NHFs | [66] |
| Ichthyosis Vulgaris | Manual Deposition | siRNA Filaggrin Knockdown Keratinocytes, NHFs | [67,68] |
| Harlequin Ichthyosis | Manual Deposition | CRISPR/Cas9 Knockdown ABCA12 N/TERT Keratinocytes, NHFs, THP-1 | [69] |
| Atopic Dermatitis | n/a | Th2 Cytokine-Treated NHKs, Th2 Cytokine-Treated NHFs | [70] |
| Atopic Dermatitis | 3D Bioprinting, | IL-4-Treated NHKs, iPSCs, NHFs, Pericytes | [71] |
| Atopic Dermatitis | Manual Deposition | IL-4- and IL-3-Treated NHFs, IL-4- and IL-3-Treated NHKs | [15] |
| Scleroderma | Manual Deposition | Patient-Derived SSc Fibroblasts, Patient-Derived SSc Keratinocytes, NHKs, NHFs | [72] |
| Scleroderma | Manual Deposition | pDCs, NHFs, NHKs | [73] |
| Melanoma | Manual Deposition | NHFs, NHKs, 451-LU | [74] |
| Melanoma | Manual Deposition | WM35, SK-MEL-28, NHFs, NHKs | [75] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stanton, D.N.; Ganguli-Indra, G.; Indra, A.K.; Karande, P. Bioengineered Efficacy Models of Skin Disease: Advances in the Last 10 Years. Pharmaceutics 2022, 14, 319. https://doi.org/10.3390/pharmaceutics14020319
Stanton DN, Ganguli-Indra G, Indra AK, Karande P. Bioengineered Efficacy Models of Skin Disease: Advances in the Last 10 Years. Pharmaceutics. 2022; 14(2):319. https://doi.org/10.3390/pharmaceutics14020319
Chicago/Turabian StyleStanton, Diana Nicole, Gitali Ganguli-Indra, Arup Kumar Indra, and Pankaj Karande. 2022. "Bioengineered Efficacy Models of Skin Disease: Advances in the Last 10 Years" Pharmaceutics 14, no. 2: 319. https://doi.org/10.3390/pharmaceutics14020319
APA StyleStanton, D. N., Ganguli-Indra, G., Indra, A. K., & Karande, P. (2022). Bioengineered Efficacy Models of Skin Disease: Advances in the Last 10 Years. Pharmaceutics, 14(2), 319. https://doi.org/10.3390/pharmaceutics14020319







