Antiviral Activity of Rhamnolipids Nano-Micelles Against Rhinoviruses—In Silico Docking, Molecular Dynamic Analysis and In-Vitro Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methodology
2.2.1. Computational Studies
In Silico Docking
Standard Dynamic Simulations
2.2.2. Production and Characterization of Rhamnolipids
2.2.3. Preparation and Characterization of RMN
2.2.4. Cytotoxicity of RMN
Cell Culture
RMN Cytotoxicity on HeLa Cells
2.2.5. RVs Propagation and Tissue Culture Infectious Dose 50% (TCID50) Determination
2.2.6. Antiviral Activity of RMN
2.2.7. Statistical Analysis
3. Results and Discussion
3.1. In-Silico Studies
3.1.1. Molecular Docking Study
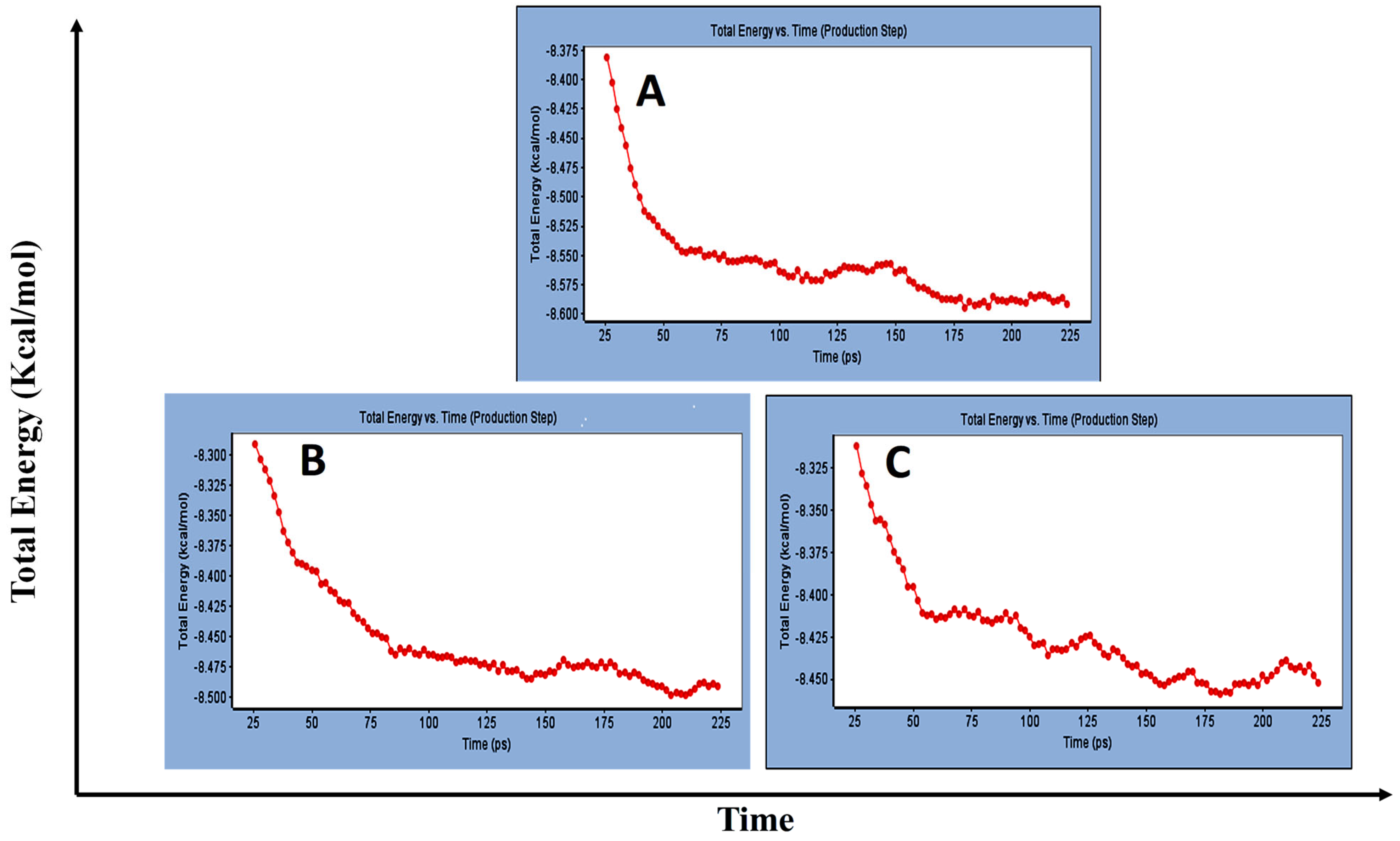
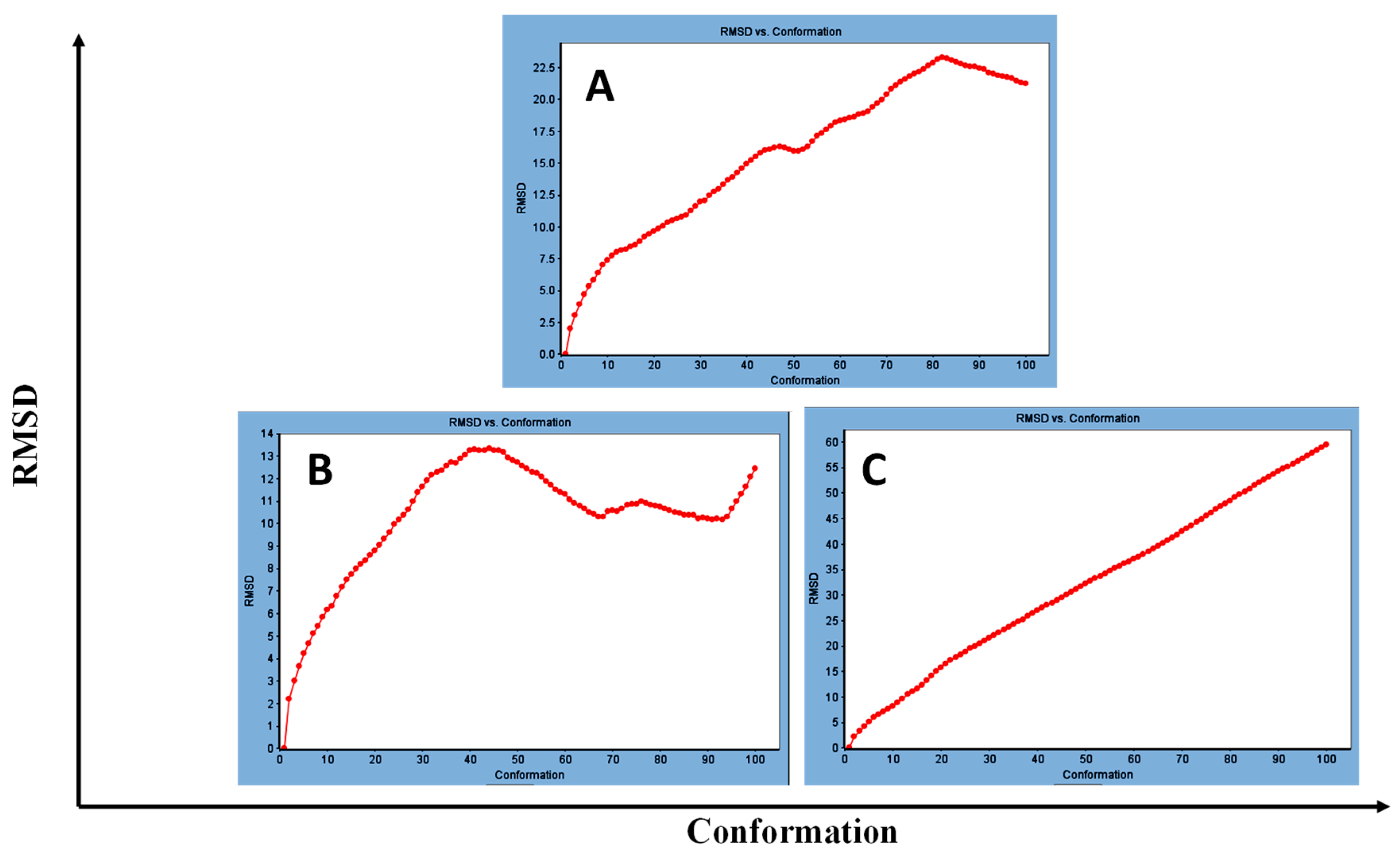
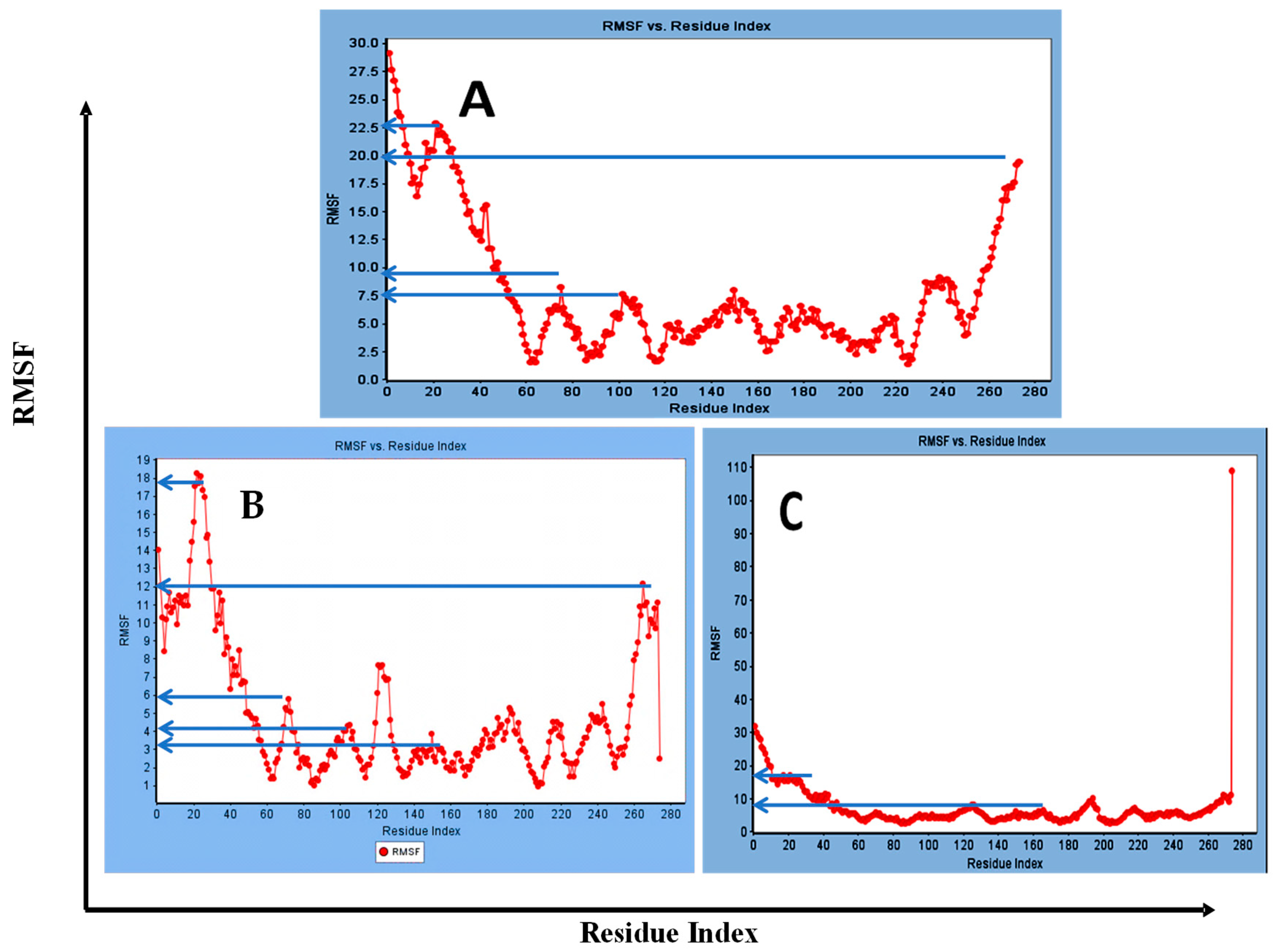
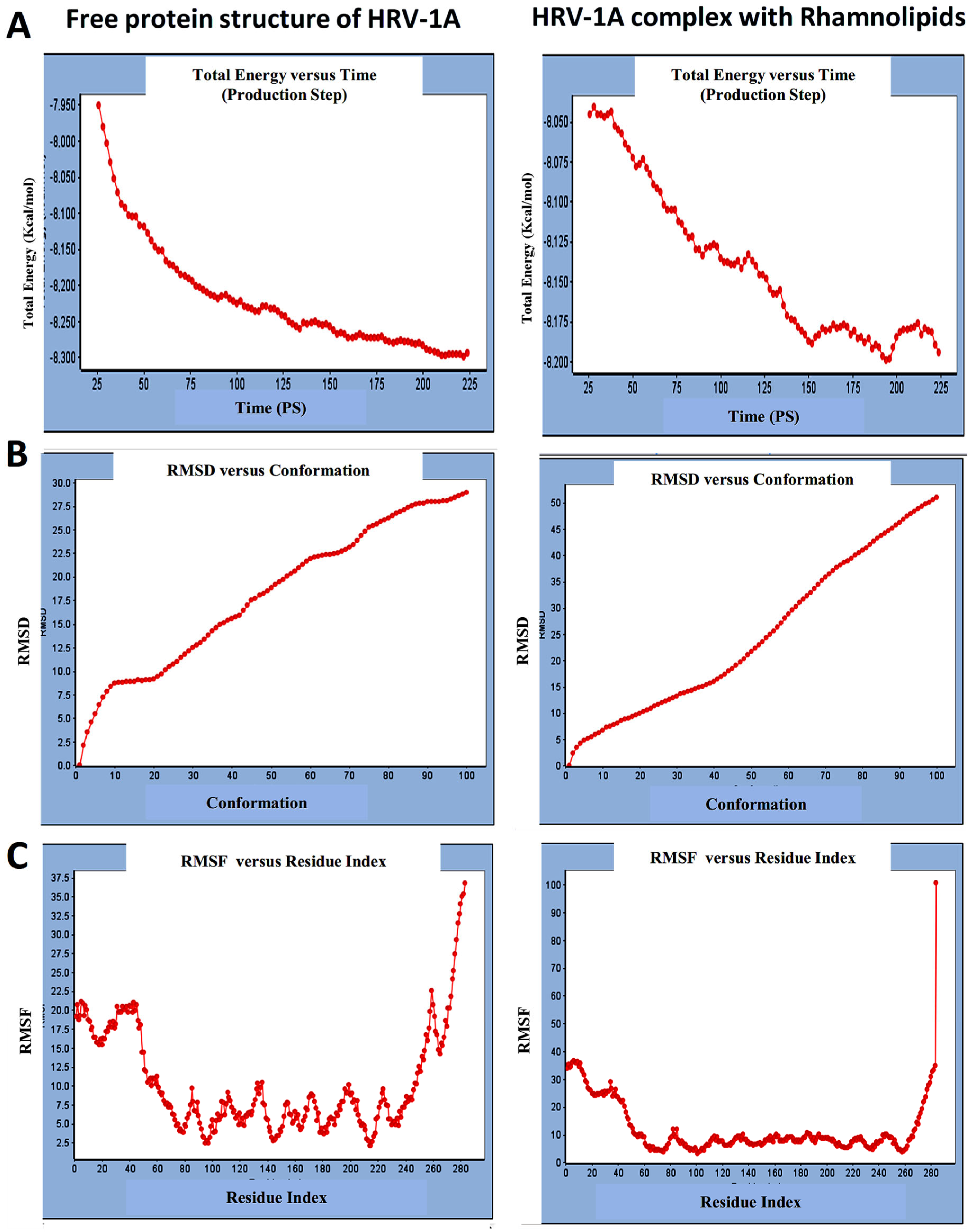
3.1.2. Standard Dynamic Simulation
3.2. Production of Rhamnolipids
3.3. Characterization of Prepared RMN
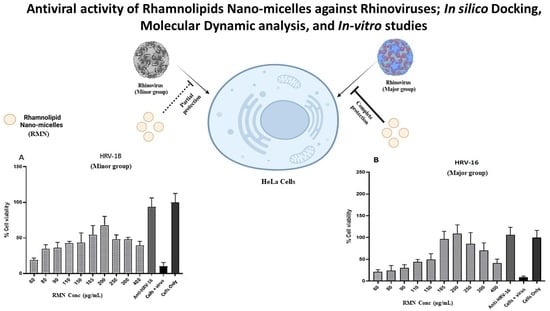
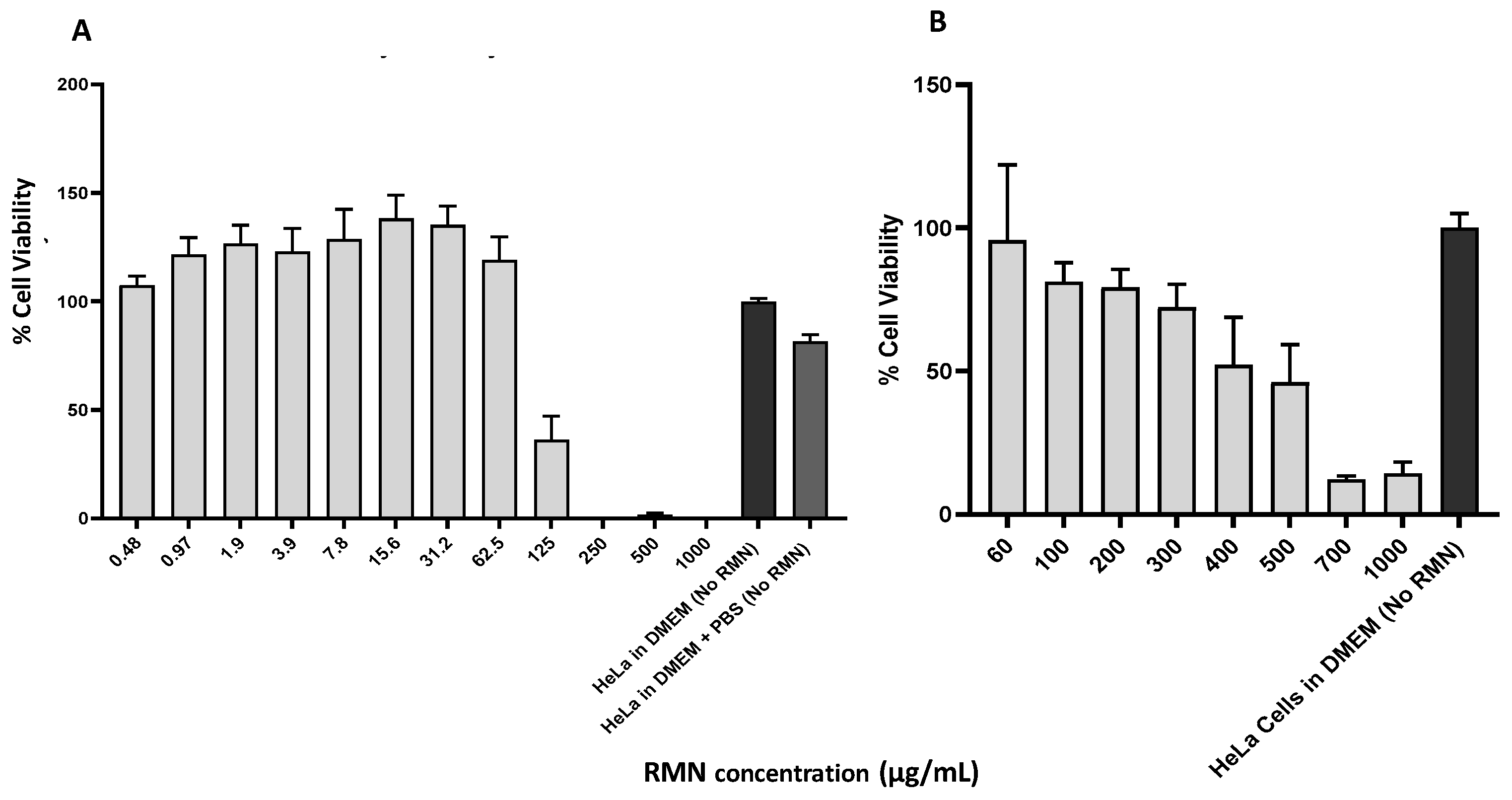
3.4. Cytotoxicity of RMN on HeLa Cells
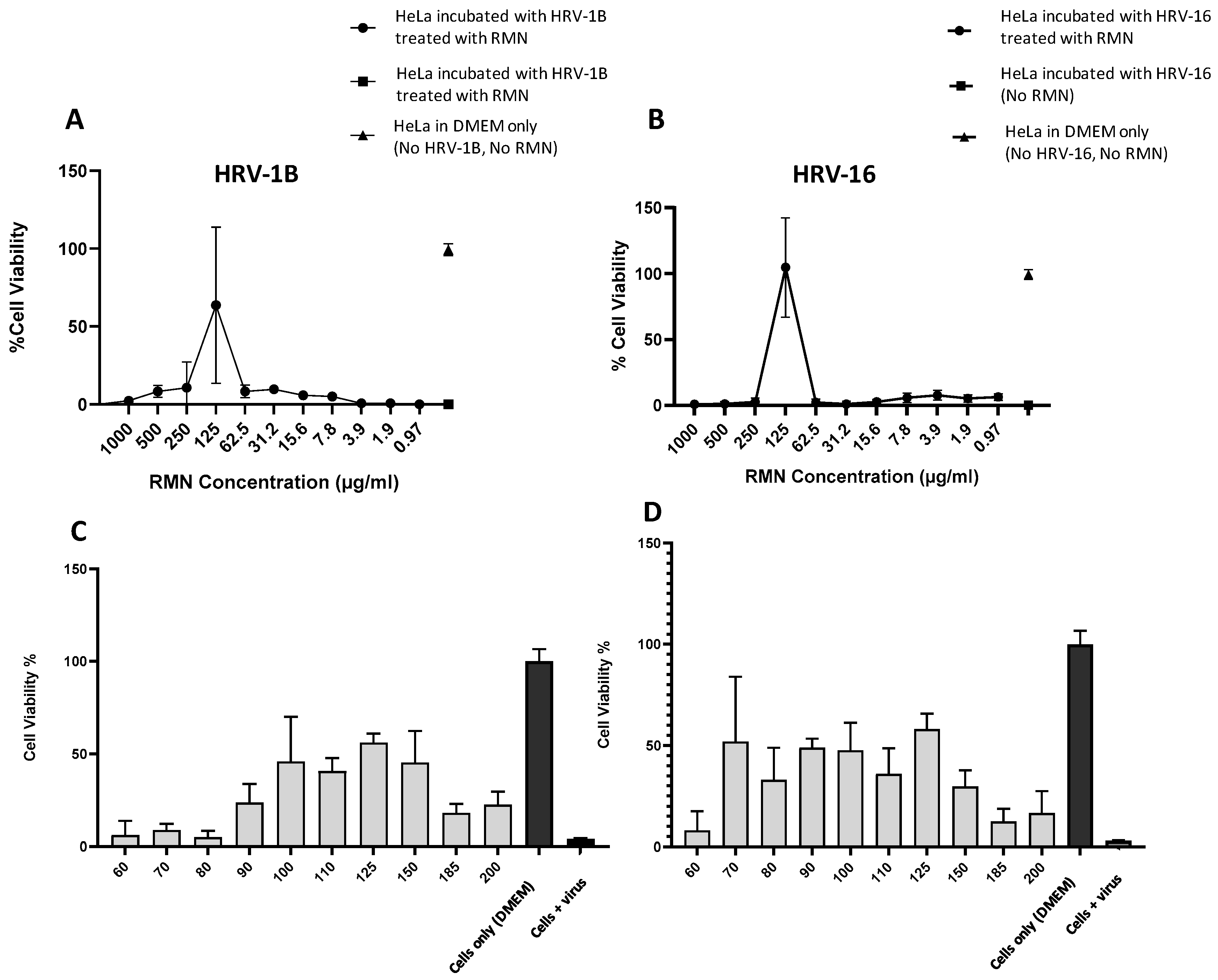
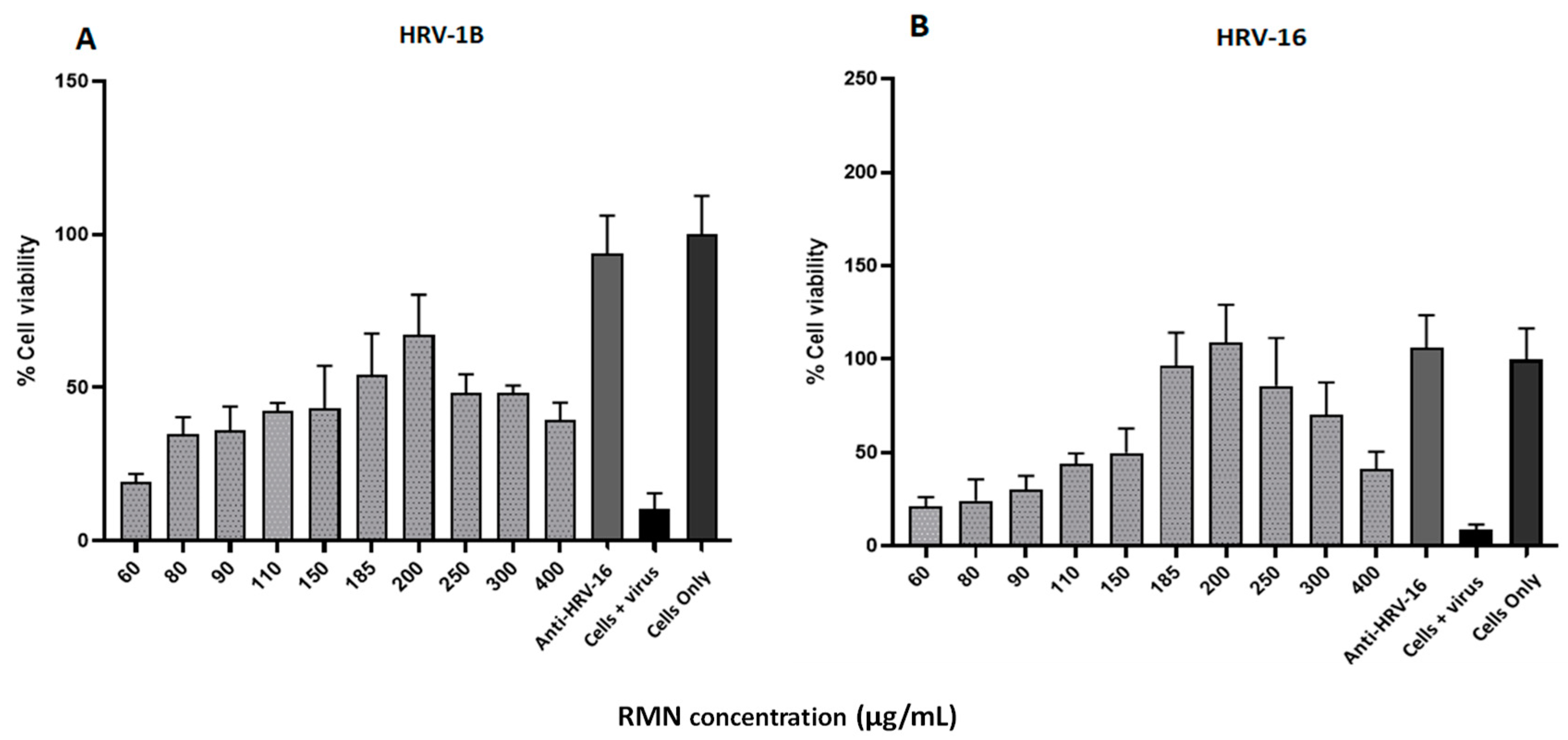
3.5. Antiviral Activity of RMN, Virus Infection Neutralization Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abo-zeid, Y.; Bakkar, M.R.; Elkhouly, G.E.; Raya, N.R.; Zaafar, D. Rhamnolipid Nano-Micelles Versus Alcohol-Based Hand Sanitizer : A Comparative Study for Antibacterial Activity against Hospital-Acquired Infections and Toxicity Concerns. Antibiotics 2022, 11, 605. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.; Raya, N.R.; Orabi, A.; Ali, A.M.; Abo-zeid, Y. Investigating the Antibacterial Activity and Safety of Zinc Oxide Nanoparticles versus a Commercial Alcohol-Based Hand-Sanitizer: Can Zinc Oxide Nanoparticles Be Useful for Hand Sanitation? Antibiotics 2022, 11, 1606. [Google Scholar] [CrossRef] [PubMed]
- Abo-zeid, Y.; Amer, A.; El-Houssieny, B.; Bakkar, M.; Sakran, W. Overview on Bacterial Resistance and Nanoparticles to Overcome Bacterial Resistance. J. Adv. Pharm. Res. 2021, 5, 312–326. [Google Scholar] [CrossRef]
- Ali, A.M.; Hill, H.J.; Elkhouly, G.E.; Raya, N.R.; Tawfik, N.F.; Bakkar, M.R.; El-basaty, A.B.; Stamataki, Z.; Abo-zeid, Y. Green and Chemical Synthesis of Iron Oxide Nanoparticles : Comparative Study for Antimicrobial Activity and Toxicity Concerns. J. Drug Deliv. Sci. Technol. 2025, 103, 106434. [Google Scholar] [CrossRef]
- Ali, A.M.; Hill, H.J.; Elkhouly, G.E.; Bakkar, M.R.; Raya, N.R.; Stamataki, Z.; Abo-zeid, Y. Rhamnolipid Nano-Micelles Inhibit SARS-CoV-2 Infection and Have No Dermal or Eye Toxic Effects in Rabbits. Antibiotics 2022, 11, 1556. [Google Scholar] [CrossRef]
- Chow, E.J.; Mermel, L.A. Hospital-Acquired Respiratory Viral Infections: Incidence, Morbidity, and Mortality in Pediatric and Adult Patients. Open Forum Infect. Dis. 2017, 4, ofx006. [Google Scholar] [CrossRef]
- Gern, J.E. The ABCs of Rhinoviruses, Wheezing, and Asthma. J. Virol. 2010, 84, 7418–7426. [Google Scholar] [CrossRef]
- Johnston, S.L.; Pattemore, P.K.; Sanderson, G.; Smith, S.; Lampe, F.; Josephs, L.; Symington, P.; Toole, S.O.; Myint, S.H.; Tyrrell, D.A.J.; et al. Community Study of Role of Viral Infections in Exacerbations of Asthma in 9-11 Year Old Children. BMJ 1995, 310, 1225–1229. [Google Scholar] [CrossRef]
- Nicholson, K.G.; Kent, J.; Ireland, D.C. Respiratory Viruses and Exacerbations of Asthma in Adults. Br. Med. J. 1993, 307, 982–986. [Google Scholar] [CrossRef]
- Papi, A.; Bellettato, C.M.; Braccioni, F.; Romagnoli, M.; Casolari, P.; Caramori, G.; Fabbri, L.M.; Johnston, S.L. Infections and Airway Inflammation in Chronic Obstructive Pulmonary Disease Severe Exacerbations. Am. J. Respir. Crit. Care Med. 2006, 173, 1114–1121. [Google Scholar] [CrossRef]
- Feddema, J.J.; Claassen, E. Prevalence of Viral Respiratory Infections amongst Asthmatics: Results of a Meta-Regression Analysis. Respir. Med. 2020, 173, 106020. [Google Scholar] [CrossRef] [PubMed]
- Henquell, C.; Mirand, A.; Deusebis, A.L.; Regagnon, C.; Archimbaud, C.; Chambon, M.; Bailly, J.L.; Gourdon, F.; Hermet, E.; Dauphin, J.B.; et al. Prospective Genotyping of Human Rhinoviruses in Children and Adults during the Winter of 2009–2010. J. Clin. Virol. 2012, 53, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Kiang, D.; Yagi, S.; Kantardjieff, K.A.; Kim, E.J.; Louie, J.K.; Schnurr, D.P. Molecular Characterization of a Variant Rhinovirus from an Outbreak Associated with Uncommonly High Mortality. J. Clin. Virol. 2007, 38, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Winther, B. Rhinovirus Infections in the Upper Airway. Proc. Am. Thorac. Soc. 2011, 8, 79–89. [Google Scholar] [CrossRef]
- Stobart, C.C.; Nosek, J.M.; Moore, M.L. Rhinovirus Biology, Antigenic Diversity, and Advancements in the Design of a Human Rhinovirus Vaccine. Front. Microbiol. 2017, 8, 2412. [Google Scholar] [CrossRef]
- Touabi, L.; Aflatouni, F.; McLean, G.R. Mechanisms of Rhinovirus Neutralisation by Antibodies. Viruses 2021, 13, 360. [Google Scholar] [CrossRef]
- Egorova, A.; Ekins, S.; Schmidtke, M.; Makarov, V. Back to the Future: Advances in Development of Broad-Spectrum Capsid-Binding Inhibitors of Enteroviruses. Eur. J. Med. Chem. 2019, 178, 606–622. [Google Scholar] [CrossRef]
- Giardina, F.A.M.; Piralla, A.; Ferrari, G.; Zavaglio, F.; Cassaniti, I.; Baldanti, F. Molecular Epidemiology of Rhinovirus/Enterovirus and Their Role on Cause Severe and Prolonged Infection in Hospitalized Patients. Microorganisms 2022, 10, 755. [Google Scholar] [CrossRef]
- Vandini, S.; Biagi, C.; Fischer, M.; Lanari, M. Impact of Rhinovirus Infections in Children. Viruses 2019, 11, 521. [Google Scholar] [CrossRef]
- Savolainen-Kopra, C.; Korpela, T.; Simonen-Tikka, M.L.; Amiryousefi, A.; Ziegler, T.; Roivainen, M.; Hovi, T. Single Treatment with Ethanol Hand Rub Is Ineffective against Human Rhinovirus-Hand Washing with Soap and Water Removes the Virus Efficiently. J. Med. Virol. 2012, 84, 543–547. [Google Scholar] [CrossRef]
- Henkel, M.; Geissler, M.; Weggenmann, F.; Hausmann, R. Production of Microbial Biosurfactants: Status Quo of Rhamnolipid and Surfactin towards Large-Scale Production. Biotechnol. J. 2017, 12, 1600561. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.M.; Hörmann, B.; Syldatk, C.; Hausmann, R. Pseudomonas Aeruginosa PAO1 as a Model for Rhamnolipid Production in Bioreactor Systems. Appl. Microbiol. Biotechnol. 2010, 87, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira dos Santos, S.; Araújo Torquato, C.; de Alexandria Santos, D.; Orsato, A.; Leite, K.; Serpeloni, J.M.; Losi-Guembarovski, R.; Romão Pereira, E.; Dyna, A.L.; Lopes Barboza, M.G.; et al. Production and Characterization of Rhamnolipids by Pseudomonas Aeruginosa Isolated in the Amazon Region, and Potential Antiviral, Antitumor, and Antimicrobial Activity. Sci. Rep. 2024, 14, 4629. [Google Scholar] [CrossRef] [PubMed]
- Firdose, A.; Maeda, T.; Sukri, M.A.M.; Yasin, N.H.M.; Sabturani, N.; Aqma, W.S. Antibacterial Mechanism of Pseudomonas Aeruginosa UKMP14T Rhamnolipids against Multidrug Resistant Acinetobacter Baumannii. Microb. Pathog. 2024, 193, 106743. [Google Scholar] [CrossRef]
- Lotfabad, T.B.; Shahcheraghi, F.; Shooraj, F. Assessment of Antibacterial Capability of Rhamnolipids Produced by Two Indigenous Pseudomonas Aeruginosa Strains. Jundishapur J. Microbiol. 2013, 6, 29–35. [Google Scholar] [CrossRef]
- Sana, S.; Datta, S.; Biswas, D.R.; Auddy, B.; Gupta, M.; Chattopadhyay, H. Excision Wound Healing Activity of a Common Biosurfactant Produced by Pseudomonas sp. Wound Med. 2018, 23, 47–52. [Google Scholar] [CrossRef]
- Carrazco-Palafox, J.; Rivera-Chavira, B.E.; Adame-Gallegos, J.R.; Rodríguez-Valdez, L.M.; Orrantia-Borunda, E.; Nevárez-Moorillón, G.V. Rhamnolipids from Pseudomonas Aeruginosa Rn19a Modifies the Biofilm Formation over a Borosilicate Surface by Clinical Isolates. Coatings 2021, 11, 136. [Google Scholar] [CrossRef]
- Bharali, P.; Das, S.; Ray, A.; Pradeep Singh, S.; Bora, U.; Kumar Konwar, B.; Singh, C.B.; Sahoo, D. Biocompatibility Natural Effect of Rhamnolipids in Bioremediation Process on Different Biological Systems at the Site of Contamination. Bioremediat. J. 2018, 22, 1516616. [Google Scholar] [CrossRef]
- Giugliano, R.; Buonocore, C.; Zannella, C.; Chianese, A.; Esposito, F.P.; Tedesco, P.; De Filippis, A.; Galdiero, M.; Franci, G.; de Pascale, D. Antiviral Activity of the Rhamnolipids Mixture from the Antarctic Bacterium Pseudomonas Gessardii M15 against Herpes Simplex Viruses and Coronaviruses. Pharmaceutics 2021, 13, 2121. [Google Scholar] [CrossRef]
- Remichkova, M.; Galabova, D.; Roeva, I.; Karpenko, E.; Shulga, A.; Galabov, A.S. Anti-Herpesvirus Activities of Pseudomonas sp. S-17 Rhamnolipid and Its Complex with Alginate. Z Naturforsch. C J. Biosci. 2008, 63, 75–81. [Google Scholar] [CrossRef]
- Schuler, B.A.; Schreiber, M.T.; Li, L.Y.; Mokry, M.; Kingdon, M.L.; Raugi, D.N.; Smith, C.; Hameister, C.; Racaniello, V.R.; Hall, D.J. Major and Minor Group Rhinoviruses Elicit Differential Signaling and Cytokine Responses as a Function of Receptor-Mediated Signal Transduction. PLoS ONE 2014, 9, e0093897. [Google Scholar] [CrossRef] [PubMed]
- Ellis, B.R.; Barrett, A.D.T. The Enigma of Yellow Fever in East Africa. Rev. Med. Virol. 2008, 18, 331–346. [Google Scholar] [CrossRef] [PubMed]
- Arruda, E.; Pitkäranta, A.; Witek, T.J.; Doyle, C.A.; Hayden, F.G. Frequency and Natural History of Rhinovirus Infections in Adults during Autumn. J. Clin. Microbiol. 1997, 35, 2864–2868. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, N.W.; Slater, L.; Glanville, N.; Haas, J.J.; Caramori, G.; Casolari, P.; Clarke, D.L.; Message, S.D.; Aniscenko, J.; Kebadze, T.; et al. Defining Critical Roles for NF-ΚB P65 and Type I Interferon in Innate Immunity to Rhinovirus. EMBO Mol. Med. 2012, 4, 1244–1260. [Google Scholar] [CrossRef]
- Greve, J.M.; Davis, G.; Meyer, A.M.; Forte, C.P.; Yost, S.C.; Marlor, C.W.; Kamarck, M.E.; McClelland, A. The Major Human Rhinovirus Receptor Is ICAM-1. Cell 1989, 56, 839–847. [Google Scholar] [CrossRef]
- Chapman, M.S.; Minor, I.; Rossmann, M.G.; Diana, G.D.; Andries, K. Human Rhinovirus 14 Complexed with Antiviral Compound R 61837. J. Mol. Biol. 1991, 217, 455–463. [Google Scholar] [CrossRef]
- Kim, S.; Smith, T.J.; Chapman, M.S.; Rossmann, M.G.; Pevear, D.C.; Dutko, F.J.; Felock, P.J.; Diana, G.D.; McKinlay, M.A. Crystal Structure of Human Rhinovirus Serotype 1A (HRV1A). J. Mol. Biol. 1989, 210, 91–111. [Google Scholar] [CrossRef]
- Hu, Y.; Wu, Y.; Jiang, C.P.; Wang, Z.; Shen, C.; Zhu, Z.; Li, H.; Zeng, Q.; Xue, Y.; Wang, Y.; et al. Investigative on the Molecular Mechanism of Licorice Flavonoids Anti-Melanoma by Network Pharmacology, 3D/2D-QSAR, Molecular Docking, and Molecular Dynamics Simulation. Front. Chem. 2022, 10, 843970. [Google Scholar] [CrossRef]
- Bakkar, M.R.; Hassan, A.; Faraag, I.; Soliman, E.R.S.; Fouda, M.S.; Mahfouz, A.; Sarguos, M.; Mclean, G.R.; Hebishy, A.M.S.; Elkhouly, G.E.; et al. Rhamnolipids Nano-Micelles as a Potential Hand Sanitizer. Antibiotics 2021, 10, 751. [Google Scholar] [CrossRef]
- Rahman, P.K.S.M.; Pasirayi, G.; Auger, V.; Ali, Z. Production of Rhamnolipid Biosurfactants by Pseudomonas Aeruginosa DS10-129 in a Microfluidic Bioreactor. Biotechnol. Appl. Biochem. 2010, 55, 45–52. [Google Scholar] [CrossRef]
- Abo-zeid, Y.; Williams, G.R.; Touabi, L.; Mclean, G.R. An Investigation of Rhinovirus Infection on Cellular Uptake of Poly (Glycerol-Adipate) Nanoparticles. Int. J. Pharm. 2020, 589, 119826. [Google Scholar] [CrossRef] [PubMed]
- Glanville, N.; Mclean, G.R.; Guy, B.; Lecouturier, V.; Berry, C.; Girerd, Y.; Gregoire, C.; Walton, R.P.; Pearson, R.M.; Kebadze, T.; et al. Cross-Serotype Immunity Induced by Immunization with a Conserved Rhinovirus Capsid Protein. PLoS Pathog. 2013, 9, e1003669. [Google Scholar] [CrossRef] [PubMed]
- Arruda, E.; Crump, C.E.; Rollins, B.S.; Ohlin, A.; Hayden, F.G. Comparative Susceptibilities of Human Embryonic Fibroblasts and HeLa Cells for Isolation of Human Rhinoviruses. J. Clin. Microbiol. 1996, 34, 1277–1279. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, N.W.; Walton, R.P.; Edwards, M.R.; Aniscenko, J.; Caramori, G.; Zhu, J.; Glanville, N.; Choy, K.J.; Jourdan, P.; Burnet, J.; et al. Mouse Models of Rhinovirus-Induced Disease and Exacerbation of Allergic Airway Inflammation. Nat. Med. 2008, 14, 199–204. [Google Scholar] [CrossRef]
- Colonno, R.J.; Condra, J.H.; Mizutani, S.; Callahan, P.L.; Davies, M.E.; Murcko, M.A. Evidence for the Direct Involvement of the Rhinovirus Canyon in Receptor Binding. Proc. Natl. Acad. Sci. USA 1988, 85, 5449–5453. [Google Scholar] [CrossRef]
- Zazeri, G.; Povinelli, A.P.R.; Pavan, N.M.; Jones, A.M.; Ximenes, V.F. Solvent-Induced Lag Phase during the Formation of Lysozyme Amyloid Fibrils Triggered by Sodium Dodecyl Sulfate: Biophysical Experimental and In Silico Study of Solvent Effects. Molecules 2023, 28, 6891. [Google Scholar] [CrossRef]
- Zazeri, G.; Povinelli, A.P.R.; Le Duff, C.S.; Tang, B.; Cornelio, M.L.; Jones, A.M. Synthesis and Spectroscopic Analysis of Piperine- And Piperlongumine-Inspired Natural Product Scaffolds and Their Molecular Docking with IL-1β and NF-ΚB Proteins. Molecules 2020, 25, 2841. [Google Scholar] [CrossRef]
- Chen, A.; Leith, M.; Tu, R.; Tahim, G.; Sudra, A.; Bhargava, S. Effects of Diluents on Cell Culture Viability Measured by Automated Cell Counter. PLoS ONE 2017, 12, e0173375. [Google Scholar] [CrossRef]
- Chevalier, S.; Bouffartigues, E.; Bodilis, J.; Maillot, O.; Lesouhaitier, O.; Feuilloley, M.G.J.; Orange, N.; Dufour, A.; Cornelis, P. Structure, Function and Regulation of Pseudomonas Aeruginosa Porins. FEMS Microbiol. Rev. 2017, 41, 698–722. [Google Scholar] [CrossRef]
- Mallia, P.; Footitt, J.; Sotero, R.; Jepson, A.; Contoli, M.; Trujillo-Torralbo, M.B.; Kebadze, T.; Aniscenko, J.; Oleszkiewicz, G.; Gray, K.; et al. Rhinovirus Infection Induces Degradation of Antimicrobial Peptides and Secondary Bacterial Infection in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2012, 186, 1117–1124. [Google Scholar] [CrossRef]
- Hofer, F.; Gruenberger, M.; Kowalski, H.; Machat, H.; Huettinger, M.; Kuechler, E.; Blaas, D. Members of the Low Density Lipoprotein Receptor Family Mediate Cell Entry of a Minor-Group Common Cold Virus. Proc. Natl. Acad. Sci. USA 1994, 91, 1839–1842. [Google Scholar] [CrossRef]
| RV Serotype | Type | Group | % Identity (Polyprotein) | % Identity (Canyon Floor) | Capsid Structure PDB Code |
|---|---|---|---|---|---|
| 1A | A | Minor | 96 | 99 | 1R1A |
| 1B | A | ||||
| 14 | B | Major | <60 | 51 | 1R09 |
| 16 | A |
| Name | Binding Mode | C-Docker Interaction Energy (Kcal/mol) | Key Amino Acids Interactions |
|---|---|---|---|
| Mono-rhamnolipids | 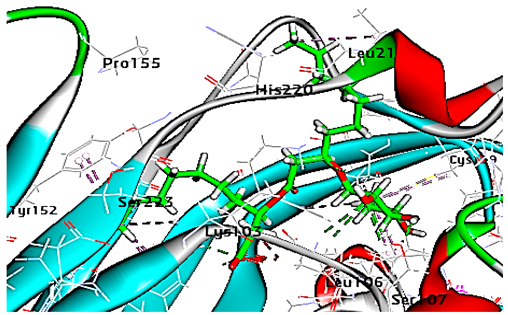 | −71.25 | 1 HBA with Lys103 2 HBA with Leu106 1 HBD with Ser107 Hydrophobic interaction with Leu218, Cys199, Ser223 |
| Di-rhamnolipids | 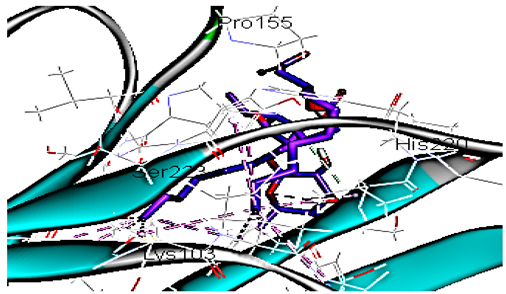 | −70.11 | 1 HBA with His220 Hydrophobic interaction with Lys103, Pro155, Ser223 |
| R 61837 | 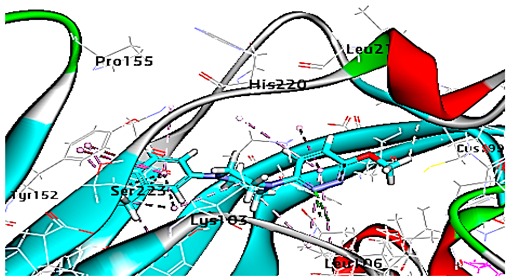 | −55.95 | 1 HBA with Lys103 Hydrophobic interactions with Leu106, Tyr152, Ser223, His220 |
| Mono-rhamnolipids (after MD simulation) |  | 74.66 | 2 HBD with Val110 and Gln111 2 HBA with Gln111 Hydrophobic interaction with Leu109, Leu106, Val177 |
| Di-rhamnolipids (after MD simulation) |  | 75.40 | 1 HBA with Arg242 2 HBD with Gln11, Cys176 Hydrophobic interaction with Leu106, Leu109, Val177, Ala175, Arg242 |
| MD = Molecular dynamic | |||
| Name | Binding Mode | C-Docker Interaction Energy (Kcal/mol) | Key Amino Acids Interactions |
|---|---|---|---|
| Mono-rhamnolipids |  | −50.4 | Hydrophobic interaction with Leu127, Tyr145, Tyr147,Ala152, Ile171, Phe182, Ile220 |
| Di-rhamnolipids |  | −53.7 | 1 HBA with Met169 Hydrophobic interaction with Tyr147, Pro149 Ile171, Phe182, Pro185, Ile220 |
| WIN 53338 | 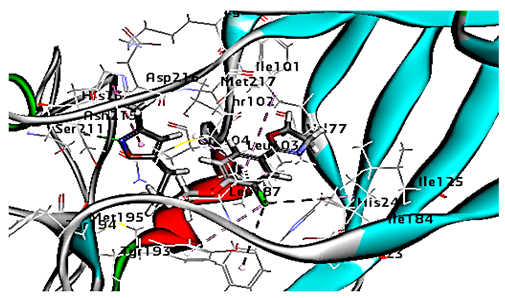 | 61.25 | 1 HBA with ASN215 Hydrophobic interaction with Ile101 Leu103 Ile125 Tyr147, PIle171, Pro185, Leu187, Ile220 Ile184 Tyr198 Met217 His268 |
| Mono-rhamnolipids (after MD simulation) | 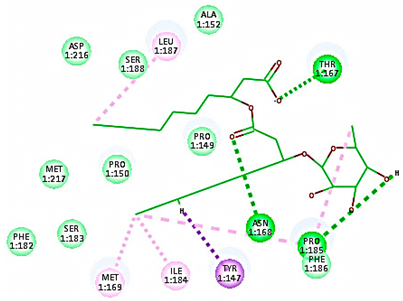 | 54.36 | 3 HBA with Thr167, Asp168, Pro185 Hydrophobic interaction with Tyr147, Ile184, Met169, Pro185, Ile187 |
| Di-rhamnolipids (after MD simulation) |  | 55.93 | 4 HBA with Met169, Tyr147, 185, Hydrophobic interaction with Tyr147, Pro149 Ile171, Phe182, Pro185, Ile220, |
| MD = Molecular dynamic | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Touabi, L.; Ismail, N.S.M.; Bakkar, M.R.; McLean, G.R.; Abo-zeid, Y. Antiviral Activity of Rhamnolipids Nano-Micelles Against Rhinoviruses—In Silico Docking, Molecular Dynamic Analysis and In-Vitro Studies. Curr. Issues Mol. Biol. 2025, 47, 333. https://doi.org/10.3390/cimb47050333
Touabi L, Ismail NSM, Bakkar MR, McLean GR, Abo-zeid Y. Antiviral Activity of Rhamnolipids Nano-Micelles Against Rhinoviruses—In Silico Docking, Molecular Dynamic Analysis and In-Vitro Studies. Current Issues in Molecular Biology. 2025; 47(5):333. https://doi.org/10.3390/cimb47050333
Chicago/Turabian StyleTouabi, Lila, Nasser S. M. Ismail, Marwa R. Bakkar, Gary R. McLean, and Yasmin Abo-zeid. 2025. "Antiviral Activity of Rhamnolipids Nano-Micelles Against Rhinoviruses—In Silico Docking, Molecular Dynamic Analysis and In-Vitro Studies" Current Issues in Molecular Biology 47, no. 5: 333. https://doi.org/10.3390/cimb47050333
APA StyleTouabi, L., Ismail, N. S. M., Bakkar, M. R., McLean, G. R., & Abo-zeid, Y. (2025). Antiviral Activity of Rhamnolipids Nano-Micelles Against Rhinoviruses—In Silico Docking, Molecular Dynamic Analysis and In-Vitro Studies. Current Issues in Molecular Biology, 47(5), 333. https://doi.org/10.3390/cimb47050333


_Kim.png)