CTC-537E7.3 as a Liver-Specific Biomarker for Hepatocellular Carcinoma: Diagnostic and Prognostic Implications
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression and Prognostic Analysis in Public Omics Datasets
2.2. qRT-PCR
2.3. Clinical Sample Collection
2.4. miRNA Expression Profiling and ceRNA Network Construction
2.5. Statistical Analysis
3. Results
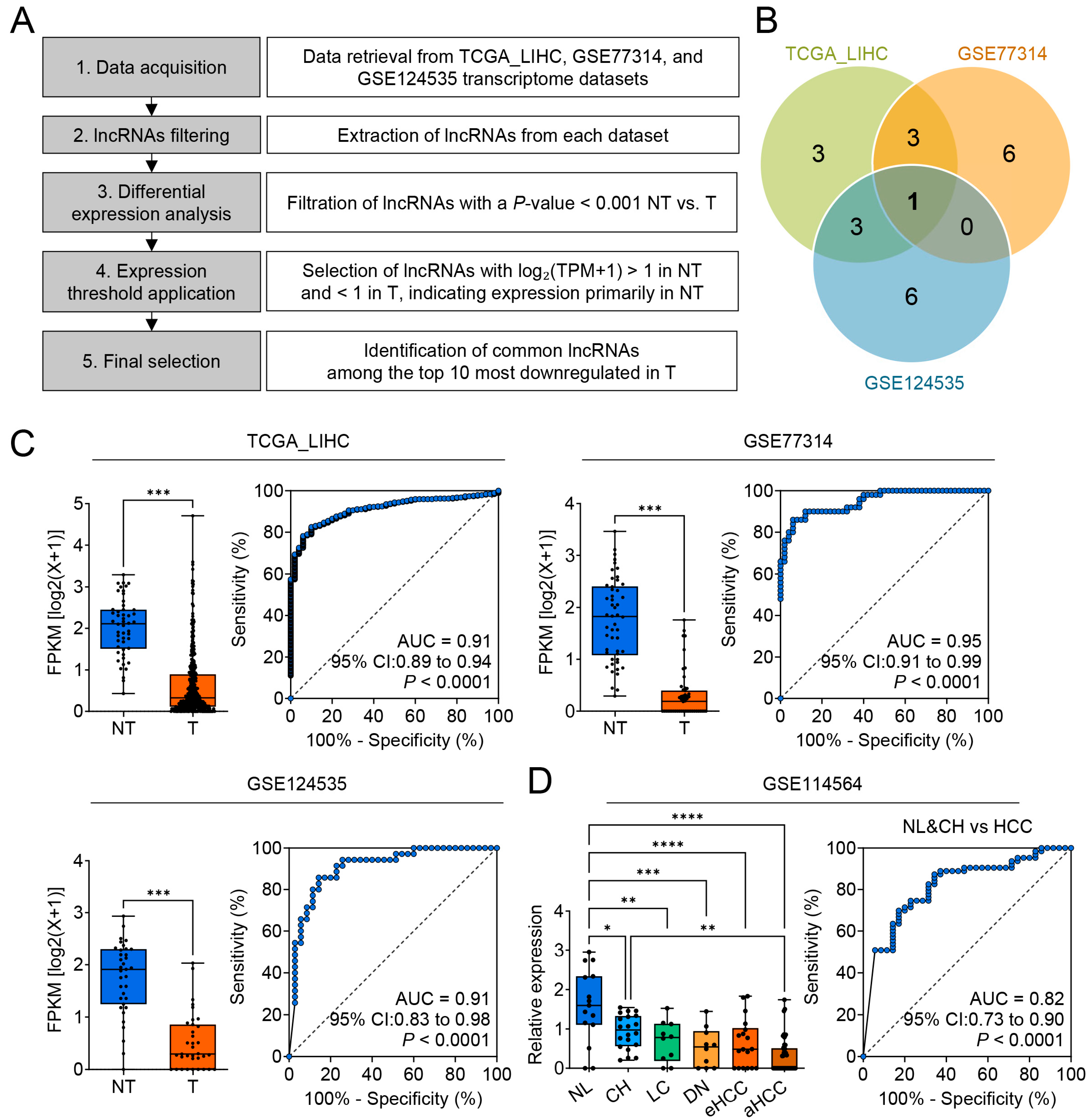
3.1. Identification of CTC-537E7.3 as a Potential Diagnostic Biomarker for Liver Cancer
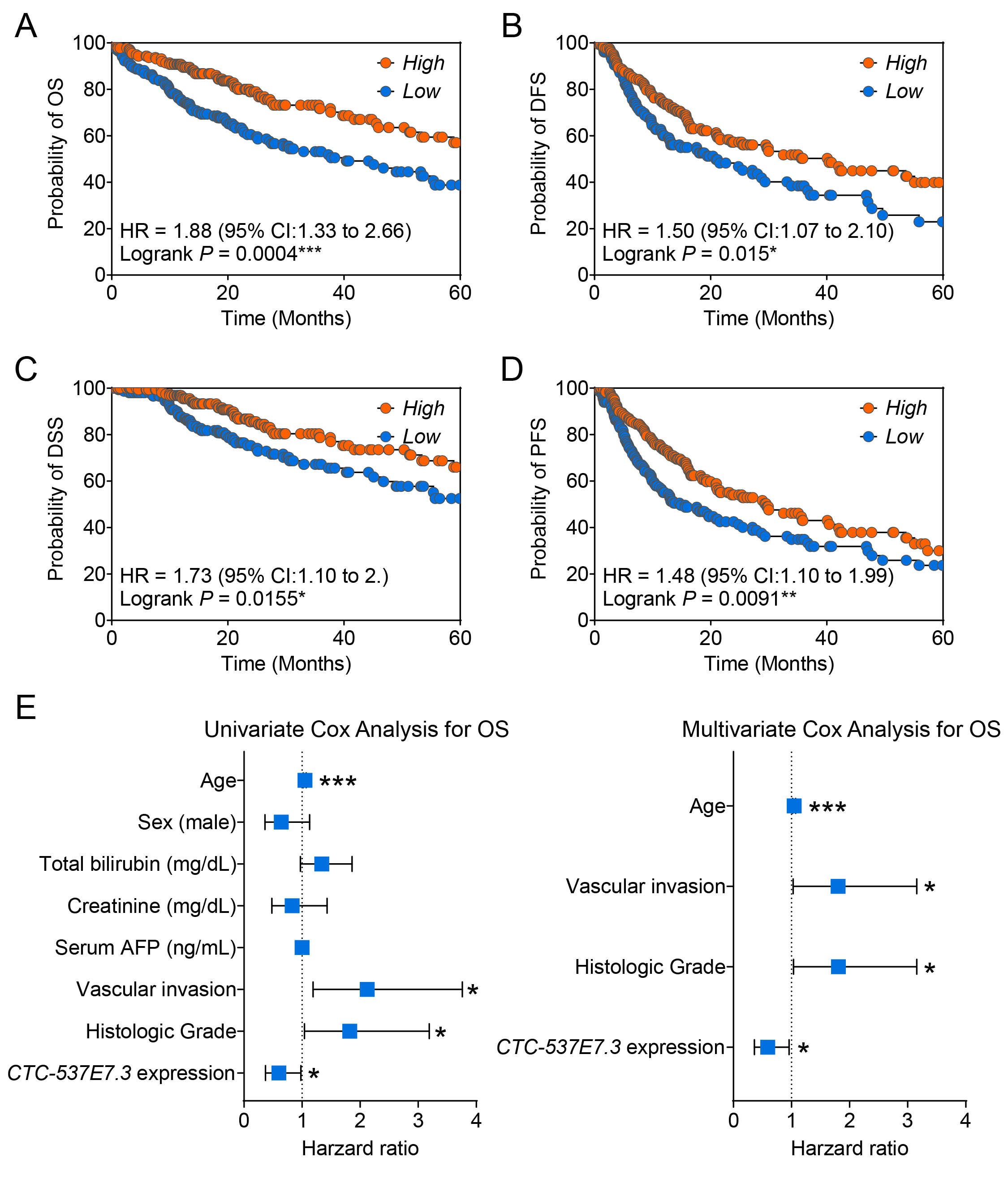
3.2. Prognostic Relevance of CTC-537E7.3 in Patients with HCC
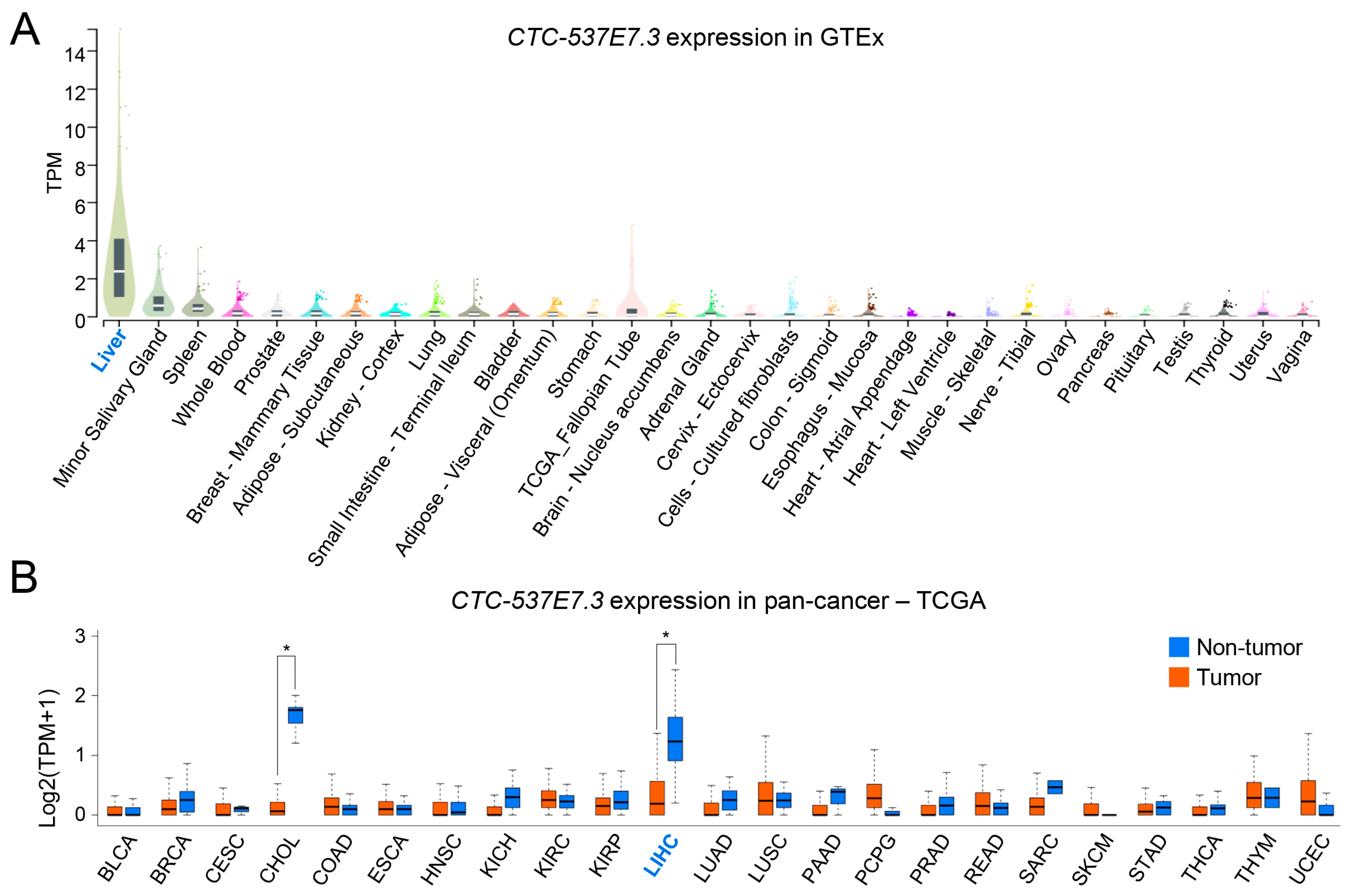
3.3. Tissue Specificity of CTC-537E7.3 in Liver and Cancer Contexts
3.4. Validation of CTC-537E7.3 as a Diagnostic Biomarker in Patients with HCC
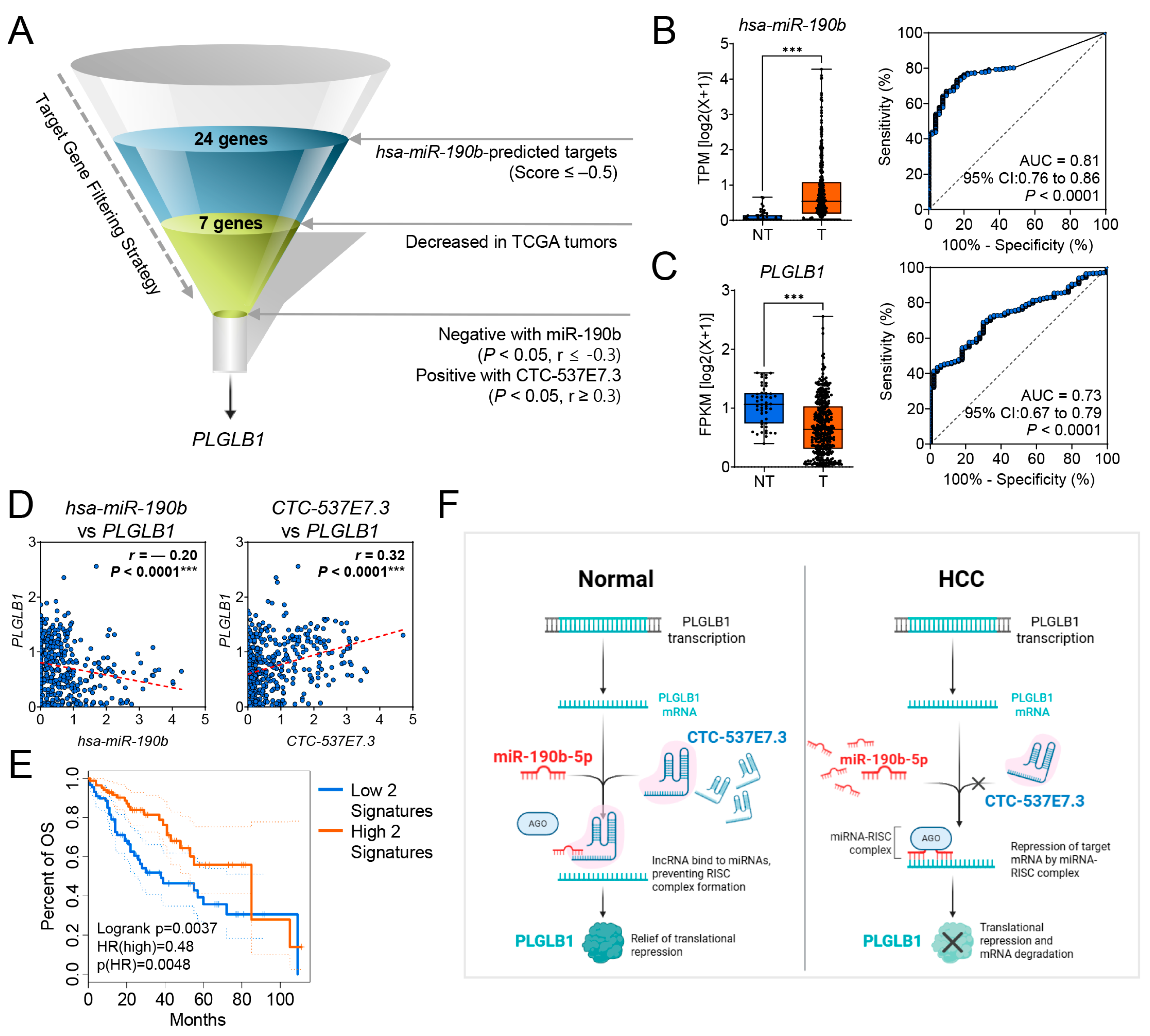
3.5. CTC-537E7.3 Acts as a ceRNA That Relieves miR-190b-5p-Mediated Repression of PLGLB1 in HCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vogel, A.; Meyer, T.; Sapisochin, G.; Salem, R.; Saborowski, A. Hepatocellular carcinoma. Lancet 2022, 400, 1345–1362. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.Q.; Mathurin, P.; Cortez-Pinto, H.; Loomba, R. Global epidemiology of alcohol-associated cirrhosis and HCC: Trends, projections and risk factors. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Samant, H.; Amiri, H.S.; Zibari, G.B. Addressing the worldwide hepatocellular carcinoma: Epidemiology, prevention and management. J. Gastrointest. Oncol. 2021, 12, S361–S373. [Google Scholar] [CrossRef] [PubMed]
- Kobeissy, A.; Merza, N.; Al-Hillan, A.; Boujemaa, S.; Ahmed, Z.; Nawras, M.; Albaaj, M.; Dahiya, D.S.; Alastal, Y.; Hassan, M. Protein Induced by Vitamin K Absence or Antagonist-II Versus Alpha-Fetoprotein in the Diagnosis of Hepatocellular Carcinoma: A Systematic Review with Meta-Analysis. J. Clin. Med. Res. 2023, 15, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M. Urgent Global Need for PIVKA-II and AFP-L3 Measurements for Surveillance and Management of Hepatocellular Carcinoma. Liver Cancer 2024, 13, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Norman, J.S.; Li, P.J.; Kotwani, P.; Shui, A.M.; Yao, F.; Mehta, N. AFP-L3 and DCP strongly predict early hepatocellular carcinoma recurrence after liver transplantation. J. Hepatol. 2023, 79, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Yarmishyn, A.A.; Kurochkin, I.V. Long noncoding RNAs: A potential novel class of cancer biomarkers. Front. Genet. 2015, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Bi, C.; Clark, B.S.; Mady, R.; Shah, P.; Kohtz, J.D. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006, 20, 1470–1484. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, Y.; Chen, S.; Li, W.; Chen, W.; Gu, W. LncRNA MACC1-AS1 sponges multiple miRNAs and RNA-binding protein PTBP1. Oncogenesis 2019, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Mouhou, E.; Genty, F.; El M’selmi, W.; Chouali, H.; Zagury, J.F.; Le Clerc, S.; Proudhon, C.; Noirel, J. High tissue specificity of lncRNAs maximises the prediction of tissue of origin of circulating DNA. Sci. Rep. 2025, 15, 12941. [Google Scholar] [CrossRef] [PubMed]
- Samuels, M.; Jones, W.; Towler, B.; Turner, C.; Robinson, S.; Giamas, G. The role of non-coding RNAs in extracellular vesicles in breast cancer and their diagnostic implications. Oncogene 2023, 42, 3017–3034. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Duan, W.; Jiang, X.; Zhao, L.; Li, J.; Wang, R.; Yan, S.; Xie, Y.; Yan, K.; Wang, Q.; et al. Cell-free lncRNA expression signatures in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. J. Cell Mol. Med. 2018, 22, 2838–2845. [Google Scholar] [CrossRef] [PubMed]
- Lemos, A.E.G.; Matos, A.D.R.; Ferreira, L.B.; Gimba, E.R.P. The long non-coding RNA PCA3: An update of its functions and clinical applications as a biomarker in prostate cancer. Oncotarget 2019, 10, 6589–6603. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Xu, J.; Chen, M.; Li, J.; Li, S.; Ye, J. A meta-analysis of UCA1 accuracy in the detection of bladder cancer. Expert Rev. Anticancer. Ther. 2024, 24, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Baek, G.O.; Ahn, H.R.; Sung, S.; Seo, C.W.; Cho, H.J.; Nam, S.W.; Cheong, J.Y.; Eun, J.W. Serum small extracellular vesicle-derived LINC00853 as a novel diagnostic marker for early hepatocellular carcinoma. Mol. Oncol. 2020, 14, 2646–2659. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Hou, G.; Li, L.; Li, Y.; Zhou, W.; Liu, L. Potential diagnostic and prognostic marker dimethylglycine dehydrogenase (DMGDH) suppresses hepatocellular carcinoma metastasis in vitro and in vivo. Oncotarget 2016, 7, 32607–32616. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Sun, A.; Zhao, Y.; Ying, W.; Sun, H.; Yang, X.; Xing, B.; Sun, W.; Ren, L.; Hu, B.; et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature 2019, 567, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef] [PubMed]
- Mounir, M.; Lucchetta, M.; Silva, T.C.; Olsen, C.; Bontempi, G.; Chen, X.; Noushmehr, H.; Colaprico, A.; Papaleo, E. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput. Biol. 2019, 15, e1006701. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Wu, Y.; Yang, Y.; Yang, Y.T.; Wang, Z.; Yuan, J.; Yang, Y.; Hua, C.; Fan, X.; Niu, G.; et al. Comprehensive analysis of long non-coding RNAs highlights their spatio-temporal expression patterns and evolutional conservation in Sus scrofa. Sci. Rep. 2017, 7, 43166. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Kim, V.N. Small and long non-coding RNAs: Past, present, and future. Cell 2024, 187, 6451–6485. [Google Scholar] [CrossRef] [PubMed]
- Tauber, H.; Huttelmaier, S.; Kohn, M. POLIII-derived non-coding RNAs acting as scaffolds and decoys. J. Mol. Cell Biol. 2019, 11, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lei, Y.; Wu, M.; Li, N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. Int. J. Mol. Sci. 2018, 19, 2958. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhao, W.; Pu, R.; Lv, Z.; Xie, H.; Li, Y.; Zhang, Z. Long non-coding RNAs as diagnostic and prognostic biomarkers for colorectal cancer (Review). Oncol. Lett. 2024, 28, 486. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.S.; Cui, Z.Q.; Gong, L.; Han, H.; Chen, P.C.; Guo, L.M.; Yu, X.; Wei, Y.H.; Ha, S.A.; Kim, J.W.; et al. Simultaneous measurements of serum AFP, GPC-3 and HCCR for diagnosing hepatocellular carcinoma. Hepatogastroenterology 2011, 58, 1718–1724. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Luo, R.; Chen, L.; Wang, H.; Huang, W.; Song, Q.; Jiang, D.; Xu, L.; Su, J.; Xu, C.; et al. CPZ: A Highly Sensitive Diagnostic Biomarker for the Differentiation Between Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma, Particularly in Poorly Differentiated Hepatocellular Carcinoma. Am. J. Surg. Pathol. 2025, 49, 711–729. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Zhang, Y.; Wang, Y.; Xu, F.; Liang, J.; Wang, W. Diagnostic accuracy and prognostic significance of Glypican-3 in hepatocellular carcinoma: A systematic review and meta-analysis. Front. Oncol. 2022, 12, 1012418. [Google Scholar] [CrossRef] [PubMed]
- Song, J.Y.; Li, X.P.; Qin, X.J.; Zhang, J.D.; Zhao, J.Y.; Wang, R. A fourteen-lncRNA risk score system for prognostic prediction of patients with non-small cell lung cancer. Cancer Biomark. 2020, 29, 493–508. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Zhou, L.; Chi, Y.; Wu, H.; Shi, L. LncRNA profile study reveals a seven-lncRNA signature predicts the prognosis of patients with colorectal cancer. Biomark. Res. 2020, 8, 8. [Google Scholar] [CrossRef] [PubMed]
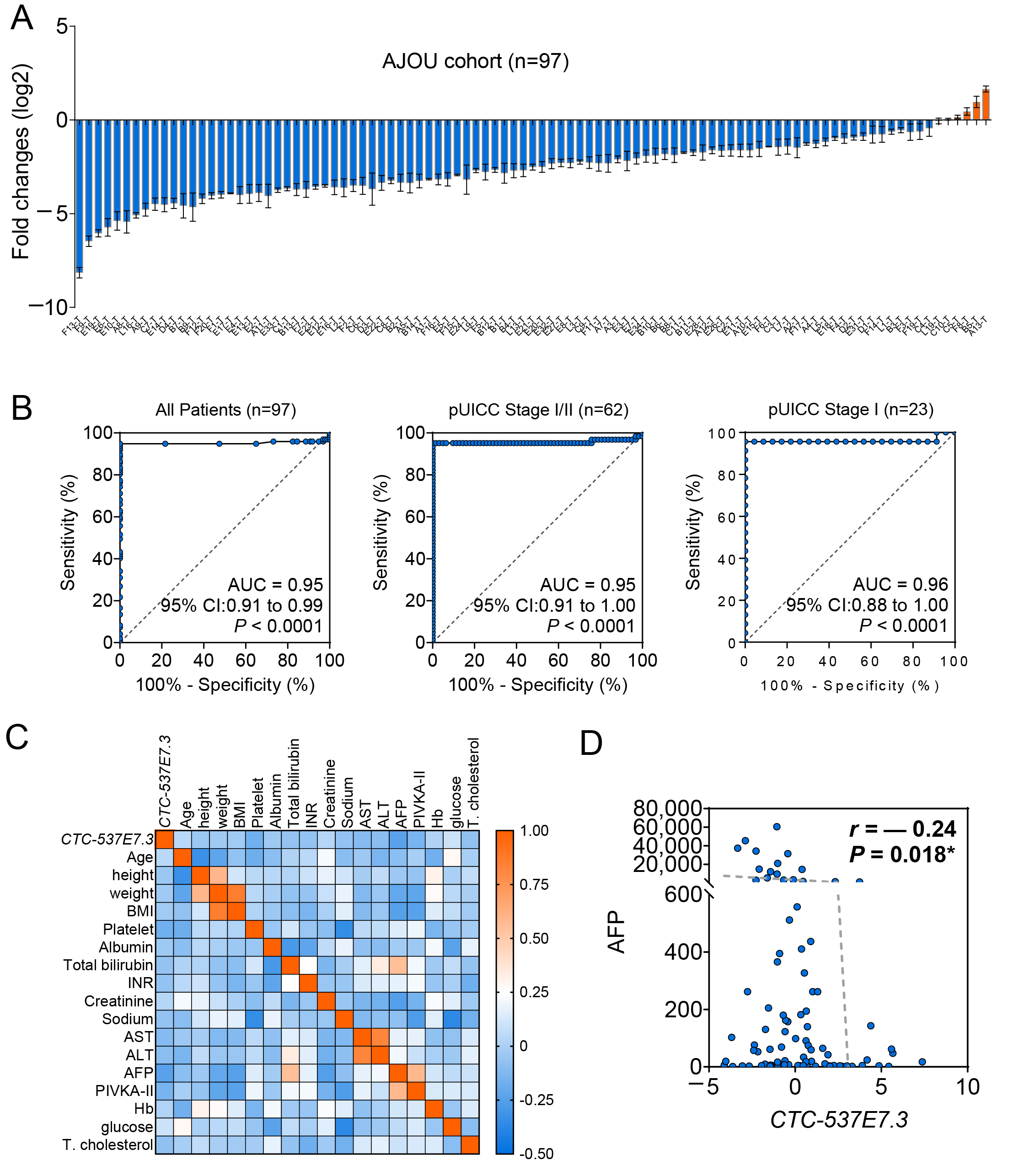
| Variables | HCC (n = 97) |
|---|---|
| Age (years), mean ± SD | 55.7 ± 10.0 |
| Male sex, n (%) | 74 (76.3%) |
| Etiology, n (%) | |
| HBV | 90 (92.8%) |
| HCV | 4 (4.1%) |
| Alcohol | 2 (2.1%) |
| HCV + Alcohol | 1 (1%) |
| Cirrhosis, n (%) | 54 (65.1) |
| Ascites, n (%) | 16 (16.3) |
| BMI | 24.2 (387) |
| Platelet, ×109/L | 179.1 (70.1) |
| Albumin (g/dL), mean ± SD | 4.4 (0.7) |
| Total bilirubin (mg/dL), mean ± SD | 0.8 (0.9) |
| INR, mean ± SD | 1.1 (0.1) |
| Creatinine (mg/dL) | 0.9 (0.3) |
| Sodium, mmol/L | 115.7 (52.3) |
| AST, U/L | 54.3 (105.3) |
| ALT, U/L | 42.5 (61.1) |
| AFP (ng/mL), mean ± SD | 94.9 (144.8) |
| PIVKA-II, mAU/mL | 8925.3 (19,566.8) |
| Hemoglobin, g/dL | 14.4 (1.6) |
| Glucose, mg/dL | 122.3 (53.6) |
| Total Cholesterol, mg/dL | 173.7 (40.4) |
| pUICC stage, n (%) | |
| pI | 24 (24.7) |
| pII | 38 (39.2) |
| pIII | 24 (24.7) |
| pIVA | 7 (7.2) |
| pIVB | 3 (3.1) |
| BCLC stage, n (%) | |
| 0 | 24 (24.7) |
| A | 53 (54.6) |
| B | 8 (8.2) |
| C | 9 (9.3) |
| D | 2 (2.1) |
| Vascular invasion, n (%) | 47 (56) |
| Metastasis, n (%) | 22 (22.7) |
| Recurrence, n (%) | 38 (39.2) |
| OS (months), mean ± SD | 46.0 (24.5) |
| Dataset | Rank | Gene Symbol | Gene ID | p-Value | Fold Change (log2) | Normal Expression | Tumor Expression |
|---|---|---|---|---|---|---|---|
| TCGA_LIHC | 1 | RP11-6B4.1 | ENSG00000243694.2 | 2.86 × 1052 | −1.749 | 2.666 | 0.917 |
| TCGA_LIHC | 2 | CTC-537E7.3 | ENSG00000248884.1 | 2.98 × 1020 | −1.402 | 2.028 | 0.626 |
| TCGA_LIHC | 3 | RP11-789C1.1 | ENSG00000250266.1 | 2.59 × 1011 | −1.148 | 1.403 | 0.254 |
| TCGA_LIHC | 4 | RP11-252E2.2 | ENSG00000261058.1 | 5.17 × 1018 | −0.981 | 1.455 | 0.474 |
| TCGA_LIHC | 5 | RP11-172E9.2 | ENSG00000230058.1 | 4.83 × 108 | −0.881 | 1.074 | 0.193 |
| TCGA_LIHC | 6 | RP11-434D9.1 | ENSG00000249364.4 | 1.87 × 1010 | −0.869 | 1.613 | 0.744 |
| TCGA_LIHC | 7 | RP11-119D9.1 | ENSG00000251637.5 | 3.45 × 1016 | −0.777 | 1.693 | 0.916 |
| TCGA_LIHC | 8 | RP11-513G11.3 | ENSG00000238097.1 | 5.94 × 1015 | −0.741 | 1.733 | 0.992 |
| TCGA_LIHC | 9 | FLJ22763 | ENSG00000241224.5 | 3.59 × 1017 | −0.681 | 1.192 | 0.511 |
| TCGA_LIHC | 10 | RP11-327J17.2 | ENSG00000259359.1 | 1.16 × 1011 | −0.639 | 1.35 | 0.71 |
| GSE77314 | 1 | LINC01093 | ENSG00000249173.5 | 3.08 × 1045 | −4.697 | 5.232 | 0.535 |
| GSE77314 | 2 | RP11-6B4.1 | ENSG00000243694.2 | 3.14 × 1023 | −3.023 | 3.871 | 0.848 |
| GSE77314 | 3 | RP11-434D9.1 | ENSG00000249364.5 | 3.74 × 1022 | −2.594 | 3.594 | 1 |
| GSE77314 | 4 | FLJ22763 | ENSG00000241224.6 | 1.87 × 1022 | −2.379 | 3.061 | 0.682 |
| GSE77314 | 5 | AC016768.1 | ENSG00000232451.1 | 2.16 × 1022 | −1.586 | 2.355 | 0.769 |
| GSE77314 | 6 | RP4-568C11.4 | ENSG00000274173.1 | 2.46 × 1017 | −1.516 | 2.109 | 0.593 |
| GSE77314 | 7 | RP11-439C15.4 | ENSG00000253764.1 | 1.02 × 1022 | −1.49 | 1.679 | 0.19 |
| GSE77314 | 8 | RP11-153K11.3 | ENSG00000233590.1 | 1.99 × 1012 | −1.467 | 1.979 | 0.512 |
| GSE77314 | 9 | CTC-537E7.3 | ENSG00000248884.1 | 5.63 × 1018 | −1.456 | 1.762 | 0.306 |
| GSE77314 | 10 | RP11-789C1.2 | ENSG00000251061.2 | 1.60 × 1014 | −1.439 | 1.506 | 0.066 |
| GSE124535 | 1 | RP11-789C1.1 | ENSG00000250266 | 4.30 × 109 | −1.232 | 1.555 | 0.322 |
| GSE124535 | 2 | CTC-537E7.3 | ENSG00000248884 | 1.05 × 1011 | −1.228 | 1.745 | 0.517 |
| GSE124535 | 3 | RP11-290F20.3 | ENSG00000224397 | 1.09 × 109 | −1.096 | 2.034 | 0.939 |
| GSE124535 | 4 | CTC-297N7.9 | ENSG00000264016 | 2.19 × 1010 | −0.986 | 1.947 | 0.961 |
| GSE124535 | 5 | RP11-327J17.2 | ENSG00000259359 | 9.98 × 109 | −0.978 | 1.693 | 0.715 |
| GSE124535 | 6 | CTB-167B5.2 | ENSG00000264868 | 5.36 × 1010 | −0.873 | 1.601 | 0.728 |
| GSE124535 | 7 | RP11-252E2.2 | ENSG00000261058 | 1.71 × 107 | −0.822 | 1.344 | 0.522 |
| GSE124535 | 8 | RP11-557H15.4 | ENSG00000232310 | 9.63 × 107 | −0.774 | 1.413 | 0.639 |
| GSE124535 | 9 | RP11-863P13.2 | ENSG00000261816 | 3.03 × 105 | −0.722 | 1.068 | 0.347 |
| GSE124535 | 10 | RP11-88H9.2 | ENSG00000231437 | 4.12 × 107 | −0.636 | 1.135 | 0.498 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.S.; Jang, S.H.; Baek, G.O.; Yoon, M.G.; Shim, J.; Han, J.E.; Kim, S.S.; Cheong, J.Y.; Eun, J.W. CTC-537E7.3 as a Liver-Specific Biomarker for Hepatocellular Carcinoma: Diagnostic and Prognostic Implications. Curr. Issues Mol. Biol. 2025, 47, 563. https://doi.org/10.3390/cimb47070563
Kim HS, Jang SH, Baek GO, Yoon MG, Shim J, Han JE, Kim SS, Cheong JY, Eun JW. CTC-537E7.3 as a Liver-Specific Biomarker for Hepatocellular Carcinoma: Diagnostic and Prognostic Implications. Current Issues in Molecular Biology. 2025; 47(7):563. https://doi.org/10.3390/cimb47070563
Chicago/Turabian StyleKim, Hyung Seok, Se Ha Jang, Geum Ok Baek, Moon Gyeong Yoon, Jaewon Shim, Ji Eun Han, Soon Sun Kim, Jae Youn Cheong, and Jung Woo Eun. 2025. "CTC-537E7.3 as a Liver-Specific Biomarker for Hepatocellular Carcinoma: Diagnostic and Prognostic Implications" Current Issues in Molecular Biology 47, no. 7: 563. https://doi.org/10.3390/cimb47070563
APA StyleKim, H. S., Jang, S. H., Baek, G. O., Yoon, M. G., Shim, J., Han, J. E., Kim, S. S., Cheong, J. Y., & Eun, J. W. (2025). CTC-537E7.3 as a Liver-Specific Biomarker for Hepatocellular Carcinoma: Diagnostic and Prognostic Implications. Current Issues in Molecular Biology, 47(7), 563. https://doi.org/10.3390/cimb47070563







