The Nutritional Gene Expression Regulation Potential of a Lysolecithin-Based Product
Abstract
1. Introduction
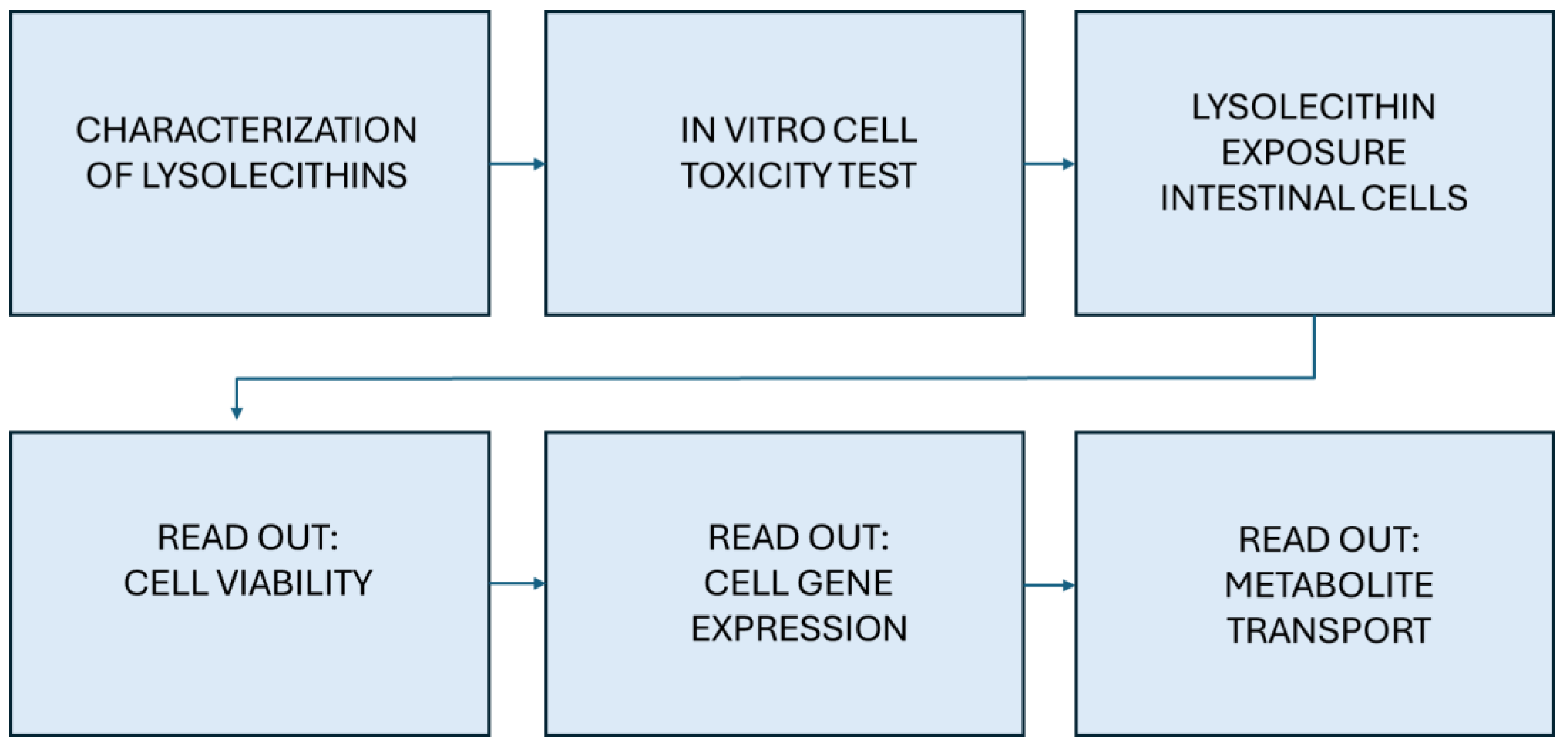
2. Materials and Methods
2.1. Lysolecithin Characterization
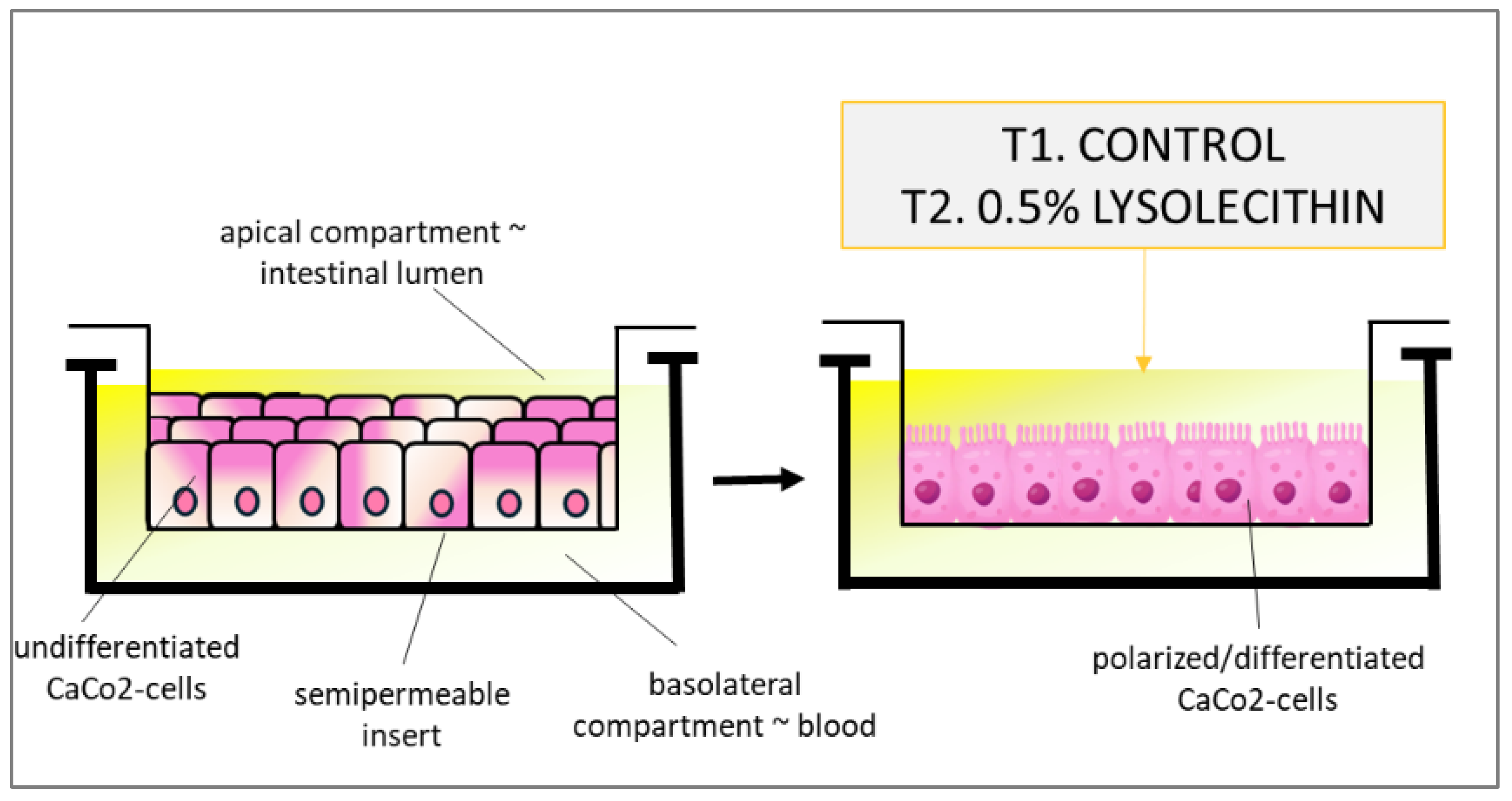
2.2. Experimental Design
2.3. Toxicity Screening
Evaluation of the Effects of Lysolecithins on Intestinal Cell Viability
2.4. Studying the Effects of Lysolecithins on Intestinal Gene Expression and Metabolite Transport/Production
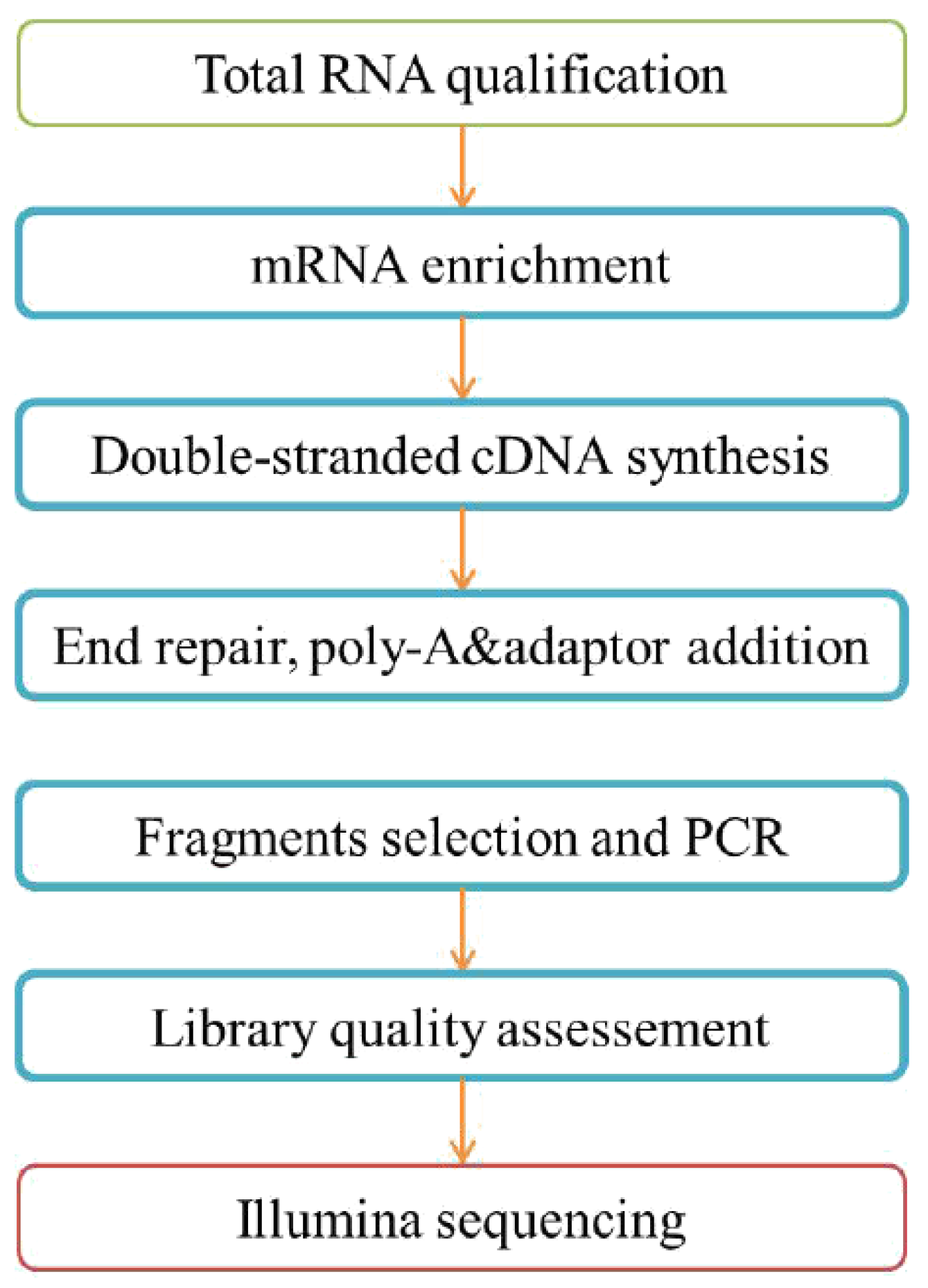
2.5. RNA Sequencing
2.5.1. Total RNA Extraction
2.5.2. Library Preparation and Sequencing
2.5.3. Total RNA Sample QC
- (1)
- NanoDrop (ThermoFisher Scientific, Waltham, MA, USA): tests RNA purity (OD260/OD280);
- (2)
- Agarose gel electrophoresis: tests RNA degradation and potential contamination;
- (3)
- Agilent 2100 (Agilent Technologies, Digem, Belgium): checks RNA integrity.
2.5.4. Data Quality Control
2.5.5. Readsmapping to the Reference Genome
2.5.6. Quantification of Gene Expression Level
2.5.7. Differential Expression Analysis
2.5.8. Enrichment Analysis of Differentially Expressed Genes
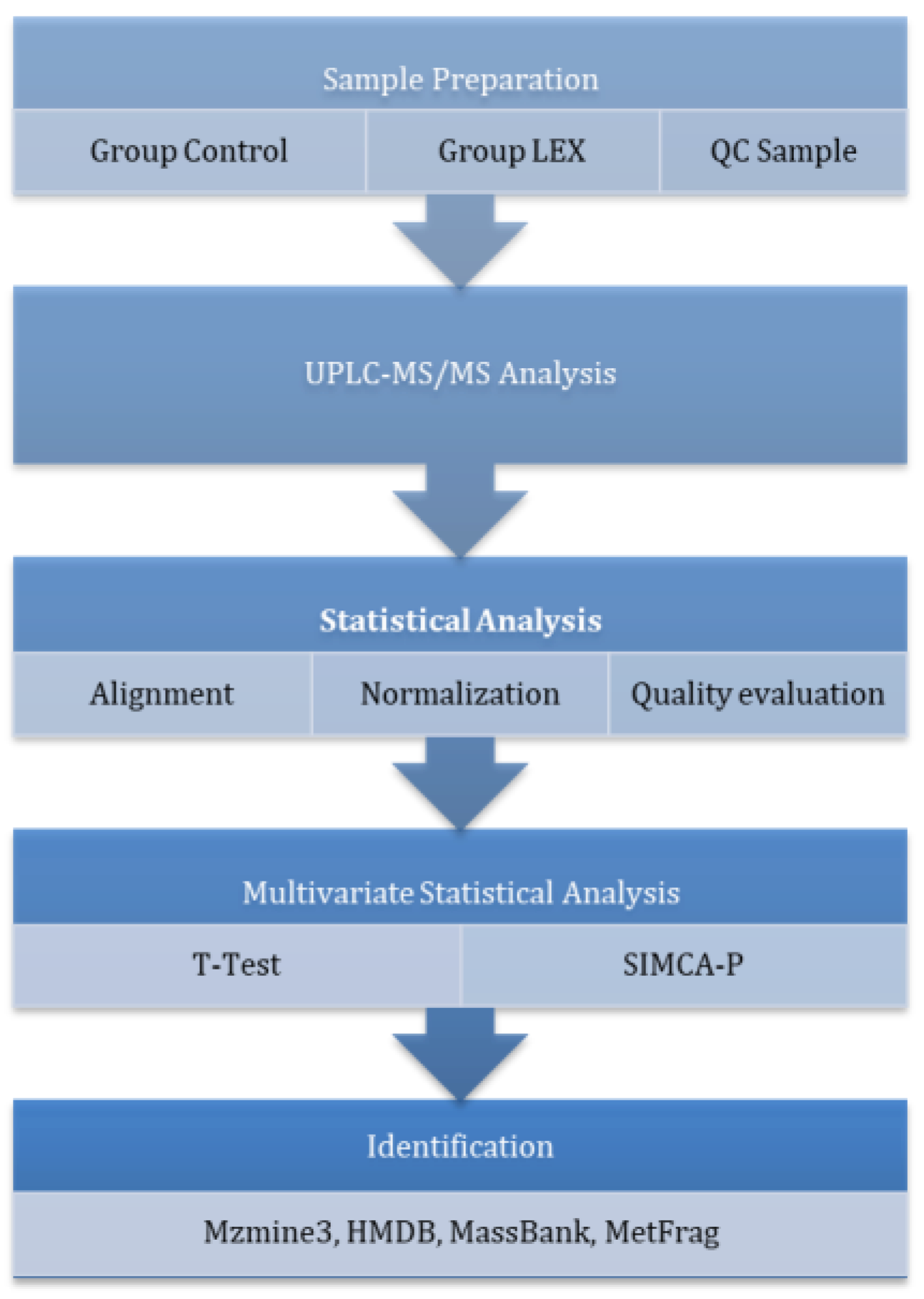
2.6. Untargeted Metabolomics
2.6.1. Sample Preparation
2.6.2. Instrumental Analysis (UPLC-MS)
2.6.3. Data Processing and Quality Control
2.6.4. Statistical and Multivariate Analysis
2.6.5. Metabolite Identification and Annotation
3. Results
3.1. Lysolecithin Characterization
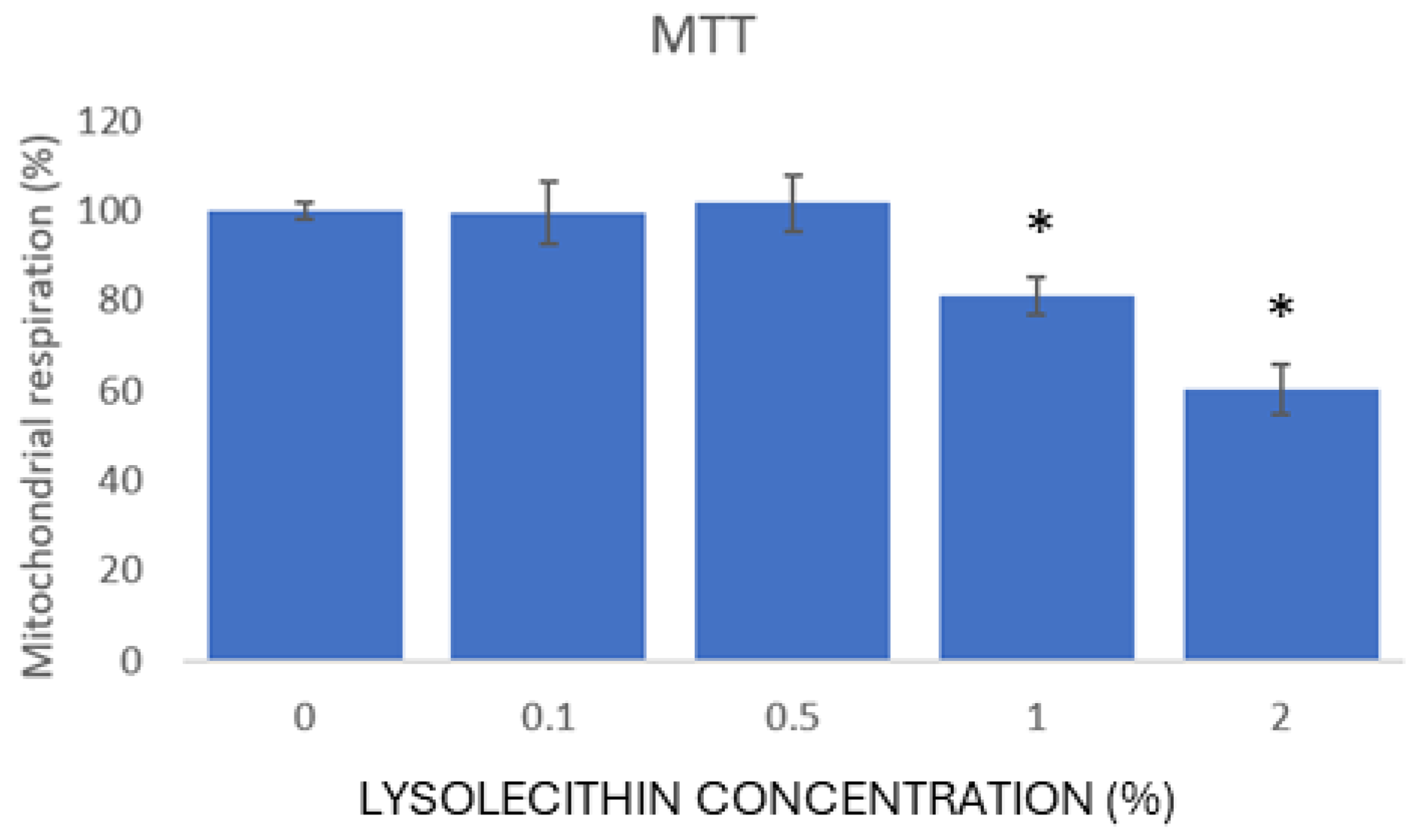
3.2. The Toxicity Screening of Lysolecithins for Intestinal Cells
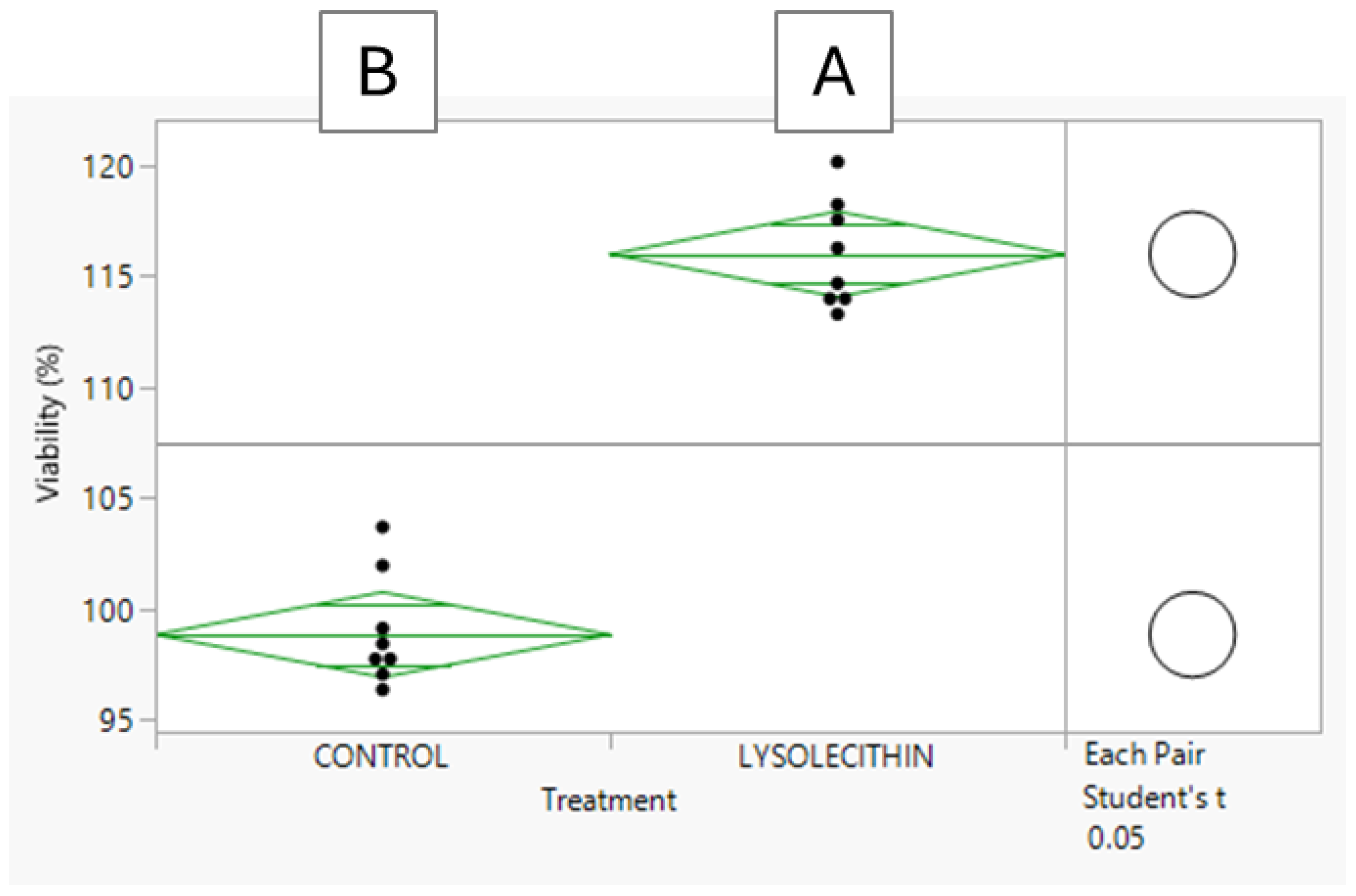
3.3. The Effect of Lysolecithins on Intestinal Cell Viability
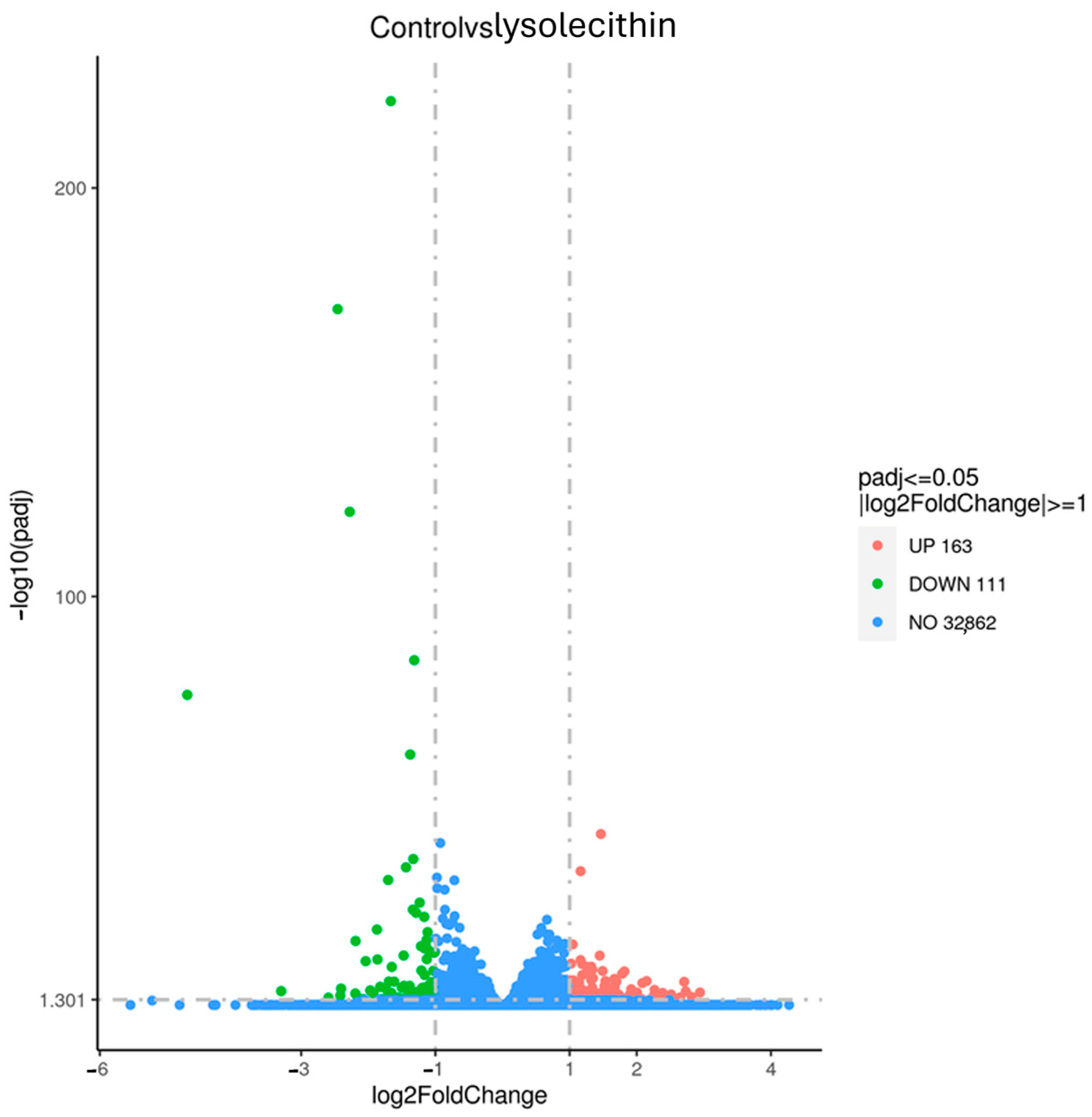
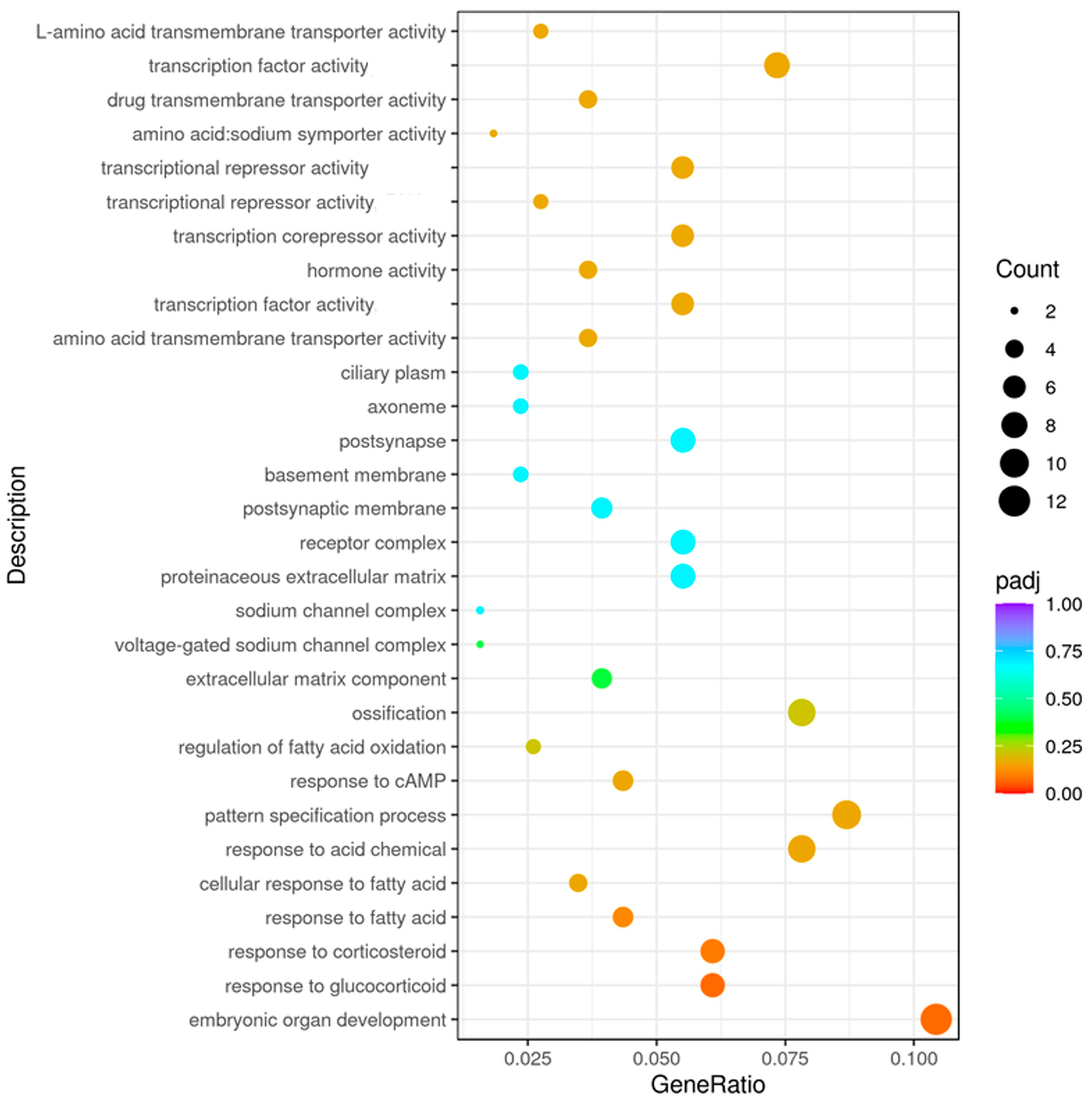
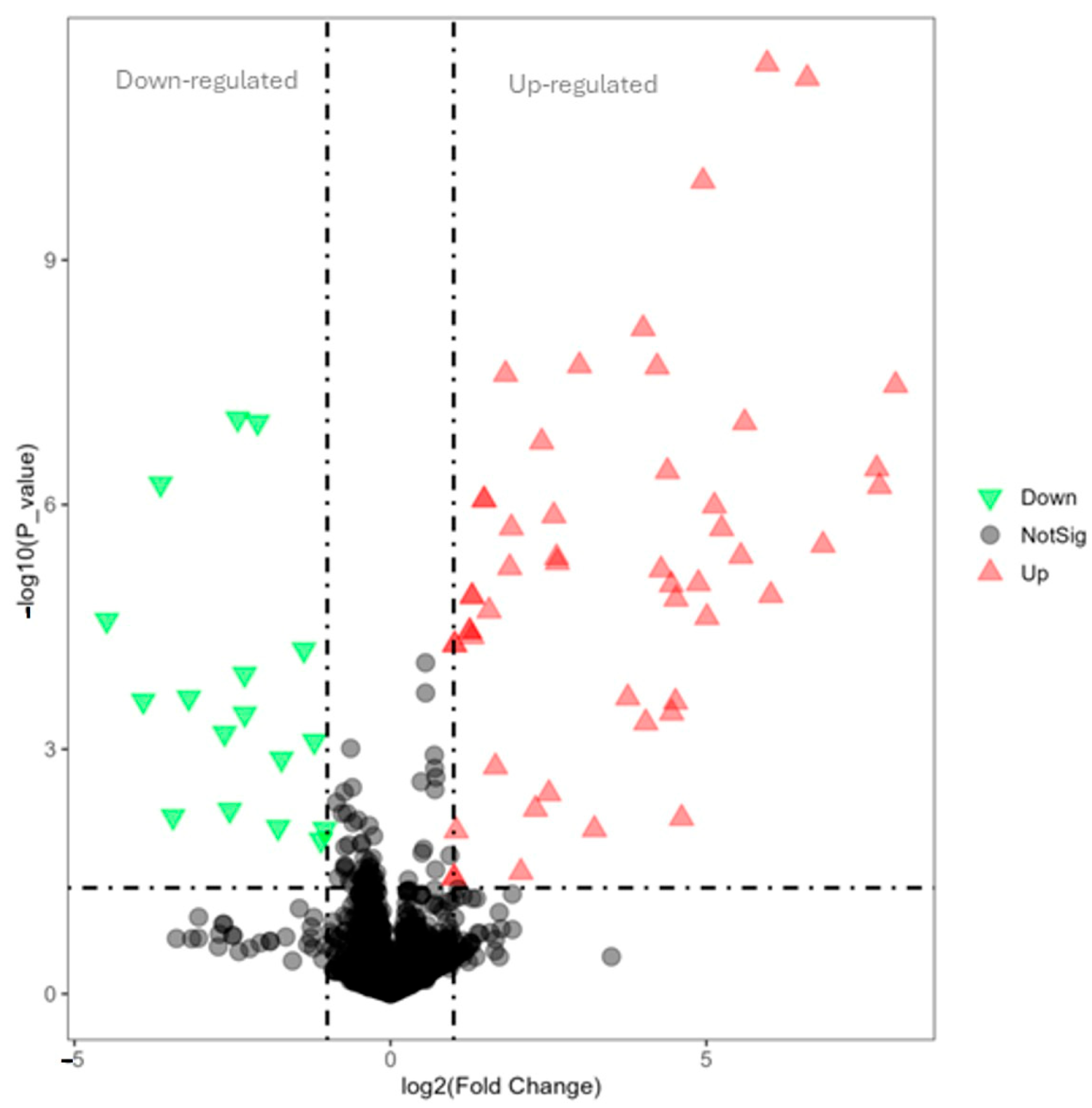
3.4. The Effects of Lysolecithins on Intestinal Gene Expression
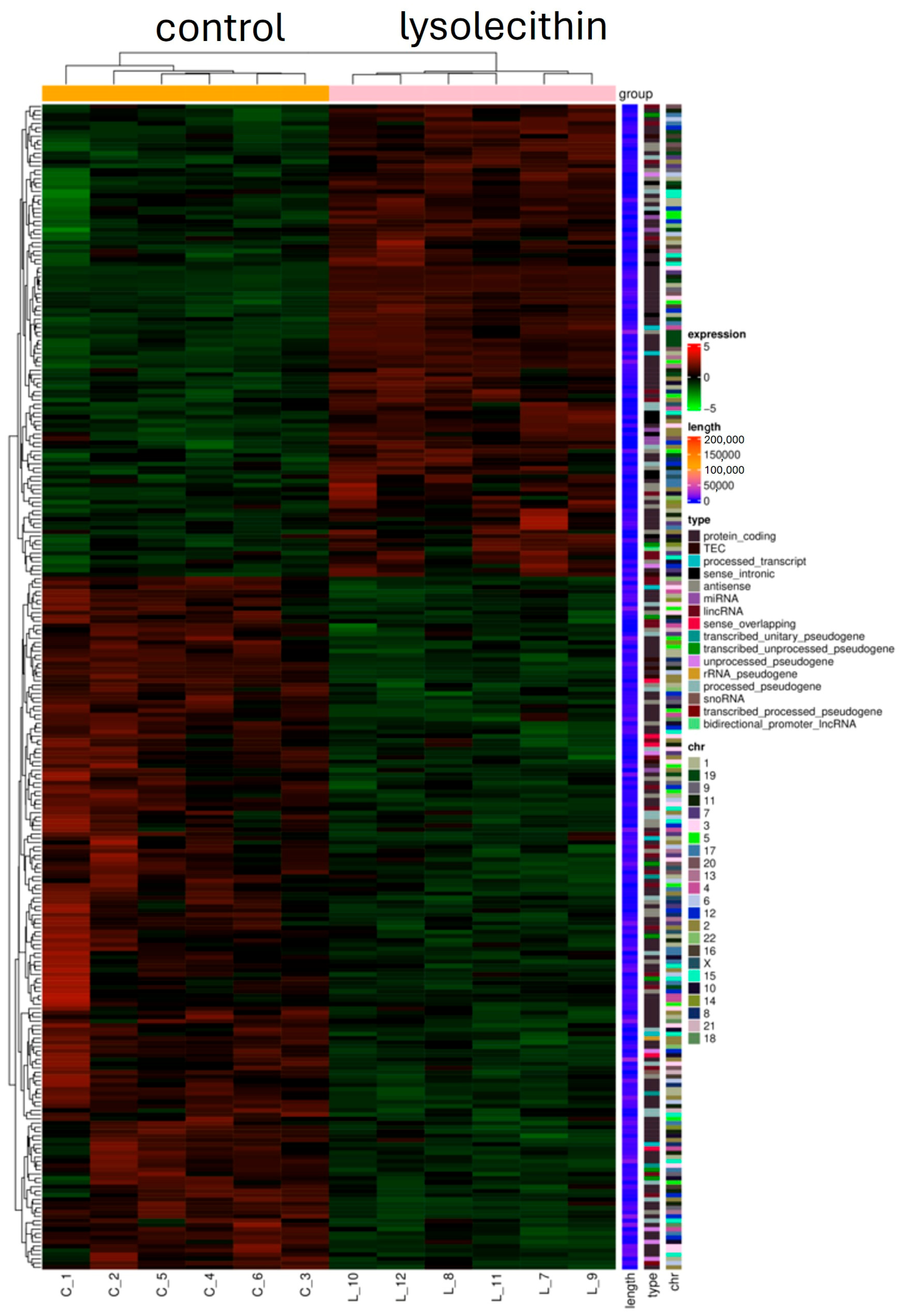
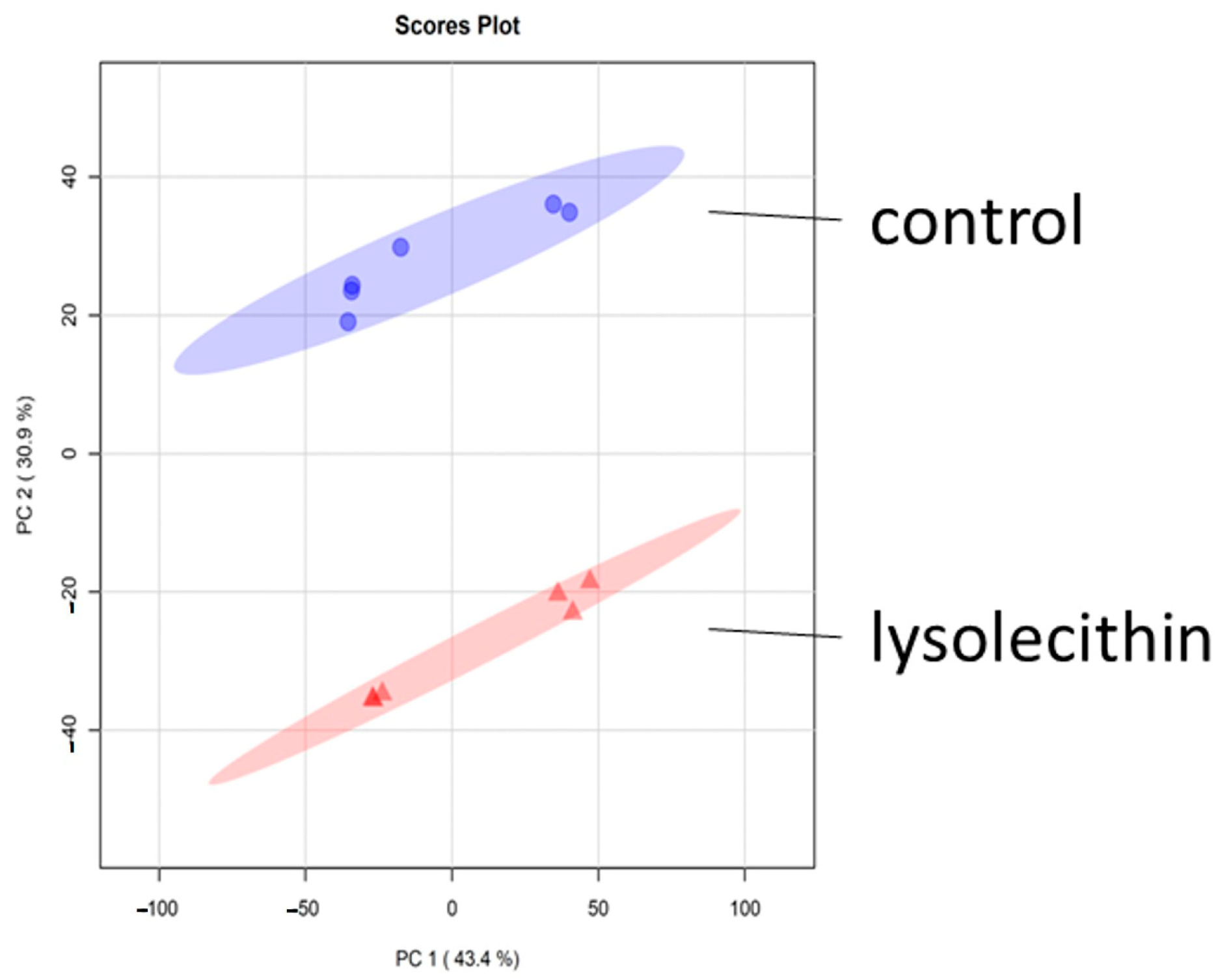
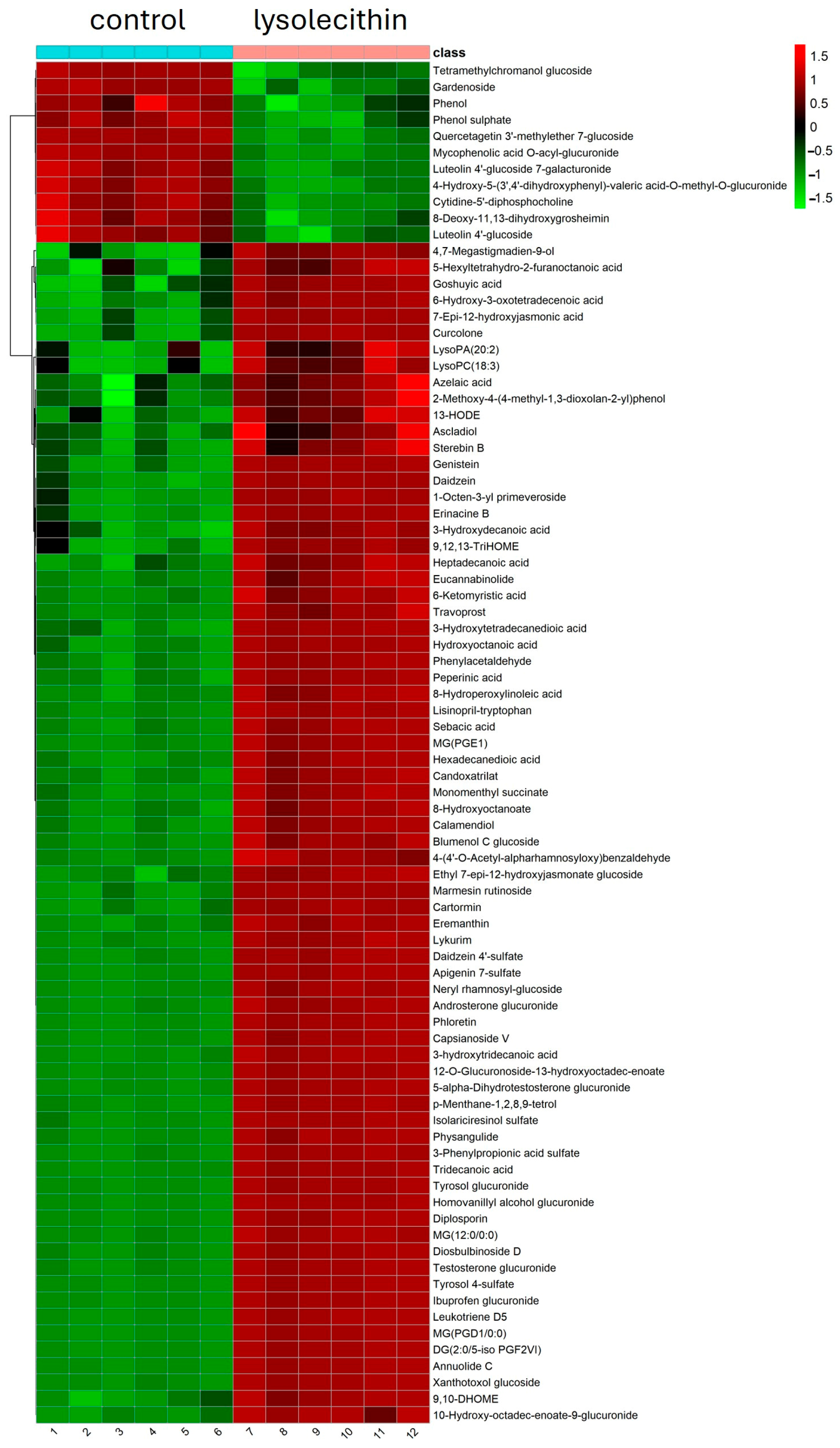
3.5. Cluster Analysis
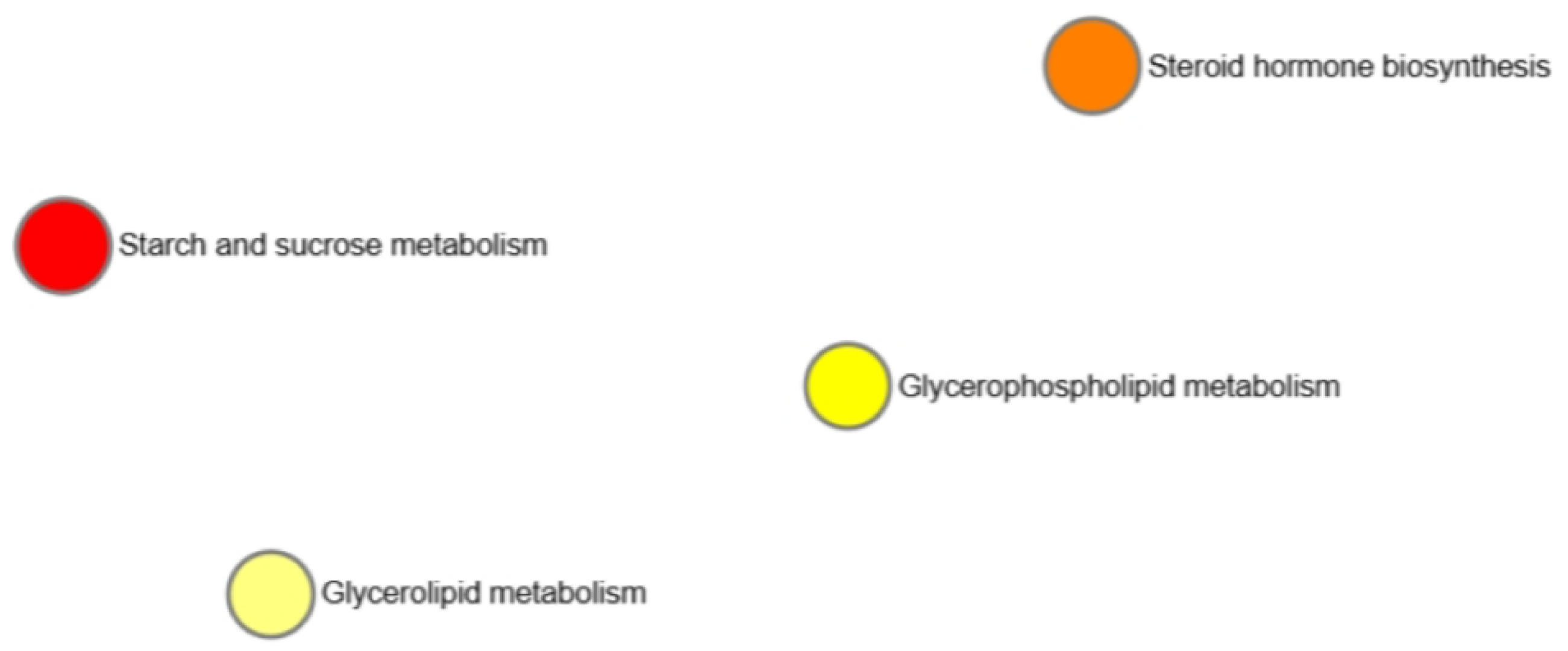
3.6. Correlation Network of Metabolites
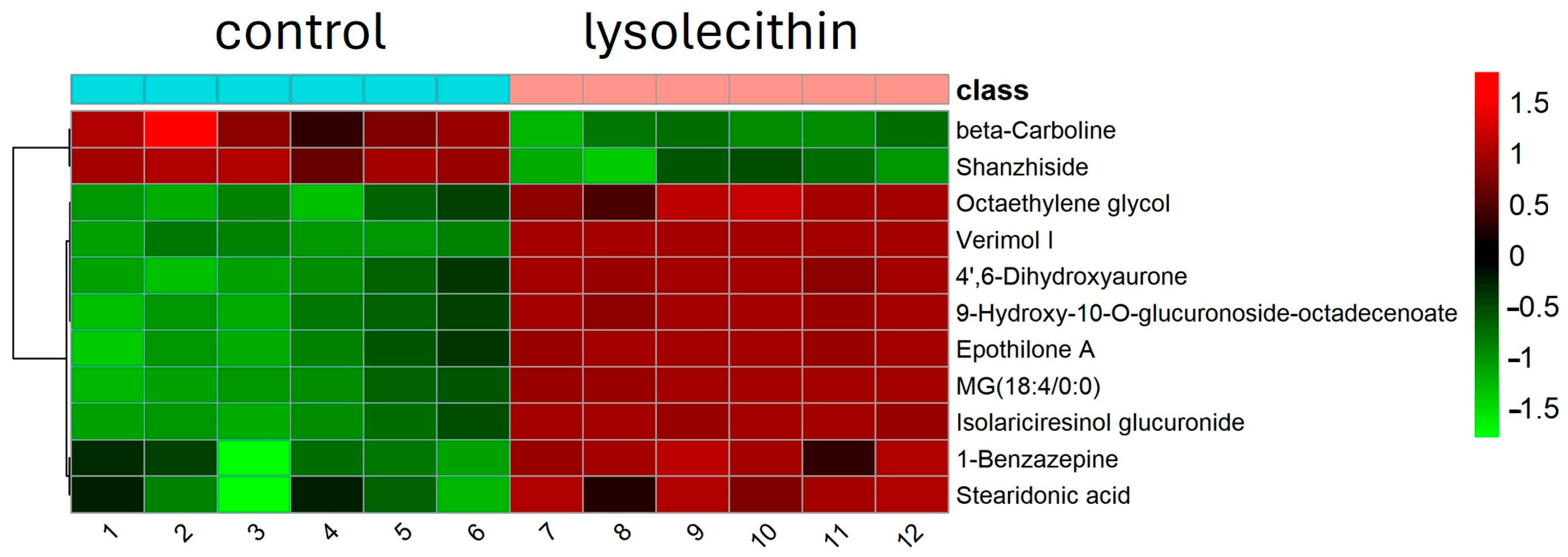
3.7. Cluster Analysis
3.8. Correlation Network of Metabolites
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LPL | Lysophospholipids |
| FA | Fatty acid |
| Caco-2 | Colorectal adenocarcinoma-2 cells |
| LPC | Lysophosphatidylcholine |
| LPE | Lysophosphatidylethanolamine |
| LPI | Lysophosphatidylinositol |
| LPA | Lysophosphatidic acid |
| TEER | Transepithelial electrical resistance |
| RNA-Seq | Ribonucleic acid sequencing |
| NCBI | National Center for Biotechnology Information |
| GEO | Gene Expression Omnibus |
| cAMP | Cyclic adenosine monophosphate |
| GO | Gene Ontology |
| RSD | Relative standard deviation |
| LC-MS | Liquid chromatography–mass spectrometry |
| PCA | Principal component analysis |
| FC | Fold-change |
| HCA | Hierarchical clustering analysis |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| AA | Amino acid |
| Thr | Threonine |
| Phe | Phenylalanine |
| Ile | Isoleucine |
| His | Histidine |
| Arg | Arginine |
| Tyr | Tyrosine |
| Glu | Glutamic acid |
| Pro | Proline |
| Gly | Glycine |
| Ala | Alanine |
| GC | Gas chromatography |
| HPLC | High-performance liquid chromatography |
| DMEM | Dulbecco’s minimum essential medium |
| FBS | Fetal bovine serum |
| PBS | Phosphate-buffered saline |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| DMSO | Dimethyl sulfoxide |
| NAD(P)H | Nicotinamide adenine dinucleotide phosphate |
| dNTP | Deoxynucleotide triphosphate |
| PCR | Polymerase chain reaction |
| FKPM | Fragments per kilobase of transcript sequence per million base pairs sequenced |
| DEG | Differentially expressed gene |
| UPLC | Ultra-performance liquid chromatography |
| MS | Mass spectrometry |
| ESI-MS | Electrospray ionization–mass spectrometry |
| PLS-DA | Partial least squares discriminant analysis |
| OPLS-DA | Orthogonal partial least squares discriminant analysis |
References
- Haetinger, V.S.; Dalmoro, Y.K.; Godoy, G.L.; Lang, M.B.; de Souza, O.F.; Aristimunha, P.; Stefanello, C. Optimizing cost, growth performance, and nutrient absorption with a bio-emulsifier based on lysophospholipids for broiler chickens. Poult. Sci. 2021, 4, 101025. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, G.A.; Wealleans, A.L.; Delis, G.A.; Janssens, G.P.J.; di Benedetto, M.; Fortomaris, P. Effects of Dietary Lysolecithin Supplementation during Late Gestation and Lactation on Sow Reproductive Performance, Sow Blood Metabolic Parameters and Piglet Performance. Animals 2022, 12, 623. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zeng, J.; Wang, M.; Liu, H.; Zhou, X.; Zhang, S.; He, J. Effects of lysolecithins on performance, egg quality, blood profiles and liver histopathology in late-phase laying hens. Br. Poult. Sci. 2023, 64, 718–725. [Google Scholar] [CrossRef]
- Kinh, L.V.; Vasanthakumari, B.L.; Sugumar, C.; Thanh, H.L.T.; Thanh, N.V.; Wealleans, A.L.; Ngoan, L.D.; Loan, N.V.T.H. Effect of a Combination of Lysolecithin, Synthetic Emulsifier and Monoglycerides on the Apparent Ileal Digestibility, Metabolizable Energy and Growth Performance of Growing Pigs. Animals 2023, 13, 88. [Google Scholar] [CrossRef]
- Ravindran, V.; Tancharoenrat, P.; Zaefarian, F.; Ravindran, G. Fats in poultry nutrition: Digestive physiology and factors influencing their utilization. Anim. Feed Sci. Technol. 2016, 213, 1–21. [Google Scholar] [CrossRef]
- Jansen, M. Modes of Action of Lysophospholipids as Feed Additives on Fat Digestion in Broilers. Ph.D. Thesis, Catholic University of Leuven, Leuven, Belgium, 2015. [Google Scholar]
- Jansen, M.; Nuyens, F.; Buyse, J.; Leleu, S.; Van Campenhout, L. Interaction between fat type and lysolecithin supplementation in broiler feeds. Poult. Sci. 2015, 94, 10.3382. [Google Scholar] [CrossRef] [PubMed]
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef]
- Tan, S.T.; Ramesh, T.; Toh, X.R.; Nguyen, L.N. Emerging roles of lysophospholipids in health and disease. Prog. Lipid Res. 2020, 80, 101068. [Google Scholar] [CrossRef]
- Li, X.; Abdel-Moneim, A.-M.E.; Mesalam, N.M.; Yang, B. Effects of Lysophosphatidylcholine on Jejuna Morphology and Its Potential Mechanism. Front. Vet. Sci. 2022, 9, 911496. [Google Scholar] [CrossRef]
- Mead, M.N. Nutrigenomics: The genome-food interface. Env. Health Perspect. 2007, 115, A582–A589. [Google Scholar] [CrossRef]
- Park, J.H.; Nguyen, D.H.; Kim, I.H. Effects of exogenous lysolecithin emulsifier supplementation on the growth performance, nutrient digestibility, and blood lipid profiles of broiler chickens. J. Poult. Sci. 2018, 55, 190–194. [Google Scholar] [CrossRef]
- Wealleans, A.L.; Buyse, J.; Scholey, D.; Van Campenhout, L.; Burton, E.; Pritchard, S.; Di Benedetto, M.; Nuyens, F.; Jansen, M. Lysolecithin but not lecithin improves nutrient digestibility and growth rates in young broilers. Br. Poult. Sci. 2020, 61, 414–423. [Google Scholar] [CrossRef]
- Zhao, P.Y.; Kim, I.H. Effect of diets with different energy and lysophospholipids levels on performance, nutrient metabolism, and body composition in broilers. Poult. Sci. 2017, 96, 1341–1347. [Google Scholar] [CrossRef] [PubMed]
- Geraldo, L.H.; Spohr, T.C.; Amaral, R.F.; Fonseca, A.C.; Garcia, C.; Mendes, F.D.; Freitas, C.; dosSantos, M.F.; Lima, F.R. Role of lysophosphatidic acid and its receptors in health and disease: Novel therapeutic strategies. Signal Transduct. Target. Ther. 2021, 6, 45. [Google Scholar] [CrossRef]
- Liu, S.; Paknejad, N.; Zhu, L.; Kihara, Y.; Ray, M.; Chun, J.; Liu, W.; Hite, R.K.; Huang, X.Y. Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate. Nat. Commun. 2022, 13, 731. [Google Scholar] [CrossRef] [PubMed]
- Yaginuma, S.; Omi, J.; Kano, K.; Aoki, J. Lysophospholipids and their producing enzymes: Their pathological roles and potential as pathological biomarkers. Pharmacol. Ther. 2023, 246, 108415. [Google Scholar] [CrossRef]
- Khandelwal, N.; Shaikh, M.; Mhetre, A.; Singh, S.; Sajeevan, T.; Joshi, A.; Narayanaswamy Balaji, K.; Chakrapani, H.; Kamat, S.S. Fatty acid chain length drives lysophosphatidylserine-dependent immunological outputs. Cell Chem. Biol. 2021, 28, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Valentine, W.J.; Fujiwara, Y.; Tsukahara, R.; Tigyi, G. Lysophospholipid signaling: Beyond the EDGs. Biochim. Biophys. Acta Gen. Subj. 2008, 1780, 597–605. [Google Scholar] [CrossRef]
- Lin, M.-E.; Herr, D.R.; Chun, J. Lysophosphatidic acid (LPA) receptors: Signaling properties and disease relevance. Prostaglandins Other Lipid Mediat. 2010, 91, 130–138. [Google Scholar] [CrossRef]
- Brkić, L.; Riederer, M.; Graier, W.F.; Malli, R.; Frank, S. Acyl chaindependent effect of lysophosphatidylcholine on cyclooxygenase (COX)-2 expression in endothelial cells. Atherosclerosis 2012, 224, 348–354. [Google Scholar] [CrossRef]
- Rao, S.P.; Riederer, M.; Lechleitner, M.; Hermansson, M.; Desoye, G.; Hallström, S.; Frank, S. Acyl chain-dependent effect of lysophosphatidylcholine on endothelium-dependent vasorelaxation. PLoS ONE 2013, 8, e65155. [Google Scholar] [CrossRef]
- Brautigan, D.L.; Li, R.; Kubicka, E.; Turner, S.D.; Garcia, J.S.; Weintraut, M.L. Lysolecithin as feed additive enhances collagen expression and villus length in the jejuna of broiler chickens. Poult. Sci. 2017, 968, 2889–2898. [Google Scholar] [CrossRef]
- Drzazga, A.; Sowinska, A.; Krzeminska, A.; Rytczak, P.; Koziolkiewicz, M.; Gendaszewska-Darmach, E. Lysophosphatidylcholine elicits intracellular calcium signaling in a GPR55-dependent manner. Biochem. Biophys. Res. Commun. 2017, 489, 242–247. [Google Scholar] [CrossRef]
- Nguyen, H.; Mee, K.; Sok, D. 2-Polyunsaturated Acyl Lysophosphatidylethanolamine Attenuates Inflammatory Response in Zymosan A-Induced Peritonitis in Mice. Lipids 2011, 46, 893–906. [Google Scholar]
- NF EN ISO 12966-2; Animal and Vegetable Fats and Oils—Gas Chromatography of Fatty Acid Methyl Esters—Part 2: Preparation of Methyl Esters of Fatty Acids. ISO: Geneva, Switzerland, 2017.
- NF EN ISO 12966-4; Animal and Vegetable Fats and Oils—Gas Chromatography of Fatty Acid Methyl Esters—Part 4: Determination by Capillary Gas Chromatography. ISO: Geneva, Switzerland, 2015.
- Becart, I.; Chevalier, C.; Biesse, J.P. Quantitative analysis of phospholipids by HPLC with a light scattering evaporating detector—Application to raw materials for cosmetic use. J. High Resolut. Chromatogr. 1990, 13, 126–129. [Google Scholar] [CrossRef]
- Weerheim, A.M.; Kolb, A.M.; Sturk, A.; Nieuwland, R. Phospholipid composition of cell-derived microparticles determined by one-dimensional high-performance thin-layer chromatography. Anal. Biochem. 2002, 302, 191–198. [Google Scholar] [CrossRef]
- Sehl, A.; Couëdelo, L.; Fonseca, L.; Vaysse, C.; Cansell, M. A critical assessment of transmethylation procedures for n-3 long-chain polyunsaturated fatty acid quantification of lipid classes. Food Chem. 2018, 251, 1–8. [Google Scholar] [CrossRef]
- Natoli, M. Cell Growing Density Affects the Structural and Functional Properties of Caco-2 Differentiated Monolayer. J. Cell. Physiol. 2011, 226, 1531–1543. [Google Scholar] [CrossRef] [PubMed]
- Artursson, P.; Palm, K.; Luthman, K. Caco-2 monolayers in experimental and theoretical predictions of drug transport. Adv. Drug Deliv. Rev. 1996, 22, 67–84. [Google Scholar] [CrossRef]
- Sambruy, Y.; Ferruzza, S.; Ranaldi, G.; De An, I. Intestinal Cell Culture Models Applications in Toxicology and Pharmacology. Cell Biol. Toxicol. 2001, 17, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Finotti, E.; Gezzi, R.; Nobili, F.; Garaguso, I.; Friedman, M. Effect of apple, baobab, red-chicory and pear extracts on cellular energy expenditure and morphology of a Caco-2 cells using transepithelial electrical resistance (TEER) and scanning electron microscopy (SEM). RSC Adv. 2015, 5, 22490–22498. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomicfeatures. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Preced. 2010, 11, R106. [Google Scholar]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Method Gene ontology 6 analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Cao, X.; van Putten, J.P.; Wösten, M.M. Biological functions of bacterial lysophospholipids. Adv. Microb. Physiol. 2023, 82, 129–154. [Google Scholar] [PubMed]
- Anliker, B.; Chun, J. Lysophospholipid G protein-coupled receptors. J. Biol. Chem. 2004, 279, 20555–20558. [Google Scholar] [CrossRef]
- Kano, K.; Aoki, J.; Hla, T. Lysophospholipid Mediators in Health and Disease. Annu. Rev. Pathol. 2022, 24, 459–483. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.V.; Herst, P.M.; Tan, A.S. Tetrazolium dyes as tools in cell biology: New insights into their cellular reduction. Biotechnol. Annu. Rev. 2005, 11, 127–152. [Google Scholar]
- Stockert, J.C.; Horobin, R.W.; Colombo, L.L.; Blázquez-Castro, A. Tetrazolium salts and formazan products in Cell Biology: Viability assessment, fluorescence imaging, and labeling perspectives. Acta Histochem. 2018, 120, 159–167. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Satoh, A.; Tezuka, H.; Han, S.I.; Takei, K.; Iwasaki, H.; Yatoh, S.; Yahagi, N.; Suzuki, H.; Iwasaki, Y.; et al. CREB3L3 controls fatty acid oxidation and ketogenesis in synergy with PPARα. Sci. Rep. 2016, 6, 39182. [Google Scholar] [CrossRef]
- Zurkinden, L.; Sviridov, D.; Vogt, B.; Escher, G. Downregulation of Cyp7a1 by cholic acid and chenodeoxycholic acid in Cyp27a1/ApoE double. Front. Endocrinol. 2020, 11, 586980. [Google Scholar] [CrossRef]
- Wootten, D.; Christopoulos, A.; Marti-Solano, M.; Babu, M.M.; Sexton, P.M. Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 2018, 19, 638–653. [Google Scholar] [CrossRef]
- Schwarzer, K.; Adams, C.A. The influence of specific phospholipids as absorption enhancer in animal nutrition. Lipid/Fett 1996, 98, 304–308. [Google Scholar] [CrossRef]
- Nutautaite, M.; Raceviciute-Stupeliene, A.; Andalibizadeh, L.; Sasyte, V.; Bliznikas, S.; Pockevicius, A. Improving broiler chickens’health by using lecithin and lysophosphatidylcholine emulsifiers: A comparative analysis of physiological indicators. Iran. J. Vet. Res. 2021, 22, 33–39. [Google Scholar] [PubMed]
- Juntanapum, W.; Bunchasak, C.; Poeikhampha, T.; Rakangthong, C.; Poungpong, K. Effects of supplementation of lysophosphatidylcholine (LPC) to lying hens on production performance, fat digestibility, blood lipid profile, and gene expression related to nutrients transport in small intestine. J. Anim. Feed Sci. 2020, 29, 258–265. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, S.; Nie, K.; Zheng, H.; Luo, Z.; Kim, I.H. Lysolecithin Improves Broiler Growth Performance through Upregulating Growth-Related Genes and Nutrient Transporter Genes Expression Independent of Experimental Diet Nutrition Level. Animals 2022, 12, 3365. [Google Scholar] [CrossRef]
- Perl, K.; Ushakov, K.; Pozniak, Y.; Yizhar-Barnea, O.; Bhonker, Y.; Shivatzki, S.; Geiger, T.; Avraham, K.B.; Shamir, R. Reduced changes in protein compared to mRNA levels across non-proliferating tissues. BMC Genom. 2017, 18, 305. [Google Scholar] [CrossRef]
- Perin, N.; Jarocka-Cyrta, E.; Keelan, M.; Clandinin, T.; Thomson, A. Dietary lipid composition modifies intestinal morphology and nutrient transport in young rats. J. Pediatr. Gastroenterol. Nutr. 1999, 28, 46–53. [Google Scholar]
- Knuplez, E.; Marsche, G. An Updated Review of Pro- and Anti-Inflammatory Properties of Plasma Lysophosphatidylcholines in the Vascular System. Int. J. Mol. Sci. 2020, 21, 4501. [Google Scholar] [CrossRef]
- Xiang, S.; Zhao, M.; Li, Z.; Zheng, H.; Ding, Y.; Peng, F.; Xiao, J.; Xie, K.; Zhang, H.; He, X.; et al. Effects of lysolecithin on growth performance, nutrient digestibility and liver lipid metabolism of yellow-feather broilers. Ital. J. Anim. Sci. 2024, 23, 1601–1610. [Google Scholar] [CrossRef]



| Phospholipid Profile of the Lysolecithin (%) | Fatty Acid Chains on Lysophospholipids (%) | ||
|---|---|---|---|
| Phosphatidylglycerols | 0.72 | 14:0 | 0.07 |
| Phosphatidylethanolamines | 8.84 | 15:0 | <0.05 |
| Phosphatidylinositols | 7.23 | 16:0 | 15.57 |
| Phosphatidylserins | <1 | 16:1 | 0.14 |
| Lipid A phospholipids | 1.67 | 17:0 | 0.14 |
| Phosphatidylcholines | 12.06 | 17:1 | 0.05 |
| Sphingomyelins | <0.4 | 18:0 | 4.22 |
| Lysophosphatidylcholines | 5.08 | 18:1 trans | <0.05 |
| Lysophosphatidylethanolamines | 3.75 | 18:1 | 15.52 |
| Lysophosphatidylinositols | 2.19 | 18:2 trans | 0.09 |
| Total | 41.54 | 18:2 (n − 6) | 56.15 |
| 18:3 trans | <0.05 | ||
| 18:3 (n − 3) | 6.93 | ||
| 20:0 | 0.24 | ||
| 20:1 | 0.15 | ||
| 20:2 (n − 6) | <0.05 | ||
| 22:0 | 0.40 | ||
| 22:1 | <0.05 | ||
| 24:0 | 0.22 | ||
| 24:1 | <0.05 | ||
| Content (g/100 g) | 68.52 | ||
| Saturated fatty acids (%) | 20.87 | ||
| Monounsaturated fatty acids (%) | 15.86 | ||
| Polyunsaturated fatty acids (%) | 63.08 | ||
| Of which (n − 6) (%) | 56.15 | ||
| Of which (n − 3) (%) | 6.93 | ||
| Gene_ID | log2FoldChange | padj | Gene_Name | Gene_Description |
|---|---|---|---|---|
| ENSG00000004799 | −4.70 | 0.0000 | PDK4 | pyruvate dehydrogenase kinase 4 [Source: HGNC Symbol; Acc:HGNC:8812] |
| ENSG00000143333 | −3.30 | 0.0004 | RGS16 | regulator of G protein signaling 16 [Source: HGNC Symbol;Acc:HGNC:9997] |
| ENSG00000060566 | −2.46 | 0.0000 | CREB3L3 | cAMP responsive element binding protein 3 like 3 [Source: HGNC Symbol; Acc:HGNC: 18855] |
| ENSG00000279357 | −2.42 | 0.0049 | AC007224.2 | TEC |
| ENSG00000199047 | −2.41 | 0.0001 | MIR378A | microRNA 378a [Source: HGNC Symbol;Acc:HGNC:31871] |
| ENSG00000147872 | −2.28 | 0.0000 | PLIN2 | perilipin 2 [Source: HGNC Symbol;Acc:HGNC:248] |
| ENSG00000259771 | −2.19 | 0.0016 | AC092756.1 | novel transcript, sense intronic to MYO1E |
| ENSG00000268635 | −2.07 | 0.0355 | AP003680.1 | novel transcript, antisense to PAK1 |
| ENSG00000103044 | −2.04 | 0.0000 | HAS3 | hyaluronan synthase 3 [Source: HGNC Symbol; Acc:HGNC:4820] |
| ENSG00000168505 | −1.97 | 0.0003 | GBX2 | gastrulation brain homeobox 2 [Source: HGNC Symbol; Acc:HGNC:4186] |
| Gene_ID | log2FoldChange | padj | Gene_Name | Gene_Description |
|---|---|---|---|---|
| ENSG00000136546 | 2.94 | 0.0009 | SCN7A | sodium voltage-gated channel alpha subunit 7 [Source:HGNC Symbol;Acc:HGNC:10594] |
| ENSG00000109743 | 2.82 | 0.0108 | BST1 | bone marrow stromal cell antigen 1 [Source:HGNC Symbol;Acc:HGNC:1118] |
| ENSG00000167910 | 2.73 | 0.0005 | CYP7A1 | cytochrome P450 family 7 subfamily A member 1 [Source:HGNC Symbol;Acc:HGNC:2651] |
| ENSG00000256651 | 2.71 | 0.0000 | AC006518.1 | taste receptor, type 2 pseudogene |
| ENSG00000081148 | 2.54 | 0.0206 | IMPG2 | interphotoreceptor matrix proteoglycan 2 [Source:HGNC Symbol;Acc:HGNC:18362] |
| ENSG00000154263 | 2.51 | 0.0026 | ABCA10 | ATP binding cassette subfamily A member 10 [Source:HGNC Symbol;Acc:HGNC:30] |
| ENSG00000235079 | 2.47 | 0.0117 | ZRANB2-AS1 | ZRANB2 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:43594] |
| ENSG00000198939 | 2.39 | 0.0011 | ZFP2 | ZFP2 zinc finger protein [Source:HGNC Symbol;Acc:HGNC:26138] |
| ENSG00000136531 | 2.32 | 0.0235 | SCN2A | sodium voltage-gated channel alpha subunit 2 [Source:HGNC Symbol;Acc:HGNC:10588] |
| ENSG00000168542 | 2.27 | 0.0318 | COL3A1 | collagen type III alpha 1 chain [Source:HGNC Symbol;Acc:HGNC:2201] |
| HMDB_ID | Compound_Name | Chemical Formula | FC (Lysolecithin/Control) | Log2(FC) | t-Test |
|---|---|---|---|---|---|
| HMDB0000792 | Sebacic acid | C10H18O4 | 19.31 | 4.27 | 0.0001 |
| HMDB0249582 | Candoxatrilat | C20H33NO7 | 16.90 | 4.08 | 0.0002 |
| HMDB0004704 | 9,10-DHOME | C18H34O4 | 16.17 | 4.02 | 0.0002 |
| HMDB0000672 | Hexadecanedioic acid | C16H30O4 | 15.05 | 3.91 | 0.0002 |
| HMDB0004708 | 9,12,13-TriHOME | C18H34O5 | 14.64 | 3.87 | 0 |
| HMDB0302703 | Eremanthin | C15H18O2 | 13.83 | 3.79 | 0.0001 |
| HMDB0303565 | Eucannabinolide | C22H28O8 | 13.81 | 3.79 | 0.0016 |
| HMDB0000560 | Goshuyic acid | C14H24O2 | 12.26 | 3.62 | 0 |
| HMDB0062363 | 6-Hydroxy-3-oxotetradecenoic acid | C14H24O4 | 11.48 | 3.52 | 0 |
| HMDB0006236 | Phenylacetaldehyde | C8H8O | 10.78 | 3.43 | 0.0001 |
| HMDB0034673 | Calamendiol | C15H26O2 | 9.07 | 3.18 | 0.0001 |
| HMDB0004706 | 8-Hydroperoxylinoleic acid | C18H32O4 | 8.68 | 3.12 | 0.0002 |
| HMDB0010387 | LysoPC(18:3) | C26H48NO7P | 8.08 | 3.01 | 0.0242 |
| HMDB0040668 | Blumenol C glucoside | C19H32O7 | 7.30 | 2.87 | 0.0002 |
| HMDB0114758 | LysoPA(20:2) | C23H43O7P | 7.24 | 2.86 | 0.049 |
| HMDB0036143 | Monomenthyl succinate | C14H24O4 | 5.66 | 2.50 | 0.0001 |
| HMDB0002259 | Heptadecanoic acid | C17H34O2 | 5.44 | 2.44 | 0.0003 |
| HMDB0030982 | 6-Ketomyristic acid | C14H26O3 | 4.77 | 2.25 | 0.0003 |
| HMDB0038731 | 4,7-Megastigmadien-9-ol | C13H22O | 4.67 | 2.22 | 0.0001 |
| HMDB0010725 | 3-Hydroxydecanoic acid | C10H20O3 | 4.05 | 2.02 | 0 |
| HMDB0004667 | 13-HODE | C18H32O3 | 3.71 | 1.89 | 0.0027 |
| HMDB0061914 | 8-Hydroxyoctanoate | C8H16O3 | 3.58 | 1.84 | 0 |
| HMDB0031127 | 5-Hexyltetrahydro-2-furanoctanoic acid | C18H34O3 | 3.33 | 1.73 | 0.0004 |
| HMDB0036199 | 2-Methoxy-4-(4-methyl-1,3-dioxolan-2-yl)phenol | C11H14O4 | 3.31 | 1.73 | 0.0008 |
| HMDB0000784 | Azelaic acid | C9H16O4 | 3.08 | 1.62 | 0.0008 |
| HMDB0035338 | Sterebin B | C20H32O5 | 2.33 | 1.22 | 0.0015 |
| HMDB0029610 | Ascladiol | C7H8O4 | 2.02 | 1.01 | 0.0112 |
| HMDB_ID | Compound_Name | Chemical Formula | FC (Contol/Lysolecithin) | Log2(FC) | t-Test |
|---|---|---|---|---|---|
| HMDB0059972 | 4-Hydroxy-5-(3′,4′-dihydroxyphenyl)-valeric acid-O-methyl-O-glucuronide | C18H26O12 | 17.96 | 4.51 | 0.0025 |
| HMDB0060491 | Mycophenolic acid O-acyl-glucuronide | C23H28O12 | 17.31 | 4.36 | 0.0001 |
| HMDB0260340 | Cytidine-5′-diphosphocholine | C14H26N4O11P2 | 15.80 | 4.32 | 0.0033 |
| HMDB0038866 | Quercetagetin 3′-methylether 7-glucoside | C22H22O13 | 10.17 | 3.82 | 0.0000 |
| HMDB0258916 | Tetramethylchromanol glucoside | C20H30O7 | 9.86 | 3.49 | 0.0000 |
| HMDB0252642 | Gardenoside | C17H24O11 | 7.49 | 3.42 | 0.0000 |
| HMDB0038809 | Luteolin 4′-glucoside 7-galacturonide | C27H28O17 | 4.18 | 2.18 | 0.0002 |
| HMDB_ID | Compound_Name | Chemical Formula | FC(LEX/Control) | Log2(FC) | t-Test |
|---|---|---|---|---|---|
| HMDB0094680 | Octaethylene glycol | C16H34O9 | 6.23 | 2.64 | 0.0005 |
| HMDB0244865 | 1-Benzazepine | C10H9N | 3.70 | 1.89 | 0.0006 |
| HMDB0006547 | Stearidonic acid | C18H28O2 | 2.95 | 1.56 | 0.0019 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Hoeck, V.; Spaepen, R.; Forier, B. The Nutritional Gene Expression Regulation Potential of a Lysolecithin-Based Product. Curr. Issues Mol. Biol. 2025, 47, 548. https://doi.org/10.3390/cimb47070548
Van Hoeck V, Spaepen R, Forier B. The Nutritional Gene Expression Regulation Potential of a Lysolecithin-Based Product. Current Issues in Molecular Biology. 2025; 47(7):548. https://doi.org/10.3390/cimb47070548
Chicago/Turabian StyleVan Hoeck, Veerle, Riet Spaepen, and Bart Forier. 2025. "The Nutritional Gene Expression Regulation Potential of a Lysolecithin-Based Product" Current Issues in Molecular Biology 47, no. 7: 548. https://doi.org/10.3390/cimb47070548
APA StyleVan Hoeck, V., Spaepen, R., & Forier, B. (2025). The Nutritional Gene Expression Regulation Potential of a Lysolecithin-Based Product. Current Issues in Molecular Biology, 47(7), 548. https://doi.org/10.3390/cimb47070548





