Correlation of Eight (8) Polymorphisms and Their Genotypes with the Risk Factors of Cardiovascular Disease in a Black Elderly Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Sample
2.2. DNA Extraction and Genotyping
2.3. Blood Pressure Measurements
2.4. Biochemical Measurements
2.5. Data Analysis
3. Results
Association of Genotypes and CVR Factors
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gaziano, T.; Reddy, K.S.; Paccaud, F.; Horton, S.; Chaturvedi, V.; Jamison, D.T.; Breman, J.G.; Measham, A.R.; Alleyne, G.; Claeson, M.; et al. Cardiovascular Disease. In Disease Control Priorities in Developing Countries, 2nd ed.; Jamison, D.T., Breman, J.G., Measham, A.R., Alleyne, G., Claeson, M., Evans, D.B., Jha, P., Mills, A., Musgrove, P., Eds.; Chapter 33; World Bank Publications: Washington, DC, USA, 2006. Available online: https://pubmed.ncbi.nlm.nih.gov/21250342/ (accessed on 4 July 2022).
- Byrne, J.; Eksteen, G.; Crickmore, C. South Africa Cardiovascular Disease Statistics Reference Document; Heart and Stroke Foundation South Africa (HSFSA): Vlaeberg, South Africa, 2016; Available online: https://www.heartfoundation.co.za/wp-content/uploads/2017/10/CVD-Stats-Reference-Document-2016-FOR-MEDIA-1.pdf (accessed on 4 July 2022).
- Sliwa, K.; Acquah, L.; Gersh, B.J.; Mocumbi, A.O. Impact of Socioeconomic Status, Ethnicity, and Urbanization on Risk Factor Profiles of Cardiovascular Disease in Africa. Circulation 2016, 133, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Heianza, Y.; Qi, L. Impact of Genes and Environment on Obesity and Cardiovascular Disease. Endocrinology 2019, 160, 81–100. [Google Scholar] [CrossRef]
- Moreau, J.L.M.; Kesteven, S.; Martin, E.M.M.A.; Lau, K.S.; Yam, M.X.; O’reilly, V.C.; Del Monte-Nieto, G.; Baldini, A.; Feneley, M.P.; Moon, A.M.; et al. Gene-environment interaction impacts on heart development and embryo survival. Development 2019, 146, dev172957. [Google Scholar] [CrossRef]
- Brennan, L.; De Roos, B. Nutrigenomics: Lessons learned and future perspectives. Am. J. Clin. Nutr. 2021, 113, 503–516. [Google Scholar] [CrossRef]
- Chalwe, J.M.; Grobler, C.; Oldewage-Theron, W. Genetic Polymorphisms and Their Interactions with the Risk Factors of Cardiovascular Diseases: Review Chapter. In Risk Factors for Cardiovascular Disease, 1st ed.; Chahine, J., Ed.; IntechOpen: London, UK, 2021; Available online: https://www.intechopen.com/chapters/78930 (accessed on 4 July 2022).
- National Center for Biotechnology Information. APOB Apolipoprotein B [Homo Sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=338 (accessed on 18 December 2022).
- National Center for Biotechnology Information. CETP Cholesteryl Ester Transfer Protein [Homo Sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=1071 (accessed on 18 December 2022).
- National Center for Biotechnology Information. LDLR Low Density Lipoprotein Receptor [Homo Sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=3949 (accessed on 13 December 2022).
- National Center for Biotechnology Information. MTHFR Methylenetetrahydrofolate Reductase [Homo Sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=4524 (accessed on 24 December 2022).
- National Center for Biotechnology Information. PCSK9 Proprotein Convertase Subtilisin/Kexin Type 9 [Homo Sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/gene/255738 (accessed on 25 December 2022).
- Francula-Zaninovic, S.; Nola, I.A. Management of Measurable Variable Cardiovascular Disease’ Risk Factors. Curr. Cardiol. Rev. 2018, 14, 153–163. [Google Scholar] [CrossRef]
- Chalwe, J.M.; Grobler, C.; Oldewage-Theron, W. Development of a Structural Equation Model to Examine the Relationships between Genetic Polymorphisms and Cardiovascular Risk Factors. Nutrients 2023, 15, 2470. [Google Scholar] [CrossRef]
- Chalwe, J.M.; Mukherjee, U.; Grobler, C.; Mbambara, S.H.; Oldewage-Theron, W. Association between hypertension, obesity and dietary intake in post-menopausal women from rural Zambian communities. Health SA Gesondheid 2021, 26, 1–7. [Google Scholar] [CrossRef]
- Singh, K.; Gupta, J.K.; Kumar, S.; Singh, K.; Meenakshi, K.; Kumar, K. PCSK9 Inhibitors: Pharmacology and Therapeutic Potential. Preprints 2022, 1, 2022050290. [Google Scholar] [CrossRef]
- Shapiro, M.D.; Tavori, H.; Fazio, S. PCSK9: From Basic Science Discoveries to Clinical Trials. Circ. Res. 2018, 122, 1420–1438. [Google Scholar] [CrossRef]
- Cariou, B.; Maya, C.L.; Costet, P. Clinical aspects of PCSK9. Atherosclerosis 2011, 216, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Nussbaum, R.L.; Mcinnes, R.R.; Willard, H.F. The Molecular, Biochemical, and Cellular Basis of Genetic Disease. In Thompson & Thompson Genetics in Medicine, 8th ed.; Nussbaum, R.L., Ed.; Chapter 12; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 215–255. Available online: https://www.clinicalkey.com/#!/content/book/3-s2.0-B9781437706963000121 (accessed on 4 July 2022).
- Seidah, N.G.; Prat, A. The Multifaceted Biology of PCSK9. Endocr. Rev. 2022, 43, 558–582. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-W.; North, K.E.; Tin, A.; Haack, K.; Franceschini, N.; Saroja Voruganti, V.; Laston, S.; Zhang, Y.; Best, L.G.; Maccluer, J.W.; et al. Both rare and common variants in PCSK9 influence plasma low-density lipoprotein cholesterol level in American Indians. J. Clin. Endocrinol. Metab. 2015, 100, E345–E349. [Google Scholar] [CrossRef] [PubMed]
- Lakoski, S.G.; Lagace, T.A.; Cohen, J.C.; Horton, J.D.; Hobbs, H.H. Genetic and Metabolic Determinants of Plasma PCSK9 Levels. J. Clin. Endocrinol. Metab. 2009, 94, 2537–2543. [Google Scholar] [CrossRef]
- Hooper, A.J.; Marais, A.D.; Tanyanyiwa, D.M.; Burnett, J.R. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis 2007, 193, 445–448. [Google Scholar] [CrossRef]
- Huang, C.C.; Fornage, M.; Lloyd-Jones, D.M.; Wei, G.S.; Boerwinkle, E.; Liu, K. Longitudinal association of PCSK9 sequence variations with low-density lipoprotein cholesterol levels: The Coronary Artery Risk Development in Young Adults Study. Circulation. Cardiovasc. Genet. 2009, 2, 354–361. [Google Scholar] [CrossRef]
- Kent, S.T.; Rosenson, R.S.; Avery, C.L.; Chen, Y.-D.I.; Correa, A.; Cummings, S.R.; Cupples, L.A.; Cushman, M.; Evans, D.S.; Gudnason, V.; et al. PCSK9 Loss-of-Function Variants, Low-Density Lipoprotein Cholesterol, and Risk of Coronary Heart Disease and Stroke. Circulation. Cardiovasc. Genet. 2017, 10, e001632. [Google Scholar] [CrossRef]
- Krittanawong, C.; Khawaja, M.; Rosenson, R.S.; Amos, C.I.; Nambi, V.; Lavie, C.J.; Virani, S.S. Association of PCSK9 Variants With the Risk of Atherosclerotic Cardiovascular Disease and Variable Responses to PCSK9 Inhibitor Therapy. Curr. Probl. Cardiol. 2022, 47, 101043. [Google Scholar] [CrossRef]
- Chikowore, T.; Cockeran, M.; Conradie, K.R.; van Zyl, T. C679X loss-of-function PCSK9 variant lowers fasting glucose levels in a black South African population: A longitudinal study. Diabetes Res. Clin. Pract. 2018, 144, 279–285. [Google Scholar] [CrossRef]
- Chikowore, T.; Sahibdeen, V.; Hendry, L.M.; Norris, S.A.; Goedecke, J.H.; Micklesfield, L.K.; Lombard, Z. C679X loss-of-function PCSK9 variant is associated with lower fasting glucose in black South African adolescents: Birth to Twenty Plus Cohort. J. Clin. Transl. Endocrinol. 2019, 16, 100186. [Google Scholar] [CrossRef]
- Chopra, A.K. Dietary management of dyslipidemia. Indian Heart J. 2024, 76 (Suppl. 1), S65–S72. [Google Scholar] [CrossRef]
- Giglio, R.V.; Stoian, A.P.; Patti, A.M.; Rizvi, A.A.; Sukhorukov, V.; Ciaccio, M.; Orekhov, A.; Rizzo, M. Genetic and Epigenetic Biomarkers for Diagnosis, Prognosis and Treatment of Metabolic Syndrome. Curr. Pharm. Des. 2021, 27, 3729–3740. [Google Scholar] [CrossRef]
| SNP Genotypes | Risk Factors | Association (p-Value) |
|---|---|---|
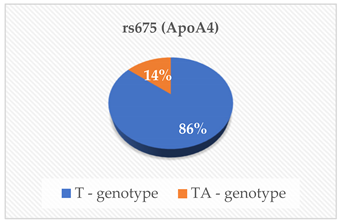 | ApoA1 (g/L) ApoB (g/L) Lipo (a) (nmol/L) | 0.971 0.857 0.536 |
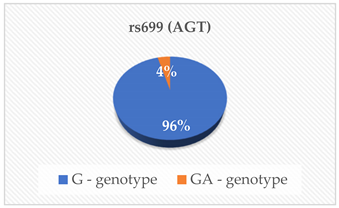 | BP (mmHg) | 0.607 |
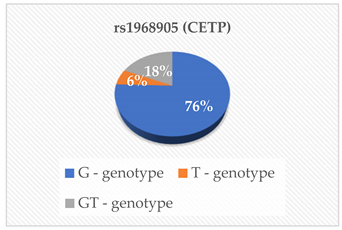 | ApoA1 (g/L) ApoB (g/L) TC (mmol/L) TG (mmol/L) LDL-C (mmol/L) HDL-C (mmol/L) Lipo (a) (nmol/L) | 0.780 0.364 0.291 0.380 0.654 0.694 0.237 |
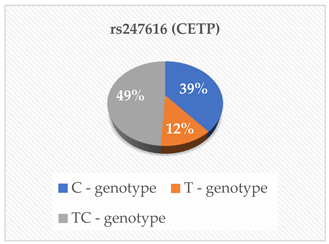 | ApoA1 (g/L) ApoB (g/L) TC (mmol/L) TG (mmol/L) LDL-C (mmol/L) HDL-C (mmol/L) Lipo (a) (nmol/L) | 0.308 0.860 0.713 0.505 0.707 0.482 0.466 |
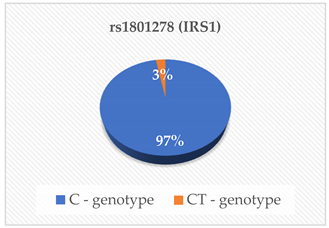 | Glucose (mmol/L) Insulin (μU/Ml) | 0.201 0.322 |
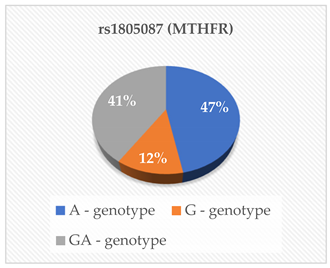 | Hcy (μmol/L) | 0.835 |
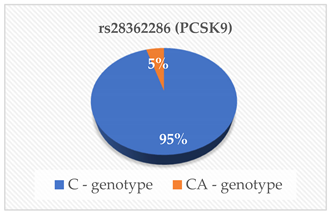 | PCSK9 (ng/mL) | 0.029 |
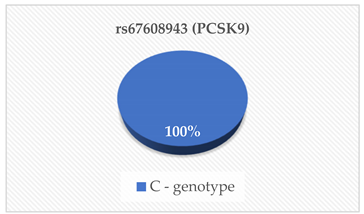 | PCSK9 (ng/mL) | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chalwe, J.M.; Grobler, C.J.; Oldewage-Theron, W.H. Correlation of Eight (8) Polymorphisms and Their Genotypes with the Risk Factors of Cardiovascular Disease in a Black Elderly Population. Curr. Issues Mol. Biol. 2024, 46, 12694-12703. https://doi.org/10.3390/cimb46110753
Chalwe JM, Grobler CJ, Oldewage-Theron WH. Correlation of Eight (8) Polymorphisms and Their Genotypes with the Risk Factors of Cardiovascular Disease in a Black Elderly Population. Current Issues in Molecular Biology. 2024; 46(11):12694-12703. https://doi.org/10.3390/cimb46110753
Chicago/Turabian StyleChalwe, Joseph Musonda, Christa Johanna Grobler, and Wilna Hendrika Oldewage-Theron. 2024. "Correlation of Eight (8) Polymorphisms and Their Genotypes with the Risk Factors of Cardiovascular Disease in a Black Elderly Population" Current Issues in Molecular Biology 46, no. 11: 12694-12703. https://doi.org/10.3390/cimb46110753
APA StyleChalwe, J. M., Grobler, C. J., & Oldewage-Theron, W. H. (2024). Correlation of Eight (8) Polymorphisms and Their Genotypes with the Risk Factors of Cardiovascular Disease in a Black Elderly Population. Current Issues in Molecular Biology, 46(11), 12694-12703. https://doi.org/10.3390/cimb46110753


_Kim.png)







