Integrated Transcriptome and Metabolome Analyses Reveal the Roles of MADS-Box Genes in Regulating Flower Development and Metabolite Accumulation in Osmanthus fragran
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources and Identification of MADS-Box Genes in Osmanthus fragrans
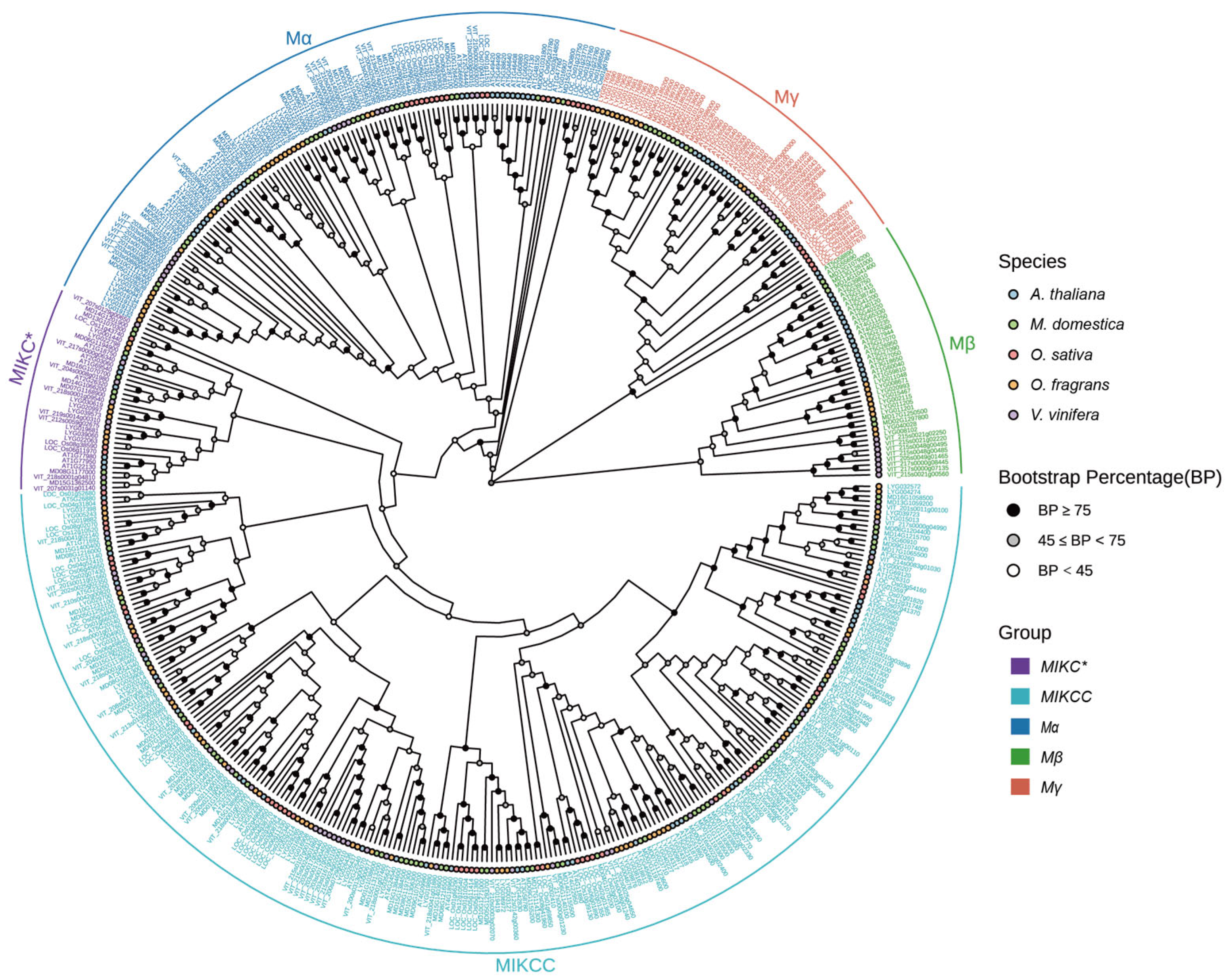
2.2. Phylogenetic Analysis of OfMADS Genes
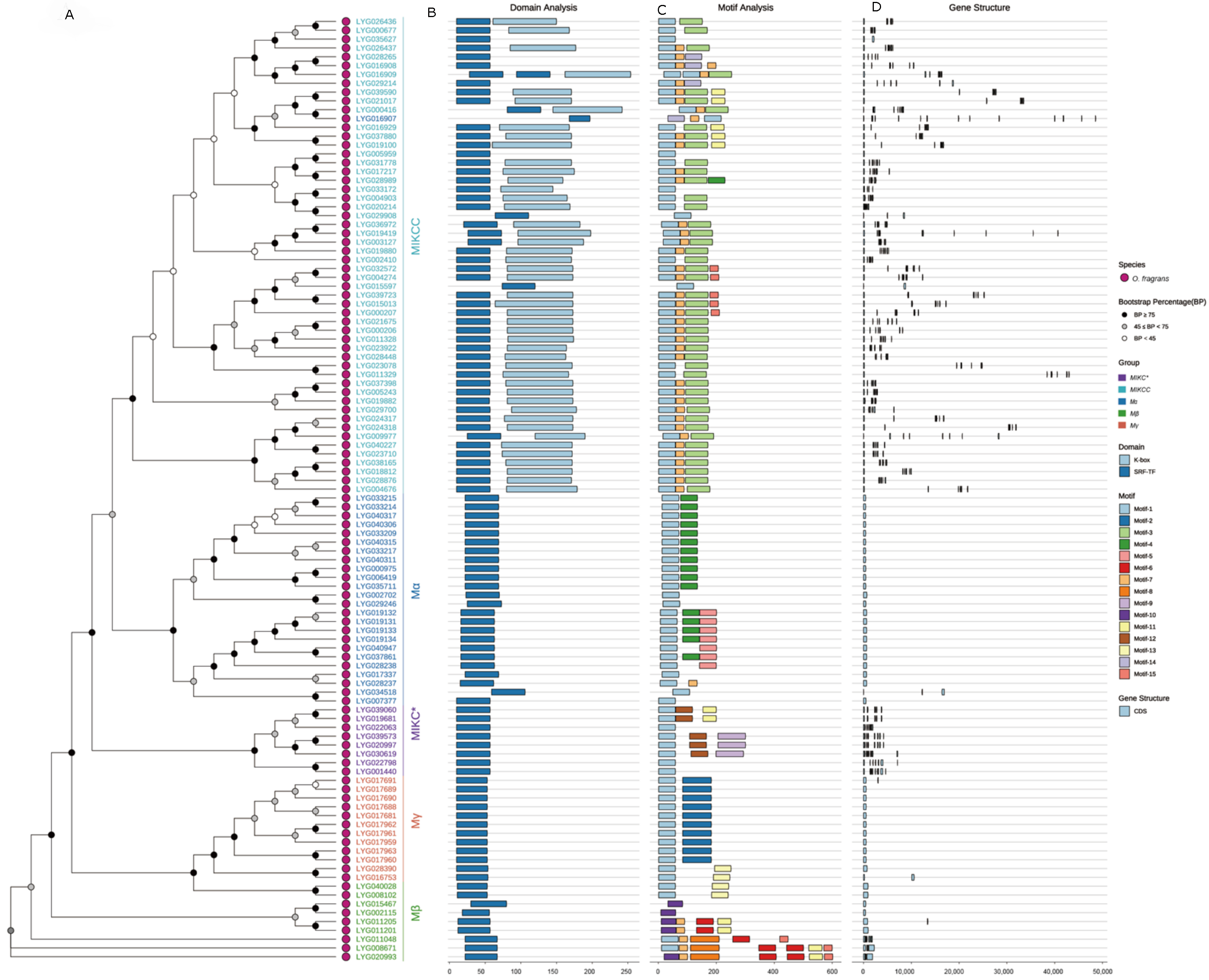
2.3. Domain, Gene Structures, Conserved Motifs Analysis of OfMADS Genes
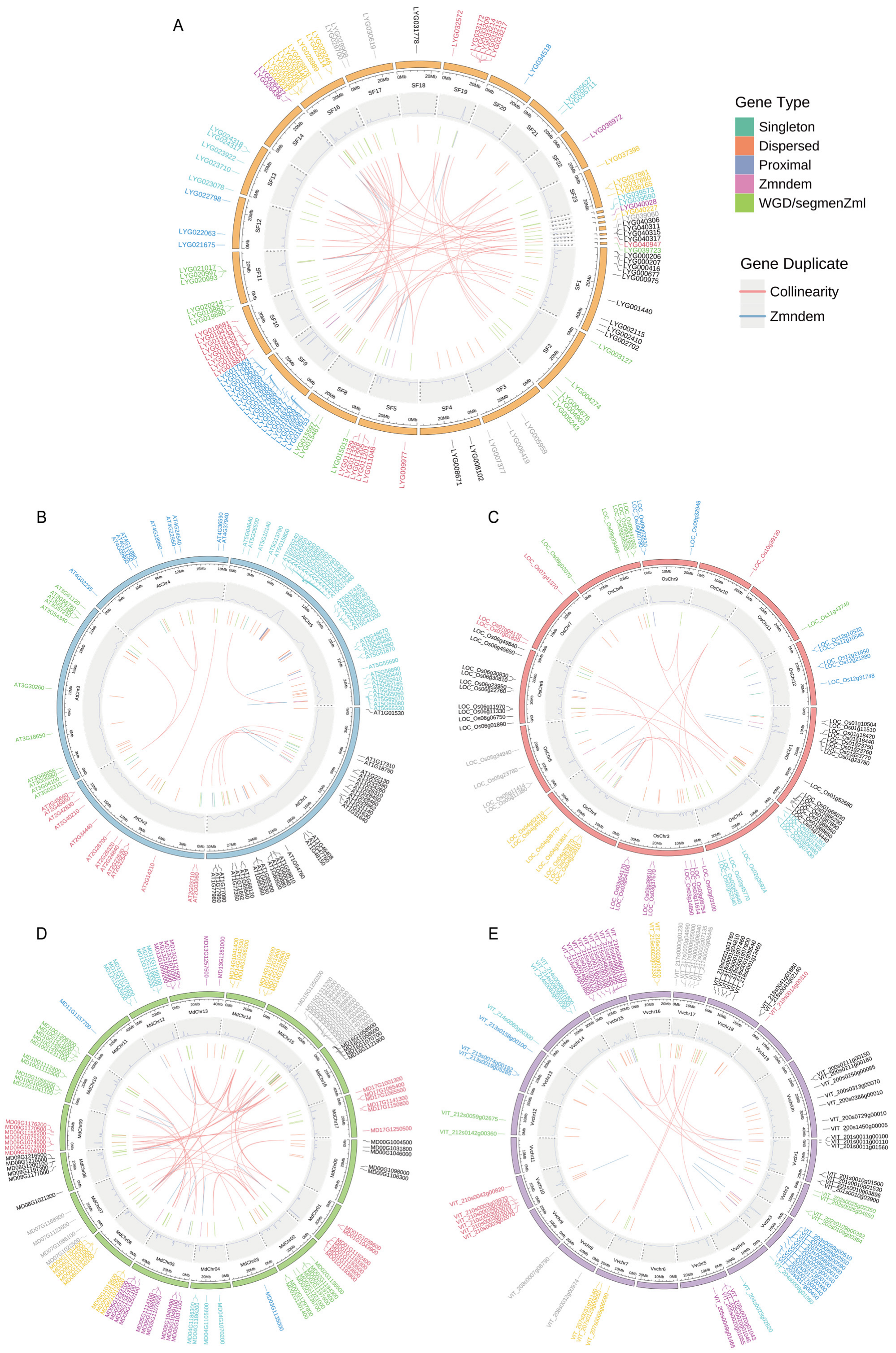
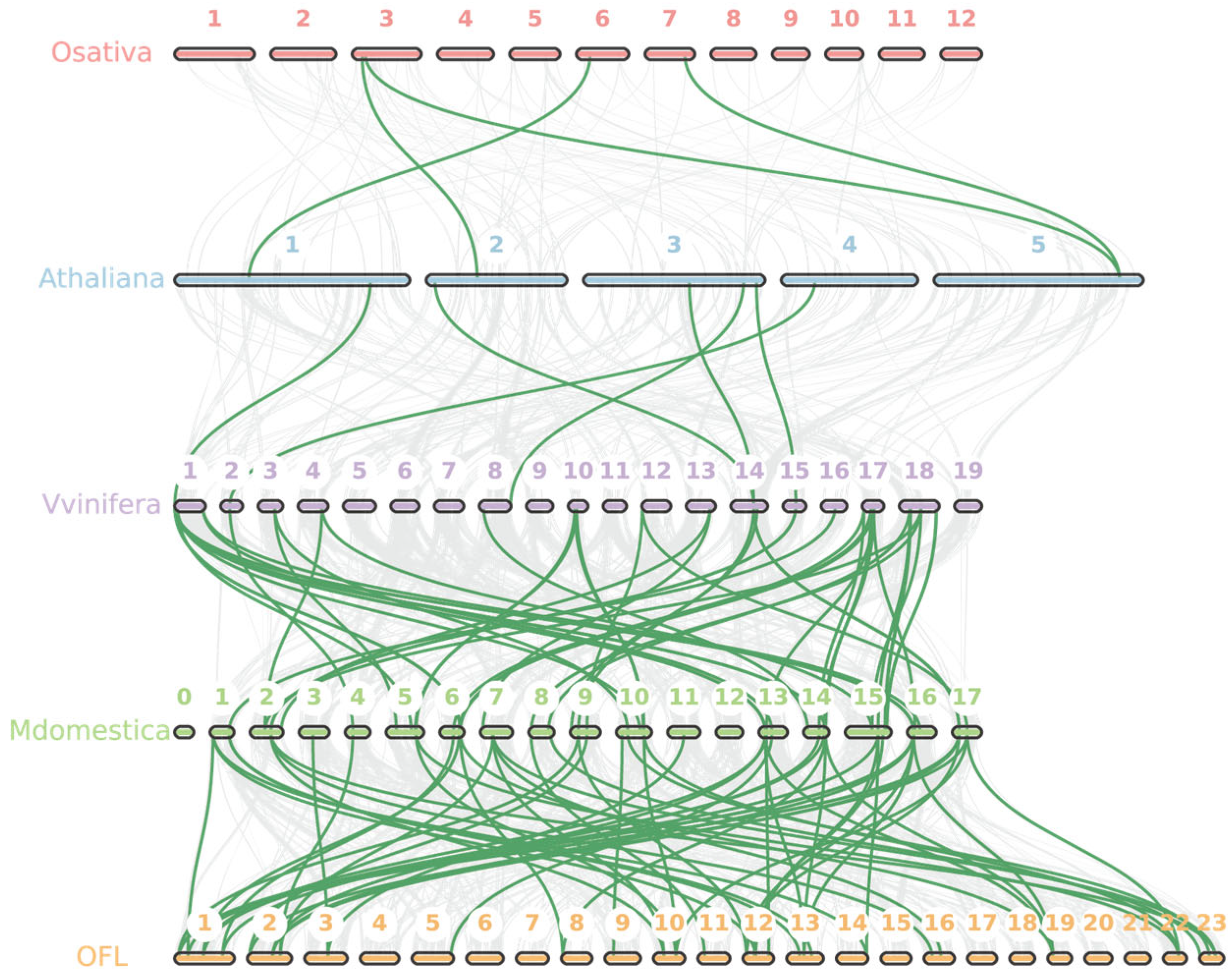
2.4. Analysis of Chromosome Distribution, Gene Duplication Events, and Selection Pressure
2.5. Analysis of Cis-Elements in the Promoter of OfMADS Genes
2.6. OfMADS Protein Interaction Network Analysis
2.7. Total RNA Extraction and RNA-Seq
2.8. Metabolomics and Data Analysis
2.9. Correlation Analysis
3. Results
3.1. Genome-Wide Identification of the OfMADS Genes
3.2. Phylogenetic Analysis of the OfMADS
3.3. Conserved Motifs, Conserved Domain and Gene Structure of the OfMADS
3.4. Gene Location and Gene Duplication Events Analysis of OfMADS Genes
3.5. Duplicated Gene Analysis
3.6. Analysis of Promoter Elements
3.7. Interaction Network of MADS-Box Proteins
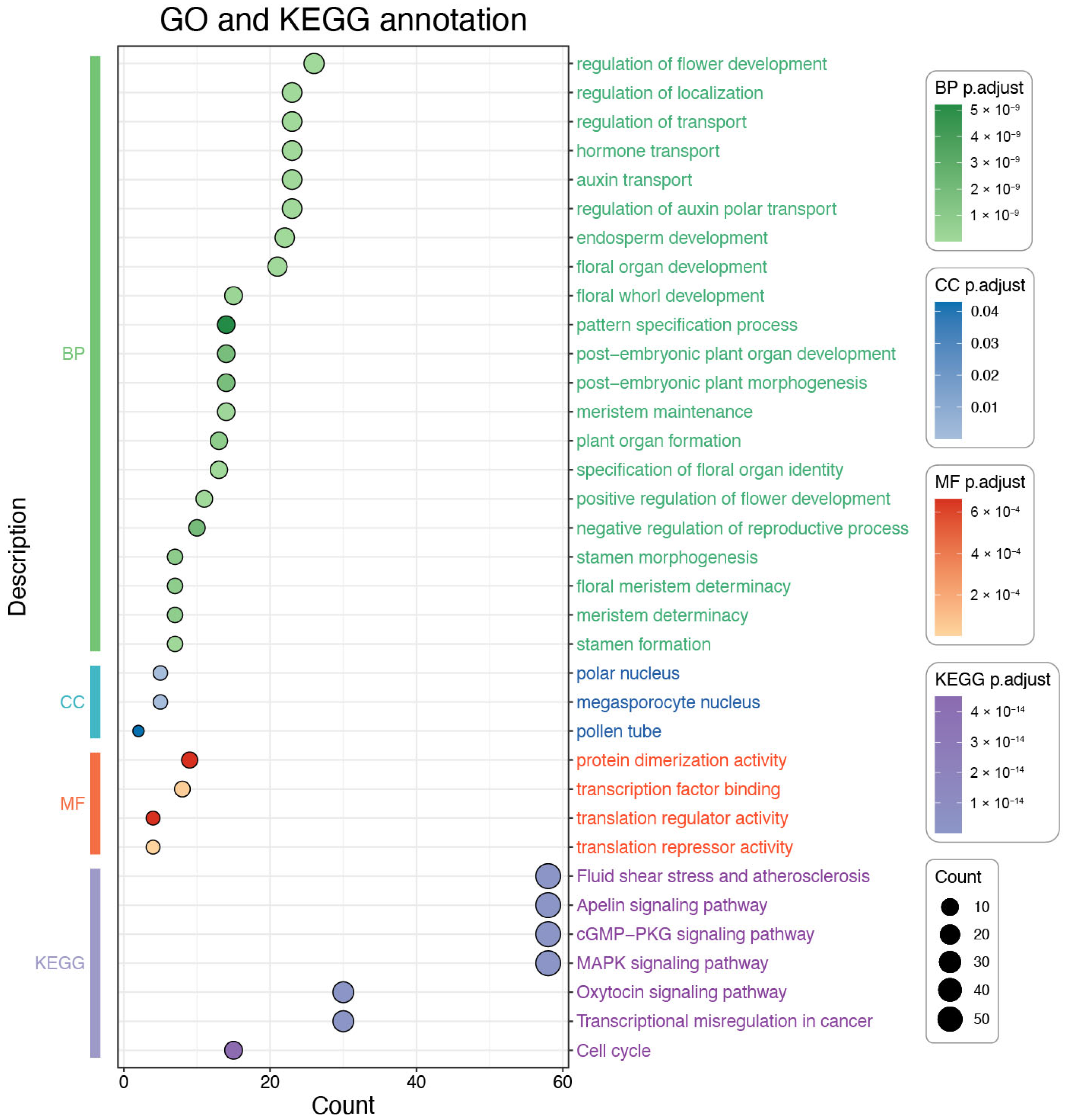
3.8. GO and KEGG Enrichment Analysis of the OfMADS Genes
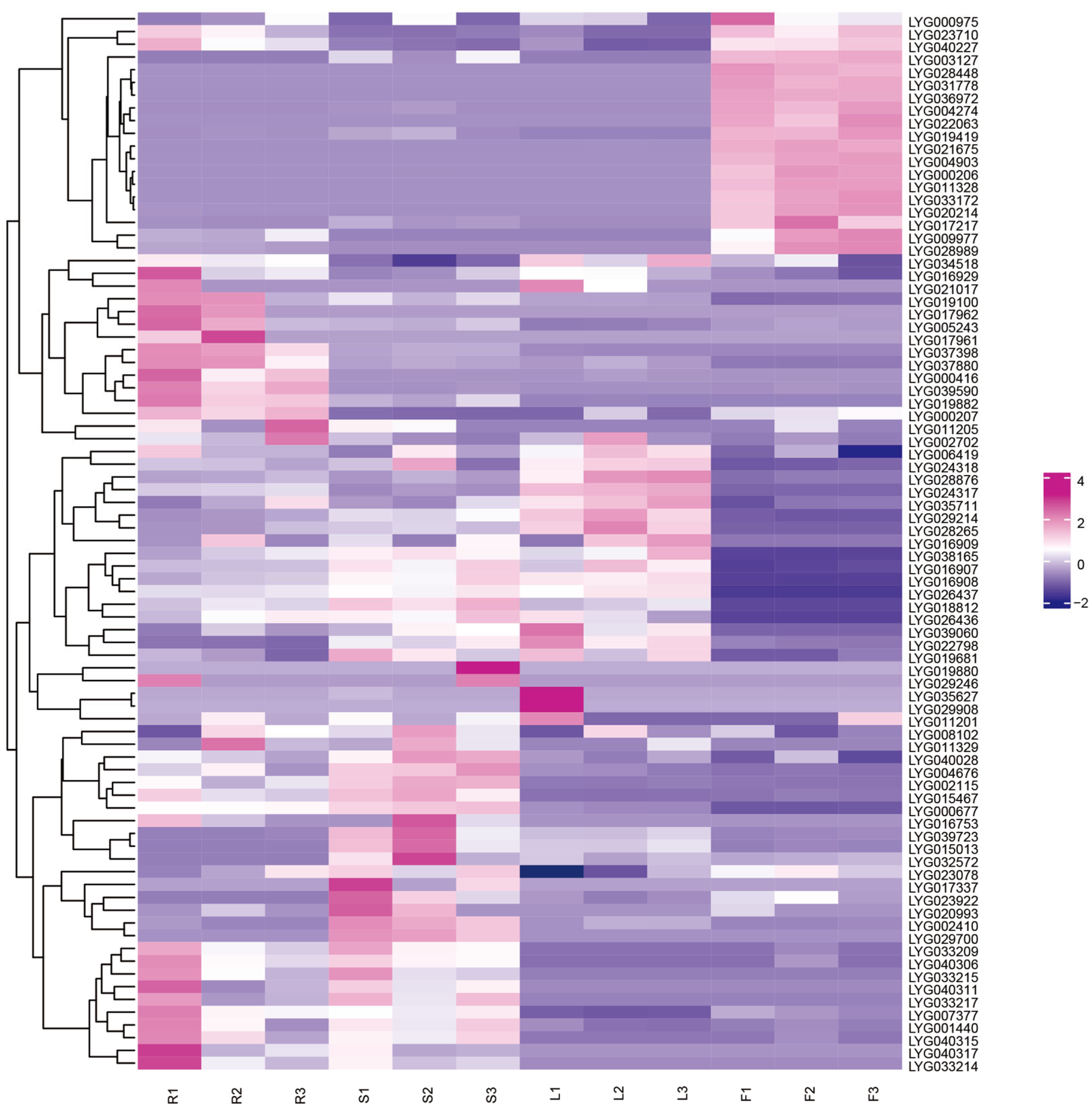
3.9. Transcriptome Data Analysis of Different Tissues
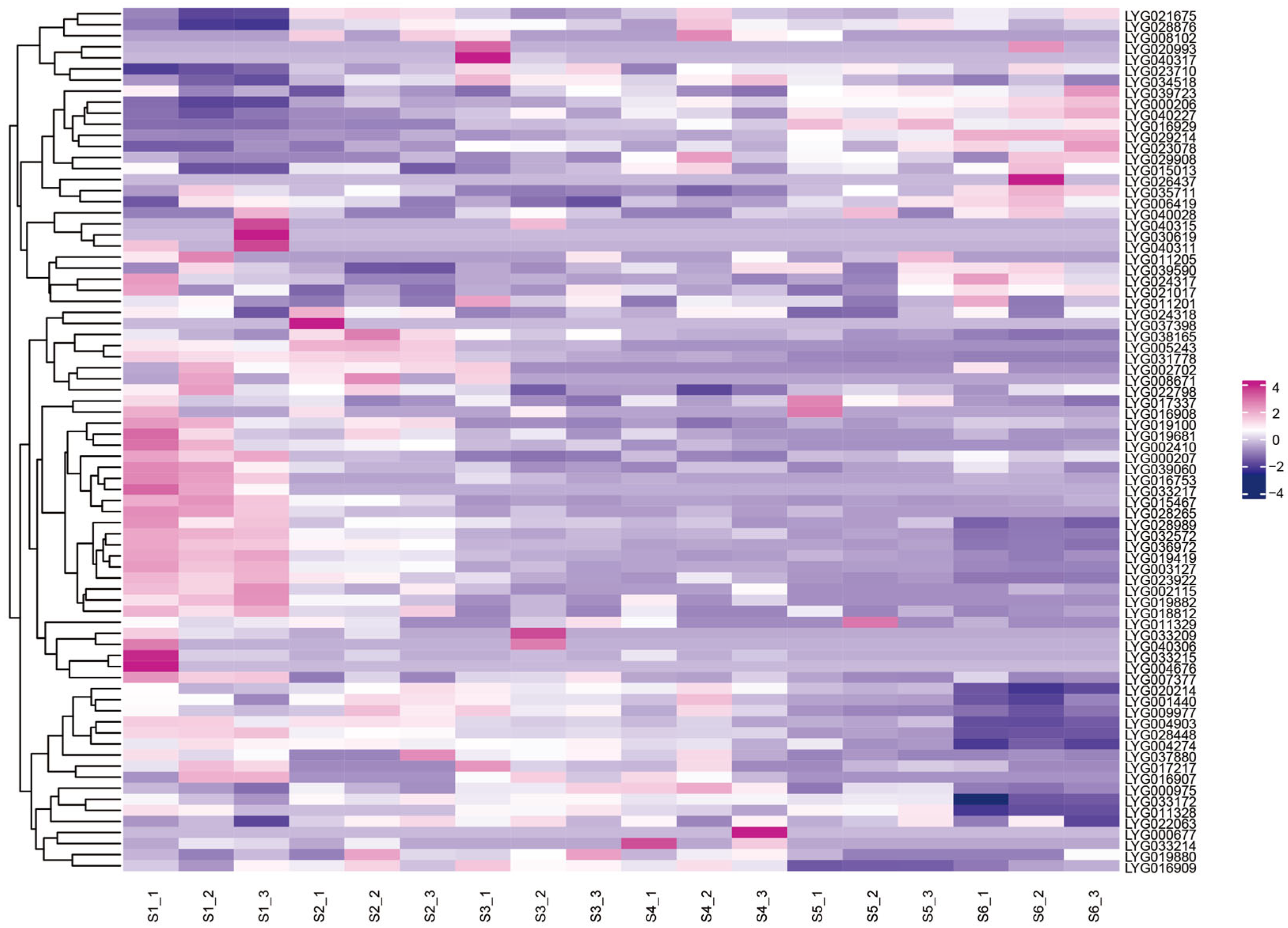
3.10. Transcriptome Analysis of Different Flowering Stages
3.11. Integrated Transcriptome and Metabolome Analysis of Different Flowering Stages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Passmore, S.; Maine, G.T.; Elble, R.; Christ, C.; Tye, B.-K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J. Mol. Biol. 1988, 204, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Sommer, H.; Beltran, J.-P.; Huijser, P.; Pape, H.; Lönnig, W.-E.; Saedler, H.; Schwarz-Sommer, Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 1990, 9, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Honma, T.; Goto, K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001, 409, 525–529. [Google Scholar] [CrossRef]
- Tahmasebi, S.; Jonoubi, P.; Majdi, M.; Majd, A.; Heidari, P. Genome-wide characterization and expression profiling of MADS-box family genes during organ development and drought stress in Camelina sativa L. Sci. Rep. 2025, 15, 9327. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; de Pouplana, L.R.; Martínez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- Allam, G.; Sakariyahu, S.K.; Shan, B.; Aung, B.; McDowell, T.; Papadopoulos, Y.; Bernards, M.A.; Hannoufa, A. Aluminum Stress Response Is Regulated Through a miR156/SPL13 Module in Medicago sativa. Genes 2025, 16, 751. [Google Scholar] [CrossRef]
- Basak, M.; Chakraborty, S.; Kundu, S.; Dey, S.; Das, M. Identification, expression analyses of APETALA1 gene homologs in Bambusa tulda and heterologous validation of BtMADS14 in Arabidopsis thaliana. Physiol. Mol. Biol. Plants 2025, 31, 389–404. [Google Scholar] [CrossRef]
- Cai, Y.; Yang, W.; Yue, J.; Chen, J.; Xing, J.; Yang, X.; Ye, D.; Tang, C.; Liu, H. Isolation and Functional Characterization of the MADS-Box Gene AGAMOUS-LIKE 24 in Rubber Dandelion Taraxacum kok-saghyz Rodin. Int. J. Mol. Sci. 2025, 26, 2271. [Google Scholar] [CrossRef]
- Cheng, Y.; Cheng, L.; Hu, G.; Guo, X.; Liu, Z.; Lan, Y. The MADS-box gene CmAP1 promotes flowering and petal development in Chinese chestnut Castanea mollissima. BMC Plant Biol. 2025, 25, 1035. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Chun, Y.; Zhang, F.; Guo, T.; Ren, M.; Zhao, J.; Yuan, S.; Wang, W.; Li, Y.; Li, X. A Novel OsMPK6-OsMADS47-PPKL1/3 Module Controls Grain Shape and Yield in Rice. Adv. Sci. 2025, 12, e01946. [Google Scholar] [CrossRef]
- Gao, X.; Li, Y.; Dai, Y.; Li, X.; Huang, C.; Zhang, S.; Li, F.; Zhang, H.; Li, G.; Sun, R.; et al. Identification of the MADS-Box Gene Family and the Key Role of BrAGL27 in the Regulation of Flowering in Chinese Cabbage Brassica rapa L. ssp. pekinensis. Int. J. Mol. Sci. 2025, 26, 2635. [Google Scholar] [CrossRef] [PubMed]
- De Bodt, S.; Raes, J.; Van de Peer, Y.; Theißen, G. And then there were many: MADS goes genomic. Trends Plant Sci. 2003, 8, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Melzer, R.; Theißen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef]
- Theißen, G.; Melzer, R.; Rümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef]
- Saedler, H.; Becker, A.; Winter, K.-U.; Kirchner, C.; Theißen, G. MADS-box genes are involved in floral development and evolution. Acta Biochim. Pol. 2001, 48, 351–358. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Krizek, B.A.; Meyerowitz, E.M. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 1996, 93, 4793–4798. [Google Scholar] [CrossRef]
- Han, J.P.; Wan, J.N.; Guan, Z.L.; Xu, H.; Wang, Q.F.; Wan, T. The origin, evolution and diversification of MADS-box transcription factors in green plants. Plant Commun. 2025, 6, 101462. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, Y.; Wang, J.; Du, M.; Yang, Y.; Cai, J.J.; Yang, E. Functional diversification of the MADS-box gene family in fine-tuning the dimorphic transition of Talaromyces marneffei. mSystems 2025, 10, e0046425. [Google Scholar] [CrossRef]
- Ren, L.; Sun, H.; Dai, S.; Feng, S.; Qiao, K.; Wang, J.; Gong, S.; Zhou, A. Identification and Characterization of MIKCc-Type MADS-Box Genes in the Flower Organs of Adonis amurensis. Int. J. Mol. Sci. 2021, 22, 9362. [Google Scholar] [CrossRef] [PubMed]
- Lyu, K.L.; Zeng, S.M.; Huang, X.Z.; Jiang, C.C. Integrated Multi-Omics Reveals DAM-Mediated Phytohormone Regulatory Networks Driving Bud Dormancy in ‘Mixue’ Pears. Plants 2025, 14, 2172. [Google Scholar] [CrossRef]
- Nobles, A.; Wendel, J.F.; Yoo, M.J. Comparative Analysis of Floral Transcriptomes in Gossypium hirsutum Malvaceae. Plants 2025, 14, 502. [Google Scholar] [CrossRef]
- Parenicova, L.; De Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Theissen, G.; Melzer, R. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann. Bot. 2007, 100, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Tian, M.; Wang, H.; Huang, Z.; Awais, M.; Xu, X.; Wang, L.; Lai, Z.; Bu, Z. Transcriptome analysis reveals that regulation network of the genes related to unique double flowers in tropical viviparous water lily. Sci. Rep. 2025, 15, 29561. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhou, Z.; Meng, F.; Wen, M.; Liu, A.; Yu, A. Characterization analyses of MADS-box genes highlighting their functions with seed development in Ricinus communis. Front. Plant Sci. 2025, 16, 1589915. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Xie, Y.; Peng, H.; Wang, M.; Zhu, W.; Xu, L.; Cheng, Y.; Wang, Y.; Zeng, J.; Fan, X.; et al. Transcriptome Profiling of Spike Development Reveals Key Genes and Pathways Associated with Early Heading in Wheat-Psathyrstachys huashanica 7Ns Chromosome Addition Line. Plants 2025, 14, 2077. [Google Scholar] [CrossRef]
- Wang, J.; Wang, L.; Usman, M.; Zhu, J.; Jiu, S.; Liu, R.; Zhang, C. ABA-Insensitive 5 ABI5 Is Involved in ABA-Induced Dormancy via Activating PavCIG1/2 Expression in Sweet Cherries. Genes 2025, 16, 596. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Zahn, L.M.; Feng, B.; Ma, H. Beyond the ABC-model: Regulation of floral homeotic genes. Adv. Bot. Res. 2006, 44, 163–207. [Google Scholar]
- Liu, E.; Zhu, S.; Du, M.; Lyu, H.; Zeng, S.; Liu, Q.; Wu, G.; Jiang, J.; Dang, X.; Dong, Z.; et al. LAX1, functioning with MADS-box genes, determines normal palea development in rice. Gene 2023, 883, 147635. [Google Scholar] [CrossRef]
- Mandel, M.A.; Gustafson-Brown, C.; Savidge, B.; Yanofsky, M.F. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 1992, 360, 273–277. [Google Scholar] [CrossRef]
- Jack, T.; Brockman, L.L.; Meyerowitz, E.M. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 1992, 68, 683–697. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; García-Ponce, B.; Sánchez, M.P.; Espinosa-Soto, C.; García-Gómez, M.L.; Piñeyro-Nelson, A.; Garay-Arroyo, A. MADS-box genes underground becoming mainstream: Plant root developmental mechanisms. New Phytol. 2019, 223, 1143–1158. [Google Scholar] [CrossRef]
- Chen, H.; Zeng, X.; Yang, J.; Cai, X.; Shi, Y.; Zheng, R.; Wang, Z.; Liu, J.; Yi, X.; Xiao, S.; et al. Whole-genome resequencing of Osmanthus fragrans provides insights into flower color evolution. Hortic. Res. 2021, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xia, H.; Cushman, S.A.; Zhao, H.; Guo, P.; Liu, Y.P.; Lin, N.; Shang, F.D. A new mechanism of flowering regulation by the competition of isoforms in Osmanthus fragrans. Ann. Bot. 2023, 132, 1089–1102. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, G.; Chen, X.; Liu, X.; Dong, B.; Wang, Y.; Zhong, S.; Deng, J.; Fang, Q.; Zhao, H. Genetic studies on continuous flowering in woody plant Osmanthus fragrans. Front. Plant Sci. 2022, 13, 1049479. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, X.; Liu, X.; Gao, G.; Dong, B.; Wang, Y.; Zhong, S.; Deng, J.; Fang, Q.; Zhao, H. OfBFT genes play an essential role in the proliferate flower formation of Osmanthus fragrans. Plant Physiol. Biochem. 2024, 20, 108463. [Google Scholar] [CrossRef]
- Zeng, Z.; Chen, S.; Xu, M.; Wang, M.; Chen, Z.; Wang, L.; Pang, J. Cloning, Expression, and Tobacco Overexpression Analyses of a PISTILLATA/GLOBOSA-like (OfGLO1) Gene from Osmanthus fragrans. Genes 2021, 12, 1748. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Q.; Jiang, G.; Wan, Q.; Dong, B.; Lu, M.; Deng, J.; Zhong, S.; Wang, Y.; Khan, I.A.; et al. Temperature-responsive module of OfAP1 and OfLFY regulates floral transition and floral organ identity in Osmanthus fragrans. Plant Physiol. Biochem. 2023, 203, 108076. [Google Scholar] [CrossRef]
- Hugouvieux, V.; Silva, C.S.; Jourdain, A.; Stigliani, A.; Charras, Q.; Conn, V.; Conn, S.J.; Carles, C.C.; Parcy, F.; Zubieta, C. Tetramerization of MADS family transcription factors SEPALLATA3 and AGAMOUS is required for floral meristem determinacy in Arabidopsis. Nucleic Acids Res. 2018, 46, 4966–4977. [Google Scholar] [CrossRef] [PubMed]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef]
- Yan, T.; Yoo, D.; Berardini, T.Z.; Mueller, L.A.; Weems, D.C.; Weng, S.; Cherry, J.M.; Rhee, S.Y. PatMatch: A program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res. 2005, 33, W262–W266. [Google Scholar] [CrossRef]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 2007, 35, D883–D887. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L.L. The Pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 2.3.1.–2.3.22. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Abdullah-Zawawi, M.R.; Ahmad-Nizammuddin, N.F.; Govender, N.; Harun, S.; Mohd-Assaad, N.; Mohamed-Hussein, Z.A. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice. Sci. Rep. 2021, 11, 19678. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, P.B.; Kasahara, R.D. An Overview on MADS Box Members in Plants: A Meta-Review. Int. J. Mol. Sci. 2024, 25, 8233. [Google Scholar] [CrossRef]
- Zhang, Z.; Zou, W.; Lin, P.; Wang, Z.; Chen, Y.; Yang, X.; Zhao, W.; Zhang, Y.; Wang, D.; Que, Y.; et al. Evolution and Function of MADS-Box Transcription Factors in Plants. Int. J. Mol. Sci. 2024, 25, 13278. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Dusi, V.; Pennisi, F.; Pandolfini, T. How Hormones and MADS-Box Transcription Factors Are Involved in Controlling Fruit Set and Parthenocarpy in Tomato. Genes 2020, 11, 1441. [Google Scholar] [CrossRef]
- Meng, L.; Zhang, S.; Chen, B.; Bai, X.; Li, Y.; Yang, J.; Wang, W.; Li, C.; Li, Y.; Li, Z. The MADS-box transcription factor GlMADS1 regulates secondary metabolism in Ganoderma lucidum. Mycologia 2021, 113, 12–19. [Google Scholar] [CrossRef]
- Saavedra Núñez, G.; González-Villanueva, E.; Ramos, P. Floral Development on Vitis vinifera Is Associated with MADS-Box Transcription Factors through the Transcriptional Regulation of VviZIP3. Plants 2023, 12, 3322. [Google Scholar] [CrossRef]
- Abraham-Juárez, M.J.; Schrager-Lavelle, A.; Man, J.; Whipple, C.; Handakumbura, P.; Babbitt, C.; Bartlett, M. Evolutionary Variation in MADS Box Dimerization Affects Floral Development and Protein Abundance in Maize. Plant Cell 2020, 32, 3408–3424. [Google Scholar] [CrossRef]
- Duan, S.F.; Yu, J.C.; Baldwin, T.C.; Yuan, Y.; Xiang, G.S.; Cui, R.; Zhao, Y.; Mo, X.C.; Lu, Y.C.; Liang, Y.L. Genome-wide identification of a MADS-box transcription factor family and their expression during floral development in Coptis teeta wall. BMC Plant Biol. 2024, 24, 1023. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Kong, W.; Wang, Q.; Fu, X. Genome-Wide Identification MIKC-Type MADS-Box Gene Family and Their Roles during Development of Floral Buds in Wheel Wingnut (Cyclocarya paliurus). Int. J. Mol. Sci. 2021, 22, 10128. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.H.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef]
- Ye, L.X.; Zhang, J.X.; Hou, X.J.; Qiu, M.Q.; Wang, W.F.; Zhang, J.X.; Hu, C.G.; Zhang, J.Z. A MADS-Box Gene CiMADS43 Is Involved in Citrus Flowering and Leaf Development through Interaction with CiAGL9. Int. J. Mol. Sci. 2021, 22, 5205. [Google Scholar] [CrossRef]
- Ferrario, S.; Busscher, J.; Franken, J.; Gerats, T.; Vandenbussche, M.; Angenent, G.C.; Immink, R.G. Ectopic expression of the petunia MADS box gene UNSHAVEN accelerates flowering and confers leaf-like characteristics to floral organs in a dominant-negative manner. Plant Cell 2004, 16, 1490–1505. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Dong, Q.; Ji, Z.; Chi, F.; Cong, P.; Zhou, Z. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene 2015, 555, 277–290. [Google Scholar] [CrossRef]
- Qian, W.; Liao, B.-Y.; Chang, A.Y.-F.; Zhang, J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010, 26, 425–430. [Google Scholar] [CrossRef]
- Meng, D.; Cao, Y.; Chen, T.; Abdullah, M.; Jin, Q.; Fan, H.; Lin, Y.; Cai, Y. Evolution and functional divergence of MADS-box genes in Pyrus. Sci. Rep. 2019, 9, 1266. [Google Scholar] [CrossRef]
- Liu, Y.; Guan, C.; Chen, Y.; Shi, Y.; Long, O.; Lin, H.; Zhang, K.; Zhou, M. Evolutionary analysis of MADS-box genes in buckwheat species and functional study of FdMADS28 in flavonoid metabolism. Plant Physiol. Biochem. 2024, 210, 108637. [Google Scholar] [CrossRef] [PubMed]
- Lucibelli, F.; Valoroso, M.C.; Theißen, G.; Nolden, S.; Mondragon-Palomino, M.; Aceto, S. Extending the Toolkit for Beauty: Differential Co-Expression of DROOPING LEAF-Like and Class B MADS-Box Genes during Phalaenopsis Flower Development. Int. J. Mol. Sci. 2021, 22, 7025. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, H.; He, Y.; Shen, X.; Lin, S.; Huang, L. MYB transcription factors and their roles in the male reproductive development of flowering plants. Plant Sci. 2023, 335, 111811. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Arya, P.; Gupta, K.; Randhawa, V.; Acharya, V.; Singh, A.K. Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malus × domestica). Sci. Rep. 2016, 6, 20695. [Google Scholar] [CrossRef]
- Jiménez, S.; Lawton-Rauh, A.L.; Reighard, G.L.; Abbott, A.G.; Bielenberg, D.G. Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biol. 2009, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zeng, X.L.; Wang, X.Q.; Pan, D.; Zhang, J.; Li, Z.Q.; Yang, J.; Zhang, Y.T.; Zeng, J.; Zhang, Q.; et al. Hormone metabolic profiling and transcriptome analysis reveal phytohormone crosstalk and the role of OfERF017 in the flowering and senescence of sweet osmanthus. Hortic. Plant J. 2025, 1, 018. [Google Scholar] [CrossRef]
- Zhang, Y.T.; Yang, J.; Zeng, X.L.; Cai, X.; Li, Z.Q.; Zeng, J.; Zhang, Q.; Chen, H.G.; Zou, J.J. Exploring miRNA-target modules regulating flower opening and senescence in Osmanthus fragrans through integrated transcriptome, miRNAome, and degradome analysis. Ind. Crops Prod. 2025, 229, 120927. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Yang, J.; Zeng, X.; Chen, H.; Zhang, Y.; Zhang, G.; Li, Z.; Cai, X.; Zou, J. Integrated Transcriptome and Metabolome Analyses Reveal the Roles of MADS-Box Genes in Regulating Flower Development and Metabolite Accumulation in Osmanthus fragran. Curr. Issues Mol. Biol. 2025, 47, 819. https://doi.org/10.3390/cimb47100819
Zhang Q, Yang J, Zeng X, Chen H, Zhang Y, Zhang G, Li Z, Cai X, Zou J. Integrated Transcriptome and Metabolome Analyses Reveal the Roles of MADS-Box Genes in Regulating Flower Development and Metabolite Accumulation in Osmanthus fragran. Current Issues in Molecular Biology. 2025; 47(10):819. https://doi.org/10.3390/cimb47100819
Chicago/Turabian StyleZhang, Qian, Jie Yang, Xiangling Zeng, Hongguo Chen, Yingting Zhang, Guifu Zhang, Zeqing Li, Xuan Cai, and Jingjing Zou. 2025. "Integrated Transcriptome and Metabolome Analyses Reveal the Roles of MADS-Box Genes in Regulating Flower Development and Metabolite Accumulation in Osmanthus fragran" Current Issues in Molecular Biology 47, no. 10: 819. https://doi.org/10.3390/cimb47100819
APA StyleZhang, Q., Yang, J., Zeng, X., Chen, H., Zhang, Y., Zhang, G., Li, Z., Cai, X., & Zou, J. (2025). Integrated Transcriptome and Metabolome Analyses Reveal the Roles of MADS-Box Genes in Regulating Flower Development and Metabolite Accumulation in Osmanthus fragran. Current Issues in Molecular Biology, 47(10), 819. https://doi.org/10.3390/cimb47100819





