Determination of the Phylogenetic Relationship of Dendrobium linawianum (Orchidaceae) Based on Comparative Analysis of Complete Chloroplast Genomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Leaf Sample from Dendrobium linawianum and DNA Sequencing
2.2. Chloroplast Genome Assembly and Annotation
2.3. Repeat and Codon Usage Analyses, Genome Comparison
2.4. Phylogenetic Analysis
3. Results
3.1. Genomic Features of Dendrobium linawianum Complete Chloroplast Genome
3.2. Repeat Sequence and Codon Usage Analyses of Complete Chloroplast Genome in D. linawianum
3.3. IR/SC Boundary Analysis of Chloroplast DNA in D. linawianum
3.4. Characteristics of the Genus Dendrobium Chloroplast Genome
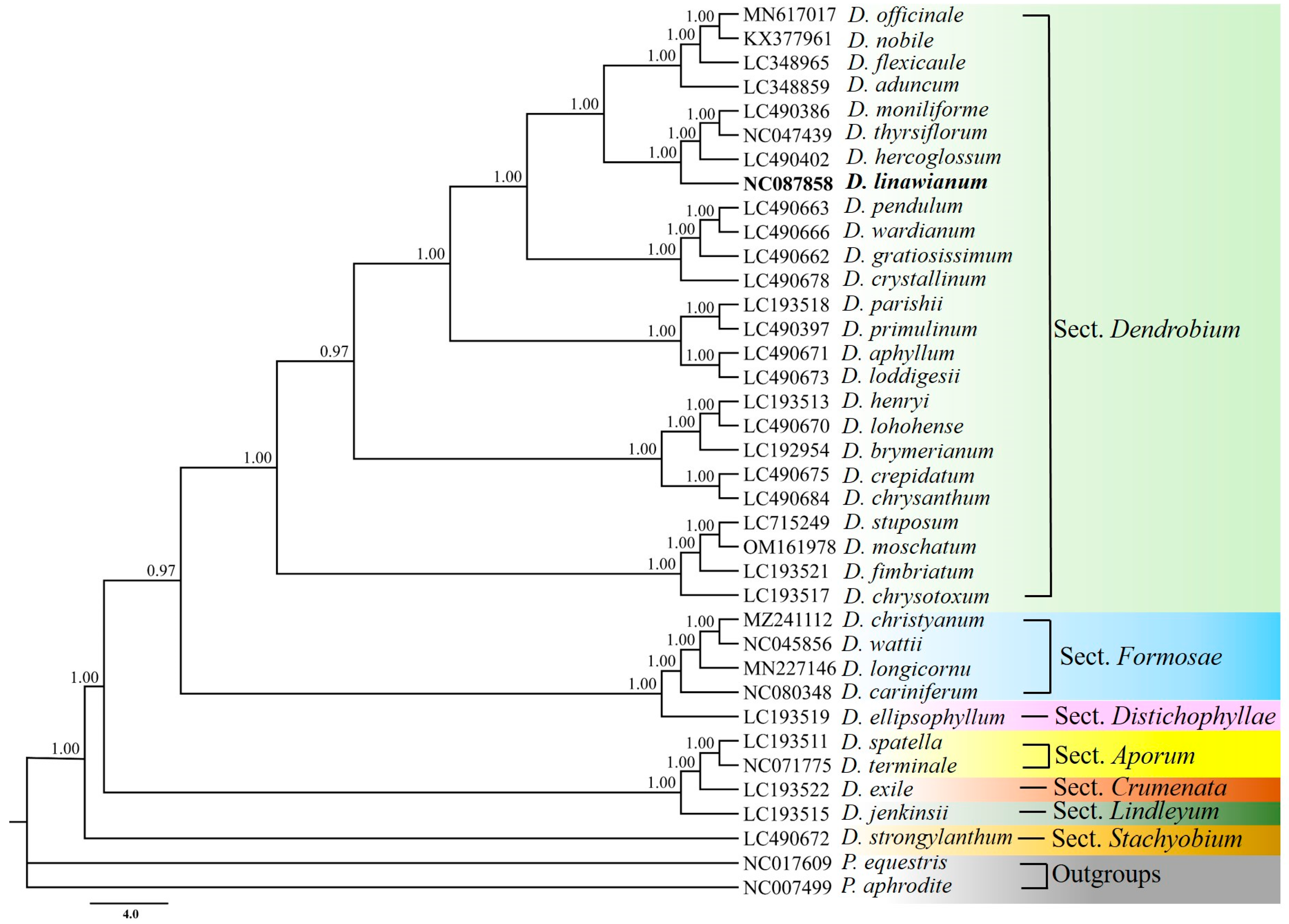
3.5. Phylogenetic Relationships
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xiang, X.G.; Schuiteman, A.; Li, D.Z.; Huang, W.C.; Chung, S.W.; Li, J.W.; Zhou, H.L.; Jin, W.T.; Lai, Y.J.; Li, Z.Y.; et al. Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid and nuclear sequences. Mol. Phylogenet. Evol. 2013, 69, 950–960. [Google Scholar] [CrossRef]
- Xiang, X.G.; Mi, X.C.; Zhou, H.L.; Li, J.W.; Chung, S.W.; Li, D.Z.; Huang, W.C.; Jin, W.T.; Li, Z.Y.; Huang, L.Q.; et al. Biogeographical diversification of mainland Asian Dendrobium (Orchidaceae) and its implications for the historical dynamics of evergreen broad-leaved forests. J. Biogeogr. 2016, 43, 1310–1323. [Google Scholar] [CrossRef]
- Cakova, V.; Bonte, F.; Lobstein, A. Dendrobium: Sources of active ingredients to treat age-related pathologies. Aging Dis. 2017, 8, 827–849. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.S.; Shun, Q.S.; Chen, L.Z. The Medicinal Plants of Dendrobium (Shi-hu) in China; Shanghai Medicinal University Press: Shanghai, China; Fudan University Press: Shanghai, China, 2001. [Google Scholar]
- Wood, H.P. The Dendrobiums; A.R.G Gantner Verlag: Ruggell, Liechtenstein, 2006. [Google Scholar]
- Zhu, G.H.; Ji, Z.H.; Wood, J.J.; Wood, H.P. Dendrobium. In Flora of China; Wu, C.Y., Raven, P.H., Hong, D.Y., Eds.; Scientific Press: Beijing, China, 2009; pp. 367–397. [Google Scholar]
- Teixeirada Silva, J.A.; Jin, X.; Dobránszki, J.; Lu, J.; Wang, H.; Zotz, G.; Cardoso, J.C.; Zeng, S. Advancesin Dendrobium molecular research: Applications in genetic variation, identification and breeding. Mol. Phylogenet. Evol. 2016, 95, 196–216. [Google Scholar] [CrossRef]
- Wang, S.; Xie, Y. The Red List of Chincese Species; Higher Education Press: Beijing, China, 2004; Volume 1. [Google Scholar]
- Ding, G.; Zhang, D.Z.; Ding, X.Y.; Zhou, Q.; Zhang, W.C.; Li, X.X. Genetic variation and conservation of the endangered Chinese endemic herb Dendrobium officinale based on SRAP analysis. Plant Syst. Evol. 2008, 276, 149–156. [Google Scholar] [CrossRef]
- Xu, X.; Liu, X.; Ge, S.; Jensen, J.D.; Hu, F.Y.; Li, X.; Dong, Y.; Gutenkunst, R.N.; Fang, L.; Huang, L.; et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2012, 30, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Yukawa, T.; Uehara, K. Vegetative diversification and radiation in subtribe Dendrobiinae (Orchidaceae): Evidence from chloroplast DNA phylogeny and anatomical characters. Plant Syst. Evol. 1996, 201, 1–14. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, Z.T.; Xu, L.S.; Zhou, K.Y. Application of mitochondrial nad 1 intron 2 sequences to molecular identification of some species of Dendrobium Sw. Chin. Tradit. Herb. Drugs 2005, 36, 1059–1062. [Google Scholar]
- Niu, Z.; Pan, J.; Xue, Q.; Zhu, S.; Liu, W.; Ding, X. Plastome-wide comparison reveals new SNV resources for the authentication of Dendrobium huoshanense and its corresponding medicinal slice (Huoshan Fengdou). Acta Pharmacol. Sin. 2018, 8, 466–477. [Google Scholar] [CrossRef]
- Yukawa, T.; Kita, K.; Handa, T. DNA phylogeny and morphological diversification of Australian Dendrobium (Orchidaceae). In Monocots: Systematics and Evolution; Wilson, K.L., Mossison, D.A., Eds.; CSIRO Publishing: Melbourne, VIC, Australia, 2000; pp. 465–471. [Google Scholar]
- Clements, M.A. Molecular phylogenetic systematics in the Dendrobiinae (Orchidaceae), with emphasis on Dendrobium section Pedilonum. Telopea 2003, 10, 247–298. [Google Scholar] [CrossRef]
- Clements, M.A. Molecular phylogenetic systematics in Dendrobieae (Orchidaceae). Aliso 2006, 22, 465–480. [Google Scholar] [CrossRef]
- Burke, J.M.; Bayly, M.J.; Adams, P.B.; Ladiges, P.Y. Molecular phylogenetic analysis of Dendrobium (Orchidaceae), with emphasis on the Australian section Dendrocoryne, and implications for generic classification. Aust. Sys.Bot. 2008, 21, 1–14. [Google Scholar] [CrossRef]
- Adams, P.B. Systematics of Dendrobiinae (Orchidaceae), with special reference to Australian taxa. Bot. J. Linn. Soc. 2011, 166, 105–126. [Google Scholar] [CrossRef]
- Schuiteman, A. Dendrobium (Orchidaceae): To split or not to split? Gard. Bull. Singap. 2011, 63, 245–257. [Google Scholar]
- Zha, X.Q.; Luo, J.P.; Wei, P. Identification and classification of Dendrobium candidum species by fingerprint technology with capillary electrophoresis. S. Afr. J. Bot. 2009, 75, 276–282. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Z.; Xu, L. Simultaneous determination of phenols (bibenzyl, phenanthrene, and fluorenone). Dendrobium species by high-performance liquid chromatography with diode array detection. J. Chromatogr. A 2006, 1104, 230–237. [Google Scholar]
- Takamiya, T.; Wongsawad, P.; Sathapattayanon, A.; Tajima, N.; Suzuki, S.; Kitamura, S.; Iijima, H.; Yukawa, T. Molecular phylogenetics and character evolution of morphologically diverse groups, Dendrobium section Dendrobium and allies. AoB Plants 2014, 6, plu045. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.W.; Luo, J.; Zhang, Y.S.; Niu, Z.T.; Xue, Q.Y.; Ding, X.Y. Iteration expansion and regional evolution: Phylogeography of Dendrobium officinale and four related taxa in southern China. Sci. Rep. 2017, 7, 43525. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Ding, X.; Liu, D.; Ding, G.; He, J.; Li, X.; Tang, F.; Chu, B. Intersimple sequence repeats (ISSR) molecular finger printing markers for authenticating populations of Dendrobium officinale Kimura et Migo. Biol. Pharm. Bull. 2006, 29, 420–422. [Google Scholar] [CrossRef]
- Feng, S.G.; Lu, J.J.; Gao, L.; Liu, J.J.; Wang, H.Z. Molecular phylogeny analysis and species identification of Dendrobium (Orchidaceae) in China. Biochem. Genet. 2014, 52, 127–136. [Google Scholar] [CrossRef]
- Kang, J.Y.; Lu, J.J.; Qiu, S.; Chen, Z.; Liu, J.J.; Wang, H.Z. Dendrobium SSR markers play a good role in genetic diversity and phylogenetic analysis of Orchidaceae species. Sci. Hortic. 2015, 183, 160–166. [Google Scholar] [CrossRef]
- Tsai, C.C.; Peng, C.I.; Huang, S.C.; Huang, P.L.; Chou, C.H. Determination of the genetic relationship of Dendrobium species (Orchidaceae) in Taiwan based on the sequence of the internal transcribed spacer of ribosomal DNA. Sci. Hortic. 2004, 101, 315–325. [Google Scholar] [CrossRef]
- Xu, S.Z.; Li, D.Z.; Li, J.W.; Xiang, X.G.; Jin, W.T.; Huang, W.C.; Jin, X.H.; Huang, L.Q. Evaluation of the DNA barcodes in Dendrobium (Orchidaceae) from mainland Asia. PLoS ONE 2015, 10, e0115168. [Google Scholar] [CrossRef]
- Zhu, B.; Gan, C.C.; Wang, H.C. Characteristics of the complete chloroplast genome of Dendrobium thyrsiflorum and its phylogenetic relationship analysis. Biotechnol. Bull. 2021, 37, 38–47. [Google Scholar]
- Li, Z.W.; Qiu, Q.; Lang, J.Q.; Wu, Y.M.; Du, H.H.; Zhou, N. Sequence analysis of complete chloroplast genome of Dendrobium heterocarpum Lindl. and Dendrobium trigonopus Rchb. f. Chin. Tradit. Herb. Drugs 2022, 53, 5159–5169. [Google Scholar]
- Shang, M.Y.; Wang, J.L.; Zhou, Y.; Zhang, M.C.; Liu, Y.L.; Duan, B.Z. Analysis of chloroplast genome structure and phylogeny of endangered Dendrobium devonianum. Chin. Tradit. Herb. Drug 2023, 54, 6424–6433. [Google Scholar]
- Wicke, S.; Schneeweiss, G.M.; de Pamphilis, C.W.; Muller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ezpeleta, N.; Brinkmann, H.; Burey, S.C.; Roure, B.; Burger, G.; Löffelhardt, W.; Bohnert, H.J.; Philippe, H.; Lang, B.F. Monophyly of primary photosynthetic eukaryotes: Green plants, red algae, and glaucophytes. Curr. Biol. 2005, 15, 1325–1330. [Google Scholar] [CrossRef] [PubMed]
- Du, X.Y.; Zeng, T.; Feng, Q.; Hu, L.; Luo, X.; Weng, Q.; He, J.; Zhu, B. The complete chloroplast genome sequence of yellow mustard (Sinapis alba L.) and its phylogenetic relationship to other Brassicaceae species. Gene 2020, 731, 144340. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.F.; Nalawade, S.M.; Mulabagal, V.; Matthew, S.; Chen, C.L.; Kuo, C.L.; Tsay, H.S. In vitro propagation by asymbiotic seed germination and 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical scavenging activity studies of tissue culture raised plants of three medicinally important species of Dendrobium. Biol. Pharm. Bull. 2004, 27, 731–735. [Google Scholar] [CrossRef]
- Ma, L.; Chen, S.Q.; Zhuang, L.B. Evaluation on Ornamental Characters of 35 Species of Dendrobium. Subtop. Plant Sci. 2019, 48, 269–273. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; De Pamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Shi, L.C.; Chen, H.M.; Jiang, M.; Wang, L.Q.; Wu, X.; Huang, L.F.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Kim, K.J.; Lee, H.L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004, 11, 247–261. [Google Scholar] [CrossRef]
- Park, S.; An, B.; Park, S. Reconfiguration of the plastid genome in Lamprocapnos spectabilis: IR boundary shifting, inversion, and intraspecific variation. Sci. Rep. 2018, 8, 13568. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.C.; Chen, S.Y.; Zhang, X.Z. Whole-genome comparison reveals divergent IR borders and mutation hotspots in chloroplast genomes of herbaceous bamboos (bambusoideae: Olyreae). Molecules 2018, 23, 1537. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, P.; Basak, S.; Ghosh, T.C. Nature of selective constraints on synonymous codon usage of rice differs in GC-poor and GC-rich genes. Gene 2007, 400, 71–81. [Google Scholar] [CrossRef]
- Gao, Y.X.; Zhou, Y.Y.; Xie, Y.; Feng, L.; Shen, S.G. The complete chloroplast genome sequence of an endangered Orchidaceae species Dendrobium monilforme and its phylogenetic implications. Conserv. Genet. Resour. 2018, 10, 397–399. [Google Scholar] [CrossRef]
- Zhu, A.; Guo, W.; Gupta, S.; Fan, W.; Mower, J.P. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016, 209, 1747–1756. [Google Scholar] [CrossRef]
- Saha, M.C.; Cooper, J.D.; Mian, M.A.; Chekhovskiy, K.; May, G.D. Tall fescue genomic SSR markers: Development and transferability across multiple grass species. Theor. Appl. Genet. 2006, 113, 1449–1458. [Google Scholar] [CrossRef]
- Curci, P.L.; De Paola, D.; Danzi, D.; Vendramin, G.G.; Sonnante, G. Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other Asteraceae. PLoS ONE 2015, 10, e0120589. [Google Scholar] [CrossRef]
- Yap, J.Y.; Rohner, T.; Greenfield, A.; Van Der Merwe, M.; McPherson, H.; Glenn, W.; Kornfeld, G.; Marendy, E.; Pan, A.Y.; Wilton, A.; et al. Complete chloroplast genome of the Wollemi pine (Wollemia nobilis): Structure and evolution. PLoS ONE 2015, 10, e0128126. [Google Scholar] [CrossRef]
- Park, I.; Kim, W.J.; Yeo, S.M.; Choi, G.; Kang, Y.M.; Piao, R.Z.; Moon, B.C. The Complete Chloroplast Genome Sequences of Fritillaria ussuriensis Maxim. and Fritillaria cirrhosa D. Don, and Comparative Analysis with Other Fritillaria Species. Molecules 2017, 22, 982. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Li, W.H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Shi, C.; Liu, Y.; Mao, S.Y.; Gao, L.Z. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: Genome structure and phylogenetic relationships. BMC Evol. Biol. 2014, 14, 151. [Google Scholar] [CrossRef]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Lin, H.C.; Lin, I.P.; Chow, T.Y.; Chen, H.H.; Chen, W.H.; Cheng, C.H.; Lin, C.Y.; Liu, S.M.; Chang, C.C.; et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): Comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 2006, 23, 279–291. [Google Scholar] [CrossRef]
- Wang, B.; Qiu, Y.L. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 2006, 16, 299–363. [Google Scholar] [CrossRef]
- Martín, M.; Sabater, B. Plastid ndh genes in plant evolution. Plant Physiol. Biochem. 2010, 48, 636–645. [Google Scholar] [CrossRef]
- Selosse, M.A.; Albert, B.; Godelle, B. Reducing the genome size of organelles favours gene transfer to the nucleus. Trends Ecol. Evol. 2001, 16, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.L.; Palmer, J.D. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 2003, 29, 380–395. [Google Scholar] [CrossRef]
- Gomez, S.M.; Brown, M.J.M.; Pironon, S.; Bureš, P.; Verde Arregoitia, L.D.; Veselý, P.; Elliott, T.L.; Zedek, F.; Pellicer, J.; Forest, F.; et al. Genome size is positively correlated with extinction risk in herbaceous angiosperms. New Phytol. 2024, 243, 2470–2485. [Google Scholar] [CrossRef]
- Yang, J.B.; Tang, M.; Li, H.T.; Zhang, Z.R.; Li, D.Z. Complete chloroplast genome of the genus Cymbidium: Lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013, 13, 84. [Google Scholar] [CrossRef]
- Hu, H.; Hu, Q.; Al-Shehbaz, I.A.; Luo, X.; Zeng, T.; Guo, X.; Liu, J. Species delimitation and interspecific relationships of the genus Orychophragmus (Brassicaceae) inferred from whole chloroplast genomes. Front. Plant Sci. 2016, 7, 1826. [Google Scholar] [CrossRef]
- Du, Y.P.; Bi, Y.; Yang, F.P.; Zhang, M.F.; Chen, X.Q.; Xue, J.; Zhang, X.H. Complete chloroplast genome sequences of Lilium: Insights into evolutionary dynamics and phylogenetic analyses. Sci. Rep. 2017, 7, 5751. [Google Scholar] [CrossRef]
- Yu, X.Q.; Drew, B.T.; Yang, J.B.; Gao, L.M.; Li, D.Z. Comparative chloroplast genomes of eleven Schima (Theaceae) species: Insights into DNA barcoding and phylogeny. PLoS ONE 2017, 12, e0178026. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.Y.; Niu, Z.T.; Xue, Q.Y.; Wang, H.; Xie, X.Z.; Ding, X.Y. Accurate authentication of Dendrobium officinale and its closely related species by comparative analysis of complete plastomes. Acta Pharm. Sin. B 2018, 8, 969–980. [Google Scholar] [CrossRef]
- Kim, Y.K.; Jo, S.; Cheon, S.H.; Kwak, M.; Kim, Y.D.; Kim, K.J. Plastome evolution and phylogeny of subtribe Aeridinae (Vandeae, Orchidaceae). Mol. Phylogenet. Evol. 2020, 144, 106721. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.K.; Tu, X.D.; Zhao, Z.; Zeng, M.Y.; Zhang, S.; Ma, L.; Zhang, G.Q.; Wang, M.M.; Liu, Z.J.; Lan, S.R.; et al. Plastid phylogenomic data yield new and robust insights into the phylogeny of Cleisostoma-Gastrochilus clades (Orchidaceae, Aeridinae). Mol. Phylogenet. Evol. 2020, 145, 106729. [Google Scholar] [CrossRef]

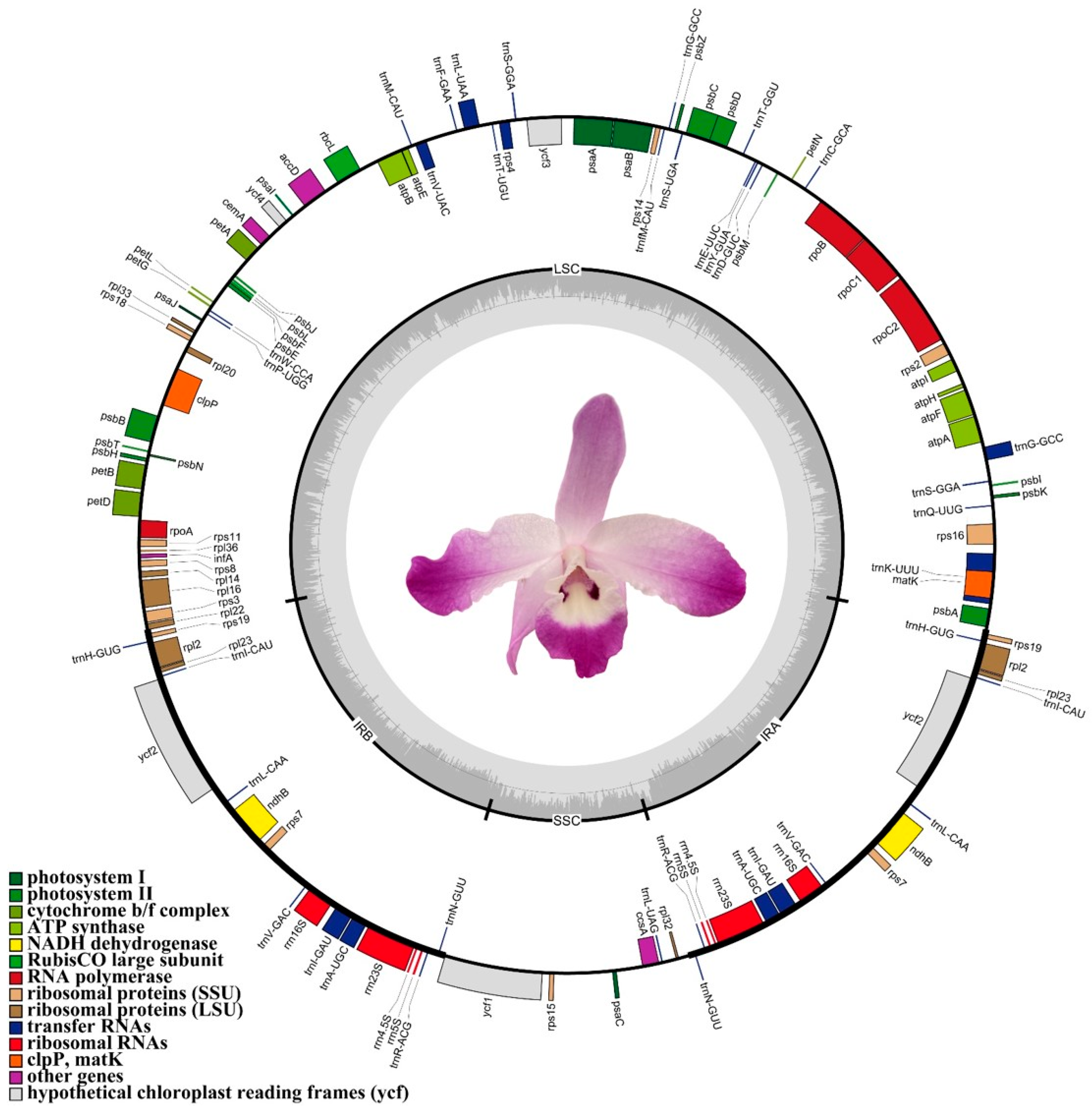
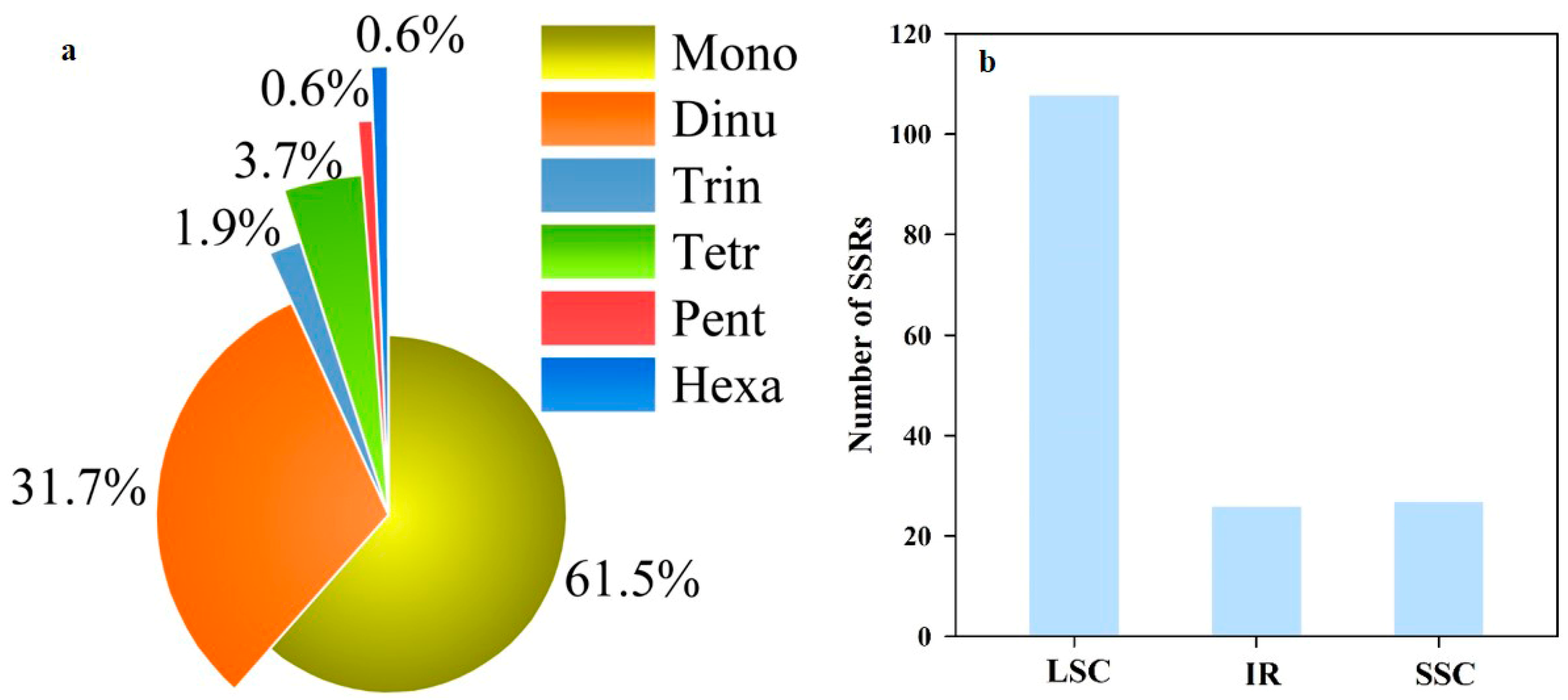

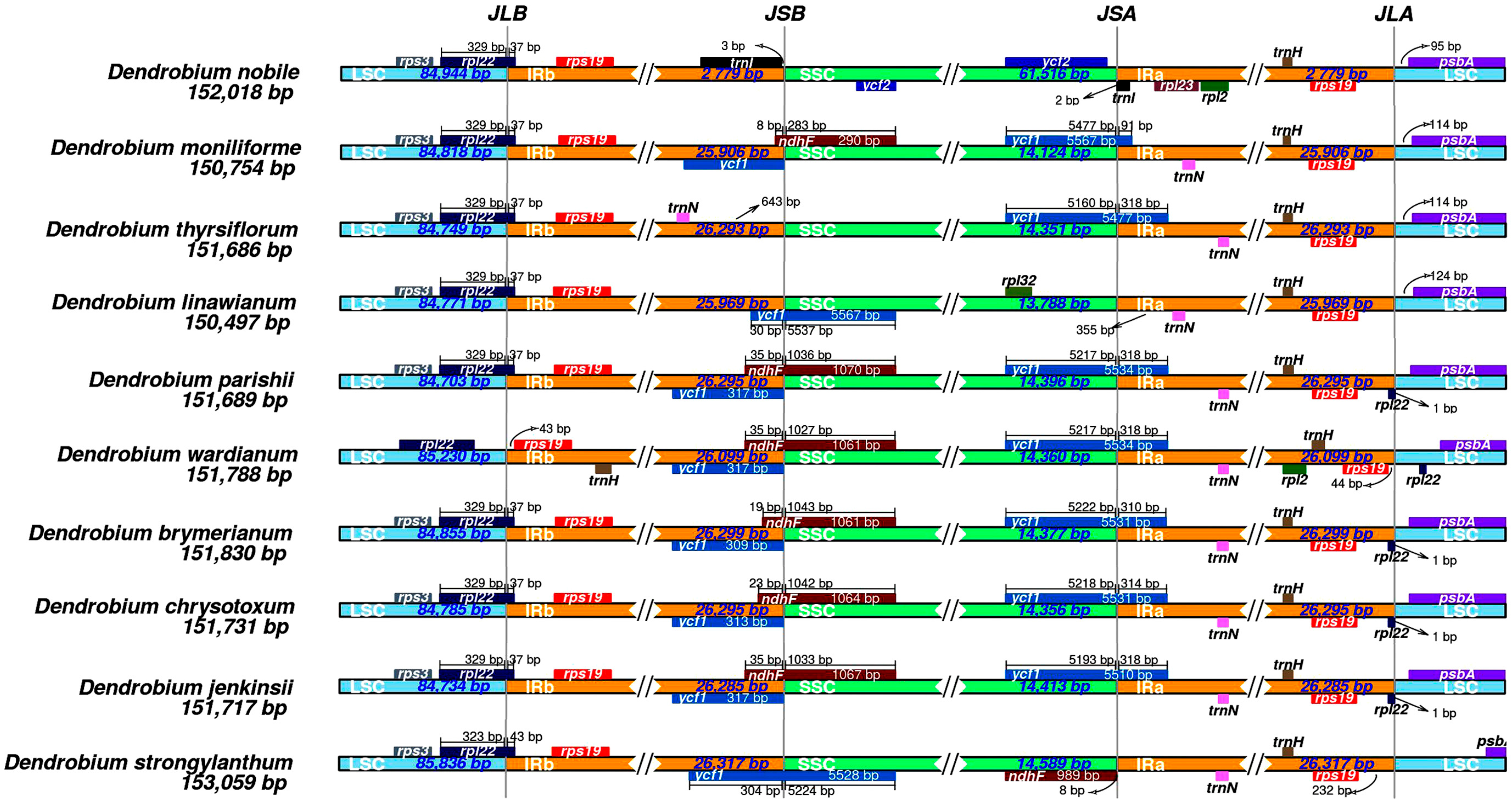
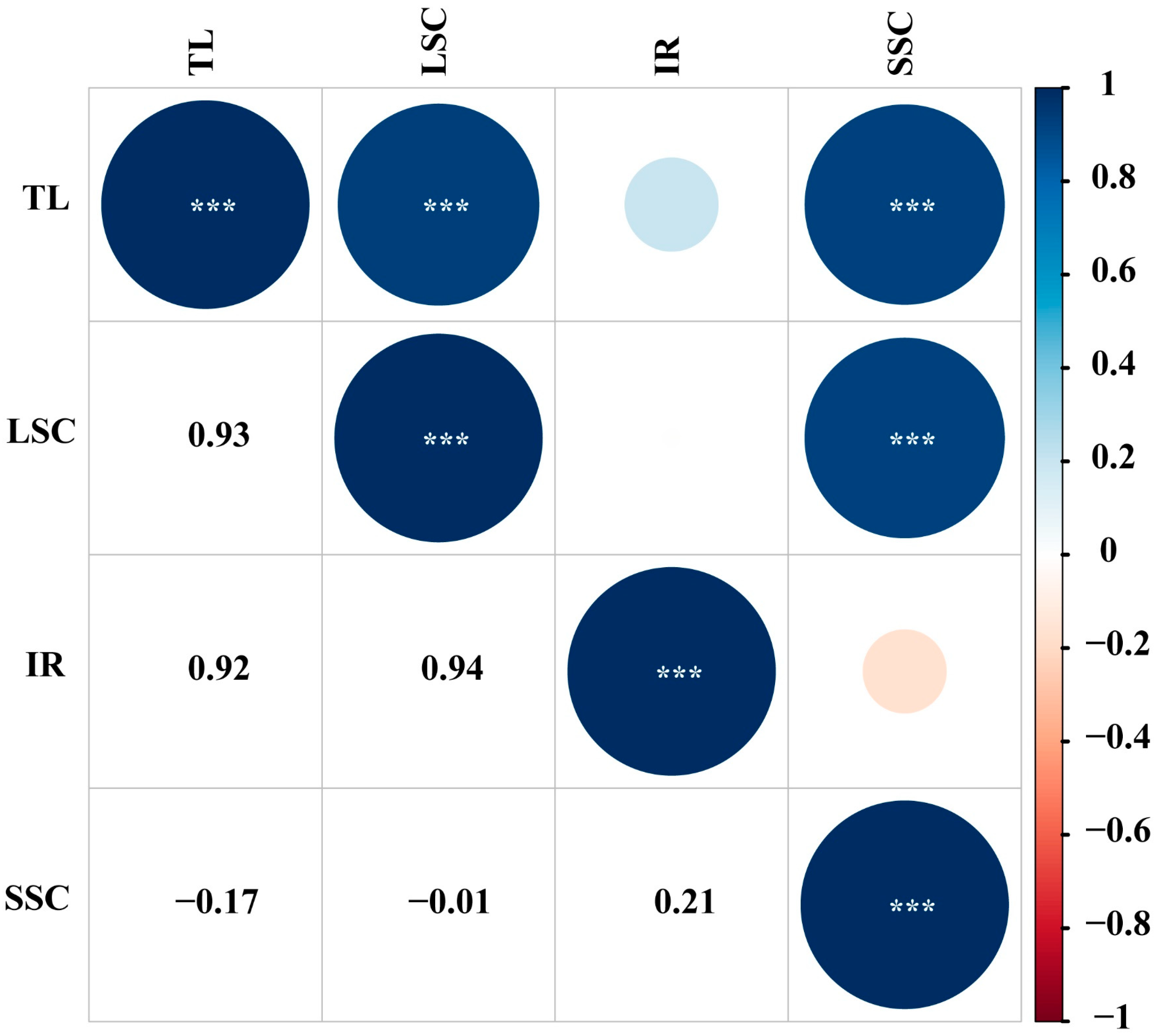

| Region | Size (bp) | T% | C% | A% | G% | A+T% | G+C% |
|---|---|---|---|---|---|---|---|
| Whole genome | 150,497 | 31.45 | 18.94 | 30.99 | 18.62 | 62.44 | 37.56 |
| LSC | 84,771 | 33.18 | 17.95 | 31.69 | 17.18 | 64.87 | 35.13 |
| SSC | 13,788 | 32.7 | 14.59 | 36.82 | 15.89 | 69.52 | 30.48 |
| IRA | 25,969 | 28.28 | 20.96 | 28.3 | 22.46 | 56.58 | 43.42 |
| IRB | 25,969 | 28.28 | 20.96 | 28.3 | 22.46 | 56.58 | 43.42 |
| Types of Genes | Group of Genes | Name of Genes |
|---|---|---|
| Photosystem I gene | psaA, psaB, psaC, psaI, psaJ | |
| Photosystem II gene | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Photosynthesis | Cytochrome b/f complex gene | petA, petB *, petD *, petG, petL, petN |
| ATP synthase gene | atpA, atpB, atpE, atpF *, atpH, atpI | |
| Rubisco gene | rbcL | |
| NADH-dehydrogenase gene | ndhB a* | |
| Self-replication genes | RNA polymerase gene | rpoA, rpoB, rpoC1 *, rpoC2 |
| Large subunit of the ribosome | rpl14, rpl16 *, rpl2 a*, rpl20, rpl22, rpl23 a, rpl32, rpl33, rpl36 | |
| Small subunit of the ribosome | rps11, rps14, rps15, rps16 *, rps18, rps19 a, rps2, rps3, rps4, rps7 a, rps8 | |
| Ribosomal RNAs gene | rrn4.5S a, rrn5S a, rrn16S a, rrn23S a | |
| Transfer RNAs gene | trnK-UUU *, trnQ-UUG, trnG-GCC a*, trnC-GCA, trnD-GUC, trnY-GUA, trnE-UUC, trnT-GGU, trnS-UGA, trnS-GGA a, trnT-UGU, trnL-UAA *, trnF-GAA, trnV-UAC *, trnW-CCA, trnP-UGG, trnH-GUG a, trnM-CAU, trnL-CAA a, trnV-GAC a, trnI-GAU a*, trnI-CAU a, trnfM-CAU, trnA-UGC a*, trnR-ACG a, trnN-GUU a, trnL-UAG | |
| Other genes | Translational initiation factor gene | infA |
| Maturase K gene | matK | |
| Subunit of the Acetyl-CoA-carboxylase gene | accD | |
| Envelope membrane protein gene | cemA | |
| c-type cytochrom synthesis gene | ccsA | |
| Protease gene | clpP ** | |
| Unkown genes | Conserved open reading frames | ycf1, ycf2 a, ycf3 **, ycf4 |
| Gene | Location | Start Codon | Stop Codon | Exon I/bp | Intron I/bp | Exon II/bp | Intron II/bp | Exon III/bp |
|---|---|---|---|---|---|---|---|---|
| trnK-UUU | LSC | GGG | CCA | 37 | 2781 | 35 | ||
| rps16 | LSC | ATG | TAA | 40 | 891 | 248 | ||
| trnG-GCC | LSC | GCG | GCT | 31 | 671 | 59 | ||
| atpF | LSC | ATG | TAG | 145 | 939 | 410 | ||
| rpoC1 | LSC | ATG | TAG | 432 | 761 | 1608 | ||
| ycf3 | LSC | ATG | TAA | 124 | 721 | 230 | 745 | 153 |
| trnL-UAA | LSC | GGG | CCA | 35 | 793 | 50 | ||
| trnV-UAC | LSC | AGG | CTA | 39 | 581 | 37 | ||
| clpP | LSC | ATG | TAA | 71 | 959 | 294 | 671 | 229 |
| petB | LSC | ATG | TAG | 6 | 728 | 642 | ||
| petD | LSC | ATG | TAA | 8 | 860 | 496 | ||
| rpl16 | LSC | ATG | TAG | 9 | 1170 | 399 | ||
| rpl2 | IR | ATA | TAG | 385 | 663 | 431 | ||
| ndhB | IR | ATG | TAG | 775 | 699 | 758 | ||
| trnI-GAU | IR | GGG | CCA | 37 | 944 | 35 | ||
| trnA-UGC | IR | GGG | TCC | 38 | 801 | 34 |
| Amino Acid | Codon | Number | RSCU | Amino Acid | Codons | Number | RSCU |
|---|---|---|---|---|---|---|---|
| Ala | GCA | 642 | 1.19 | Pro | CCA | 473 | 1.09 |
| GCC | 286 | 0.53 | CCC | 386 | 0.89 | ||
| GCG | 237 | 0.44 | CCG | 218 | 0.50 | ||
| GCU | 986 | 1.83 | CCU | 653 | 1.51 | ||
| Cys | UGC | 158 | 0.60 | Gln | CAA | 1213 | 1.48 |
| UGU | 372 | 1.40 | CAG | 425 | 0.52 | ||
| Asp | GAC | 345 | 0.38 | Arg | AGA | 854 | 1.88 |
| GAU | 1479 | 1.62 | AGG | 333 | 0.73 | ||
| Glu | GAA | 1782 | 1.46 | CGA | 590 | 1.30 | |
| GAG | 655 | 0.54 | CGC | 149 | 0.33 | ||
| Phe | UUC | 983 | 0.78 | CGG | 203 | 0.45 | |
| UUU | 1546 | 1.22 | CGU | 601 | 1.32 | ||
| Gly | GGA | 1060 | 1.55 | Ser | AGC | 213 | 0.36 |
| GGC | 283 | 0.41 | AGU | 700 | 1.18 | ||
| GGG | 503 | 0.74 | UCA | 660 | 1.12 | ||
| GGU | 889 | 1.30 | UCC | 600 | 1.01 | ||
| His | CAC | 257 | 0.45 | UCG | 328 | 0.55 | |
| CAU | 877 | 1.55 | UCU | 1050 | 1.77 | ||
| Ile | AUA | 1029 | 0.88 | Thr | ACA | 648 | 1.22 |
| AUC | 787 | 0.68 | ACC | 378 | 0.71 | ||
| AUU | 1676 | 1.44 | ACG | 251 | 0.47 | ||
| Lys | AAA | 1805 | 1.42 | ACU | 855 | 1.60 | |
| AAG | 745 | 0.58 | Val | GUA | 788 | 1.40 | |
| Leu | CUA | 617 | 0.85 | GUC | 302 | 0.54 | |
| CUC | 317 | 0.43 | GUG | 373 | 0.66 | ||
| CUG | 341 | 0.47 | GUU | 785 | 1.4 | ||
| CUU | 911 | 1.25 | Trp | UGG | 812 | 1.00 | |
| UUA | 1253 | 1.72 | Tyr | UAC | 313 | 0.42 | |
| UUG | 942 | 1.29 | UAU | 1165 | 1.58 | ||
| Met | AUG | 1001 | 1.00 | Terminater | UAA | 114 | 1.08 |
| Asn | AAC | 483 | 0.47 | UAG | 106 | 1.01 | |
| AAU | 1575 | 1.53 | UGA | 95 | 0.90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Huang, Q.; Zhang, Y.; Lǚ, D.; Chen, R.; Jia, Y.; Li, Q. Determination of the Phylogenetic Relationship of Dendrobium linawianum (Orchidaceae) Based on Comparative Analysis of Complete Chloroplast Genomes. Curr. Issues Mol. Biol. 2025, 47, 869. https://doi.org/10.3390/cimb47100869
Zhang F, Huang Q, Zhang Y, Lǚ D, Chen R, Jia Y, Li Q. Determination of the Phylogenetic Relationship of Dendrobium linawianum (Orchidaceae) Based on Comparative Analysis of Complete Chloroplast Genomes. Current Issues in Molecular Biology. 2025; 47(10):869. https://doi.org/10.3390/cimb47100869
Chicago/Turabian StyleZhang, Fengping, Qiyong Huang, Yaqiong Zhang, Dongqin Lǚ, Rui Chen, Yanshu Jia, and Qiongchao Li. 2025. "Determination of the Phylogenetic Relationship of Dendrobium linawianum (Orchidaceae) Based on Comparative Analysis of Complete Chloroplast Genomes" Current Issues in Molecular Biology 47, no. 10: 869. https://doi.org/10.3390/cimb47100869
APA StyleZhang, F., Huang, Q., Zhang, Y., Lǚ, D., Chen, R., Jia, Y., & Li, Q. (2025). Determination of the Phylogenetic Relationship of Dendrobium linawianum (Orchidaceae) Based on Comparative Analysis of Complete Chloroplast Genomes. Current Issues in Molecular Biology, 47(10), 869. https://doi.org/10.3390/cimb47100869





