TRIM25, TRIM28 and TRIM59 and Their Protein Partners in Cancer Signaling Crosstalk: Potential Novel Therapeutic Targets for Cancer
Abstract
1. Introduction
2. Key Signaling Pathways in Cancers and Their Signaling Crosstalk: A Brief Overview
2.1. NF-κB: Master Switch for Inflammation
2.2. p53: Guardian of the Genome
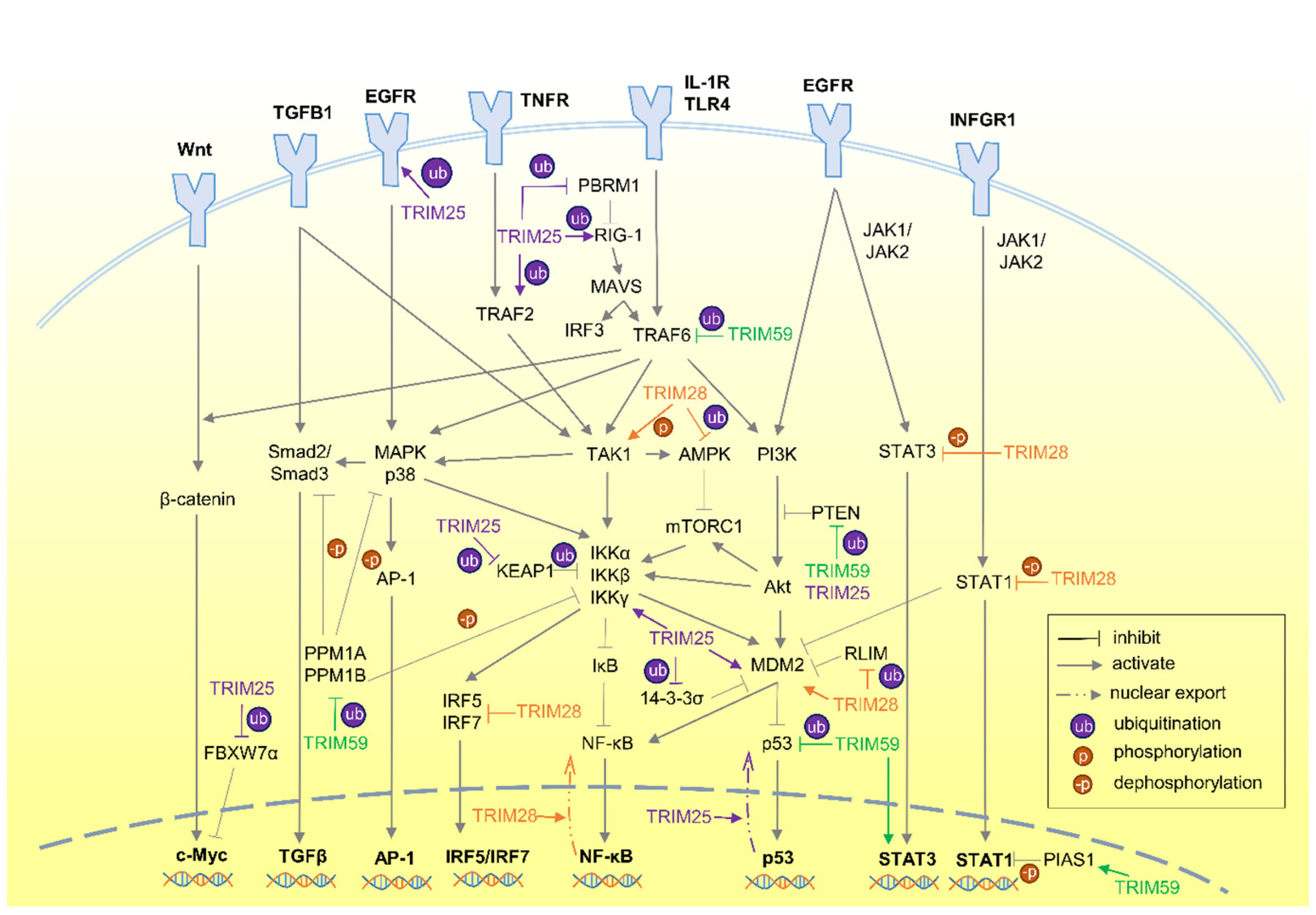
3. TRIM Proteins in Signaling Crosstalk
3.1. TRIM25
3.2. TRIM28
3.3. TRIM59
4. Summary, Limitations and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hatakeyama, S. TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends Biochem. Sci. 2017, 42, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Venuto, S.; Merla, G. E3 ubiquitin ligase TRIM proteins, cell cycle and mitosis. Cells 2019, 8, 510. [Google Scholar] [CrossRef] [PubMed]
- McNab, F.W.; Rajsbaum, R.; Stoye, J.P.; O’Garra, A. Tripartite-motif proteins and innate immune regulation. Curr. Opin. Immunol. 2011, 23, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Mallery, D.L.; McEwan, W.A.; Bidgood, S.R.; Towers, G.J.; Johnson, C.M.; James, L.C. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21). Proc. Natl. Acad. Sci. USA 2010, 107, 19985–19990. [Google Scholar] [CrossRef]
- Reymond, A.; Meroni, G.; Fantozzi, A.; Merla, G.; Cairo, S.; Luzi, L.; Riganelli, D.; Zanaria, E.; Messali, S.; Cainarca, S.; et al. The tripartite motif family identifies cell compartments. EMBO J. 2001, 20, 2140–2151. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Weinert, C.; Morger, D.; Djekic, A.; Grütter, M.G.; Mittl, P.R. Crystal structure of TRIM20 C-terminal coiled-coil/B30. 2 fragment: Implications for the recognition of higher order oligomers. Sci. Rep. 2015, 5, 10819. [Google Scholar] [CrossRef]
- James, L.C.; Keeble, A.H.; Khan, Z.; Rhodes, D.A.; Trowsdale, J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl. Acad. Sci. USA 2007, 104, 6200–6205. [Google Scholar] [CrossRef]
- Biris, N.; Yang, Y.; Taylor, A.B.; Tomashevski, A.; Guo, M.; Hart, P.J.; Diaz-Griffero, F.; Ivanov, D.N. Structure of the rhesus monkey TRIM5α PRYSPRY domain, the HIV capsid recognition module. Proc. Natl. Acad. Sci. USA 2012, 109, 13278–13283. [Google Scholar] [CrossRef]
- D’cRuz, A.A.; Kershaw, N.J.; Chiang, J.J.; Wang, M.K.; Nicola, N.A.; Babon, J.J.; Gack, M.U.; Nicholson, S.E. Crystal structure of the TRIM25 B30.2 (PRYSPRY) domain: A key component of antiviral signalling. Biochem. J. 2013, 456, 231–240. [Google Scholar] [CrossRef]
- Zhao, G.; Liu, C.; Wen, X.; Luan, G.; Xie, L.; Guo, X. The translational values of TRIM family in pan-cancers: From functions and mechanisms to clinics. Pharmacol. Ther. 2021, 227, 107881. [Google Scholar] [CrossRef] [PubMed]
- Kashimoto, K.; Komatsu, S.; Ichikawa, D.; Arita, T.; Konishi, H.; Nagata, H.; Takeshita, H.; Nishimura, Y.; Hirajima, S.; Kawaguchi, T.; et al. Overexpression of TRIM44 contributes to malignant outcome in gastric carcinoma. Cancer Sci. 2012, 103, 2021–2026. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Jin, C.; Ye, X.; Qiu, B.; Jianjun, X.; Zhu, S.; Xiang, L.; Wu, H.; Yongbing, W. TRIM44 promotes human esophageal cancer progression via the AKT/mTOR pathway. Cancer Sci. 2018, 109, 3080–3092. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Kimura, N.; Takayama, K.I.; Sato, Y.; Suzuki, T.; Azuma, K.; Fujimura, T.; Ikeda, K.; Kume, H.; Inoue, S. TRIM44 promotes cell proliferation and migration by inhibiting FRK in renal cell carcinoma. Cancer Sci. 2020, 111, 881–890. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, Y.; Ma, M.; Chang, L. Knockdown of TRIM44 inhibits the proliferation and invasion in papillary thyroid cancer cells through suppressing the Wnt/beta-catenin signaling pathway. Biomed. Pharmacother. 2017, 96, 98–103. [Google Scholar] [CrossRef]
- Kawabata, H.; Azuma, K.; Ikeda, K.; Sugitani, I.; Kinowaki, K.; Fujii, T.; Osaki, A.; Saeki, T.; Horie-Inoue, K.; Inoue, S. TRIM44 is a poor prognostic factor for breast cancer patients as a modulator of NF-kappaB signaling. Int. J. Mol. Sci. 2017, 18, 1931. [Google Scholar] [CrossRef]
- Tan, Y.; Yao, H.; Hu, J.; Liu, L. Knockdown of TRIM44 inhibits the proliferation and invasion in prostate cancer cells. Oncol. Res. 2017, 25, 1253–1259. [Google Scholar] [CrossRef]
- Yamada, Y.; Takayama, K.I.; Fujimura, T.; Ashikari, D.; Obinata, D.; Takahashi, S.; Ikeda, K.; Kakutani, S.; Urano, T.; Fukuhara, H.; et al. A novel prognostic factor TRIM44 promotes cell proliferation and migration, and inhibits apoptosis in testicular germ cell tumor. Cancer Sci. 2017, 108, 32–41. [Google Scholar] [CrossRef]
- Aierken, G.; Seyiti, A.; Alifu, M.; Kuerban, G. Knockdown of Tripartite-59 (TRIM59) inhibits cellular proliferation and migration in human cervical cancer cells. Oncol. Res. 2017, 25, 381–388. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, H.; Wang, Y.; Zhang, P.; Qi, Y. Tripartite Motif-containing Protein 59 (TRIM59) promotes epithelial ovarian cancer progression via the Focal Adhesion Kinase(FAK)/AKT/Matrix Metalloproteinase (MMP) pathway. Med. Sci. Monit. 2019, 25, 3366–3373. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, Z.; Wang, X.; Zhang, X.; Chen, Y.; Bai, J.; Di, W. TRIM59 is a novel marker of poor prognosis and promotes malignant progression of ovarian cancer by inducing Annexin A2 expression. Int. J. Mol. Sci. 2018, 14, 2073–2082. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Ye, Y.; He, L.; Xie, J.; Jing, J.; Ma, G.; Pan, H.; Han, L.; Han, W.; Zhou, Y. TRIM59 promotes breast cancer motility by suppressing p62-selective autophagic degradation of PDCD10. PLoS Biol. 2018, 16, e3000051. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ji, B.; Feng, Y.; Zhang, Y.; Ji, D.; Zhu, C.; Wang, S.; Zhang, C.; Zhang, D.; Sun, Y. TRIM59 facilitates the proliferation of colorectal cancer and promotes metastasis via the PI3K/AKT pathway. Oncol. Rep. 2017, 38, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Sui, X.; Han, D.; Gao, J.; Liu, Y.; Zhou, L. TRIM59 promotes cell proliferation, migration and invasion in human hepatocellular carcinoma cells. Pharmazie 2017, 72, 674–679. [Google Scholar] [CrossRef]
- Chen, W.; Zhao, K.; Miao, C.; Xu, A.; Zhang, J.; Zhu, J.; Su, S.; Wang, Z. Silencing Trim59 inhibits invasion/migration and epithelial-to-mesenchymal transition via TGF-beta/Smad2/3 signaling pathway in bladder cancer cells. Onco Targets Ther. 2017, 10, 1503–1512. [Google Scholar] [CrossRef]
- Lin, W.Y.; Wang, H.; Song, X.; Zhang, S.X.; Zhou, P.S.; Sun, J.M.; Li, J.S. Knockdown of tripartite motif 59 (TRIM59) inhibits tumor growth in prostate cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4864–4873. [Google Scholar]
- Bell, J.L.; Malyukova, A.; Kavallaris, M.; Marshall, G.M.; Cheung, B.B. TRIM16 inhibits neuroblastoma cell proliferation through cell cycle regulation and dynamic nuclear localization. Cell Cycle 2013, 12, 889–898. [Google Scholar] [CrossRef]
- Sutton, S.K.; Cheung, B.B.; Massudi, H.; Tan, O.; Koach, J.; Mayoh, C.; Carter, D.R.; Marshall, G.M. Heterozygous loss of keratinocyte TRIM16 expression increases melanocytic cell lesions and lymph node metastasis. J. Cancer Res. Clin. Oncol. 2019, 145, 2241–2250. [Google Scholar] [CrossRef]
- Tan, H.; Qi, J.; Chu, G.; Liu, Z. Tripartite Motif 16 inhibits the migration and invasion in ovarian cancer cells. Oncol. Res. 2017, 25, 551–558. [Google Scholar] [CrossRef]
- Kim, P.Y.; Tan, O.; Liu, B.; Trahair, T.; Liu, T.; Haber, M.; Norris, M.D.; Marshall, G.M.; Cheung, B.B. High TDP43 expression is required for TRIM16-induced inhibition of cancer cell growth and correlated with good prognosis of neuroblastoma and breast cancer patients. Cancer Lett. 2016, 374, 315–323. [Google Scholar] [CrossRef]
- Masood, R.; Hochstim, C.; Cervenka, B.; Zu, S.; Baniwal, S.K.; Patel, V.; Kobielak, A.; Sinha, U.K. A novel orthotopic mouse model of head and neck cancer and lymph node metastasis. Oncogenesis 2013, 2, e68. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.M.; Sun, R.; Luo, D.H.; Sun, J.; Zhang, M.Y.; Wang, M.H.; Yang, Y.; Wang, H.Y.; Mai, S.J. Upregulated TRIM29 promotes proliferation and metastasis of nasopharyngeal carcinoma via PTEN/AKT/mTOR signal pathway. Oncotarget 2016, 7, 13634–13650. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Xu, B.; Yao, Y.; Yu, X.; Cao, H.; Zhang, J.; Liu, J.; Sheng, H. RNA interference against TRIM29 inhibits migration and invasion of colorectal cancer cells. Oncol. Rep. 2016, 36, 1411–1418. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.X.; Ding, S.G.; Liu, L.N.; Wang, Y.; Zhang, J.; Zhang, H.J.; Zhang, Y. Predicative value of expression of TrkB and TRIM29 in biopsy tissues from preoperative gastroscopy in lymph node metastasis of gastric cancer. Chin. J. Prev. Med. 2012, 92, 376–379. [Google Scholar]
- Kosaka, Y.; Inoue, H.; Ohmachi, T.; Yokoe, T.; Matsumoto, T.; Mimori, K.; Tanaka, F.; Watanabe, M.; Mori, M. Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph node metastasis in gastric cancer. Ann. Surg. Oncol. 2007, 14, 2543–2549. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Xiong, J.P.; Deng, J.; Xiang, X.J. TRIM29 functions as an oncogene in gastric cancer and is regulated by miR-185. Int. J. Clin. Exp. Pathol. 2015, 8, 5053–5061. [Google Scholar]
- Song, X.; Fu, C.; Yang, X.; Sun, D.; Zhang, X.; Zhang, J. Tripartite motif-containing 29 as a novel biomarker in non-small cell lung cancer. Oncol. Lett. 2015, 10, 2283–2288. [Google Scholar] [CrossRef]
- Zhou, Z.Y.; Yang, G.Y.; Zhou, J.; Yu, M.H. Significance of TRIM29 and beta-catenin expression in non-small-cell lung cancer. J. Chin. Med. Assoc. 2012, 75, 269–274. [Google Scholar] [CrossRef]
- Zhan, W.; Han, T.; Zhang, C.; Xie, C.; Gan, M.; Deng, K.; Fu, M.; Wang, J.B. TRIM59 promotes the proliferation and migration of non-small cell lung cancer cells by upregulating cell cycle related proteins. PLoS ONE 2015, 10, e0142596. [Google Scholar] [CrossRef]
- Liu, C.; Huang, X.; Hou, S.; Hu, B.; Li, H. Silencing of tripartite motif (TRIM) 29 inhibits proliferation and invasion and increases chemosensitivity to cisplatin in human lung squamous cancer NCI-H520 cells. Thorac. Cancer 2015, 6, 31–37. [Google Scholar] [CrossRef]
- Kanno, Y.; Watanabe, M.; Kimura, T.; Nonomura, K.; Tanaka, S.; Hatakeyama, S. TRIM29 as a novel prostate basal cell marker for diagnosis of prostate cancer. Acta Histochem. 2014, 116, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Yanagi, T.; Watanabe, M.; Hata, H.; Kitamura, S.; Imafuku, K.; Yanagi, H.; Homma, A.; Wang, L.; Takahashi, H.; Shimizu, H.; et al. Loss of TRIM29 alters keratin distribution to promote cell invasion in squamous cell carcinoma. Cancer Res. 2018, 78, 6795–6806. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Welm, B.; Boucher, K.M.; Ebbert, M.T.; Bernard, P.S. TRIM29 functions as a tumor suppressor in nontumorigenic breast cells and invasive ER+ breast cancer. Am. J. Pathol. 2012, 180, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Caratozzolo, M.F.; Marzano, F.; Mastropasqua, F.; Sbisa, E.; Tullo, A. TRIM8: Making the right decision between the oncogene and tumour suppressor role. Genes 2017, 8, 354. [Google Scholar] [CrossRef]
- Valletti, A.; Marzano, F.; Pesole, G.; Sbisà, E.; Tullo, A. Targeting chemoresistant tumors: Could TRIM proteins-p53 axis be a possible answer? Int. J. Mol. Sci. 2019, 20, 1776. [Google Scholar] [CrossRef]
- Bahreyni-Toossi, M.T.; Zafari, N.; Azimian, H.; Mehrad-Majd, H.; Farhadi, J.; Vaziri Nezamdoust, F. Alteration in expression of TRIM29, TRIM37, TRIM44, and beta-catenin genes after irradiation in human cells with different radiosensitivity. Cancer Biother. Radiopharm. 2023, 38, 506–511. [Google Scholar] [CrossRef]
- Zhang, R.; Li, S.W.; Liu, L.; Yang, J.; Huang, G.; Sang, Y. TRIM11 facilitates chemoresistance in nasopharyngeal carcinoma by activating the beta-catenin/ABCC9 axis via p62-selective autophagic degradation of Daple. Oncogenesis 2020, 9, 45. [Google Scholar] [CrossRef]
- Zhang, Y.; Du, H.; Li, Y.; Yuan, Y.; Chen, B.; Sun, S. Elevated TRIM23 expression predicts cisplatin resistance in lung adenocarcinoma. Cancer Sci. 2020, 111, 637–646. [Google Scholar] [CrossRef]
- Pan, X.; Chen, Y.; Shen, Y.; Tantai, J. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis. 2019, 10, 429. [Google Scholar] [CrossRef]
- Horie-Inoue, K. TRIM proteins as trim tabs for the homoeostasis. J. Biochem. 2013, 154, 309–312. [Google Scholar] [CrossRef]
- Tomar, D.; Singh, R. TRIM family proteins: Emerging class of RING E3 ligases as regulator of NF-κB pathway. Biol. Cell 2015, 107, 22–40. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J. The role of Tripartite Motif family proteins in TGF-beta signaling pathway and cancer. J. Cancer Prev. 2018, 23, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, C.; Wang, X.; Hu, W.; Feng, Z. Tumor suppressor p53 cross-talks with TRIM family proteins. Genes Dis. 2021, 8, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Dolcet, X.; Llobet, D.; Pallares, J.; Matias-Guiu, X. NF-kB in development and progression of human cancer. Virchows Arch. 2005, 446, 475–482. [Google Scholar] [CrossRef]
- Xia, L.; Tan, S.; Zhou, Y.; Lin, J.; Wang, H.; Oyang, L.; Tian, Y.; Liu, L.; Su, M.; Wang, H.; et al. Role of the NFκB-signaling pathway in cancer. Onco Targets Ther. 2018, 11, 2063–2073. [Google Scholar] [CrossRef]
- Taniguchi, K.; Karin, M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Serasanambati, M.; Chilakapati, S.R. Function of Nuclear Factor Kappa B (NF-kB) in human diseases-A review. South Indian J. Biol. Sci. 2016, 2, 368. [Google Scholar] [CrossRef]
- Zinatizadeh, M.R.; Schock, B.; Chalbatani, G.M.; Zarandi, P.K.; Jalali, S.A.; Miri, S.R. The Nuclear Factor Kappa B (NF-kB) signaling in cancer development and immune diseases. Genes Dis. 2021, 8, 287–297. [Google Scholar] [CrossRef]
- Puszynski, K.; Bertolusso, R.; Lipniacki, T. Crosstalk between p53 and nuclear factor-B systems: Pro- and anti-apoptotic functions of NF-B. IET Syst. Biol. 2009, 3, 356–367. [Google Scholar] [CrossRef]
- Schneider, G.; Krämer, O.H. NFκB/p53 crosstalk—A promising new therapeutic target. Biochim. Biophys. Acta Rev. Cancer 2011, 1815, 90–103. [Google Scholar] [CrossRef]
- Bosman, M.C.J.; Schuringa, J.J.; Vellenga, E. Constitutive NF-κB activation in AML: Causes and treatment strategies. Crit. Rev. Oncol. Hematol. 2016, 98, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.J.; Liu, Z.-g. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Oeckinghaus, A.; Hayden, M.S.; Ghosh, S. Crosstalk in NF-κB signaling pathways. Nat. Immunol. 2011, 12, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Karin, M. Dangerous liaisons: STAT3 and NF-κB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010, 21, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B. Crosstalk with the Jak-STAT Pathway in Inflammation. In Jak-Stat Signaling: From Basics to Disease; Decker, T., Müller, M., Eds.; Springer: Vienna, Austria, 2012; pp. 353–370. [Google Scholar]
- Luo, K. Signaling cross talk between TGF-β/Smad and other signaling pathways. Cold Spring Harb. Perspect. Biol. 2017, 9, a022137. [Google Scholar] [CrossRef]
- Ma, B.; Hottiger, M.O. Crosstalk between Wnt/β-Catenin and NF-κB signaling pathway during inflammation. Front. Immunol. 2016, 7, 378. [Google Scholar] [CrossRef]
- Suzuki, K.; Matsubara, H. Recent advances in p53 research and cancer treatment. J. Biomed. Biotechnol. 2011, 2011, 978312. [Google Scholar] [CrossRef]
- Zhu, G.; Pan, C.; Bei, J.-X.; Li, B.; Liang, C.; Xu, Y.; Fu, X. Mutant p53 in cancer progression and targeted therapies. Front. Oncol. 2020, 10, 595187. [Google Scholar] [CrossRef]
- Niazi, S.; Purohit, M.; Niazi, J.H. Role of p53 circuitry in tumorigenesis: A brief review. Eur. J. Med. Chem. 2018, 158, 7–24. [Google Scholar] [CrossRef]
- Marei, H.E.; Althani, A.; Afifi, N.; Hasan, A.; Caceci, T.; Pozzoli, G.; Morrione, A.; Giordano, A.; Cenciarelli, C. p53 signaling in cancer progression and therapy. Cancer Cell Int. 2021, 21, 703. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef] [PubMed]
- Nag, S.; Qin, J.; Srivenugopal, K.S.; Wang, M.; Zhang, R. The MDM2-p53 pathway revisited. J. Biomed. Res. 2013, 27, 254–271. [Google Scholar] [CrossRef]
- Abraham, A.G.; O’Neill, E. PI3K/Akt-mediated regulation of p53 in cancer. Biochem. Soc. Trans. 2014, 42, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Astle, M.V.; Hannan, K.M.; Ng, P.Y.; Lee, R.S.; George, A.J.; Hsu, A.K.; Haupt, Y.; Hannan, R.D.; Pearson, R.B. AKT induces senescence in human cells via mTORC1 and p53 in the absence of DNA damage: Implications for targeting mTOR during malignancy. Oncogene 2012, 31, 1949–1962. [Google Scholar] [CrossRef] [PubMed]
- Goyal, H.; Chachoua, I.; Pecquet, C.; Vainchenker, W.; Constantinescu, S.N. A p53-JAK-STAT connection involved in myeloproliferative neoplasm pathogenesis and progression to secondary acute myeloid leukemia. Blood Rev. 2020, 42, 100712. [Google Scholar] [CrossRef]
- Elston, R.; Inman, G.J. Crosstalk between p53 and TGF-β Signalling. J. Signal Transduct. 2012, 2012, 294097. [Google Scholar] [CrossRef]
- Higgins, S.P.; Tang, Y.; Higgins, C.E.; Mian, B.; Zhang, W.; Czekay, R.-P.; Samarakoon, R.; Conti, D.J.; Higgins, P.J. TGF-β1/p53 signaling in renal fibrogenesis. Cell Signal. 2018, 43, 1–10. [Google Scholar] [CrossRef]
- Stramucci, L.; Pranteda, A.; Bossi, G. Insights of crosstalk between p53 protein and the MKK3/MKK6/p38 MAPK signaling pathway in cancer. Cancers 2018, 10, 131. [Google Scholar] [CrossRef]
- Maiuri, M.C.; Galluzzi, L.; Morselli, E.; Kepp, O.; Malik, S.A.; Kroemer, G. Autophagy regulation by p53. Curr. Opin. Cell Biol. 2010, 22, 181–185. [Google Scholar] [CrossRef]
- Dashzeveg, N.; Yoshida, K. Crosstalk between tumor suppressors p53 and PKCδ: Execution of the intrinsic apoptotic pathways. Cancer Lett. 2016, 377, 158–163. [Google Scholar] [CrossRef]
- Wu, W.K.K.; Wang, X.J.; Cheng, A.S.L.; Luo, M.X.M.; Ng, S.S.M.; To, K.F.; Chan, F.K.L.; Cho, C.H.; Sung, J.J.Y.; Yu, J. Dysregulation and crosstalk of cellular signaling pathways in colon carcinogenesis. Crit. Rev. Oncol. Hematol. 2013, 86, 251–277. [Google Scholar] [CrossRef] [PubMed]
- Zubbair Malik, M.; Ali, S.; Jahoor Alam, M.; Ishrat, R.; Brojen Singh, R.K. Dynamics of p53 and Wnt cross talk. Comput. Biol. Chem. 2015, 59, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Zhu, Z. The role of TRIM proteins in PRR signaling pathways and immune-related diseases. Int. Immunopharmacol. 2021, 98, 107813. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, G.A.; Rajsbaum, R.; Sánchez-Aparicio, M.T.; Maestre, A.M.; Valdiviezo, J.; Shi, M.; Inn, K.-S.; Fernandez-Sesma, A.; Jung, J.; García-Sastre, A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 2013, 38, 384–398. [Google Scholar] [CrossRef]
- Zhan, W.; Zhang, S. TRIM proteins in lung cancer: Mechanisms, biomarkers and therapeutic targets. Life Sci. 2021, 268, 118985. [Google Scholar] [CrossRef]
- Martín-Vicente, M.; Medrano, L.M.; Resino, S.; García-Sastre, A.; Martínez, I. TRIM25 in the regulation of the antiviral innate immunity. Front. Immunol. 2017, 8, 1187. [Google Scholar] [CrossRef]
- Choudhury, N.R.; Heikel, G.; Michlewski, G. TRIM25 and its emerging RNA-binding roles in antiviral defense. WIREs RNA 2020, 11, e1588. [Google Scholar] [CrossRef]
- Heikel, G.; Choudhury, N.R.; Michlewski, G. The role of Trim25 in development, disease and RNA metabolism. Biochem. Soc. Trans. 2016, 44, 1045–1050. [Google Scholar] [CrossRef]
- Inoue, S.; Orimo, A.; Hosoi, T.; Kondo, S.; Toyoshima, H.; Kondo, T.; Ikegami, A.; Ouchi, Y.; Orimo, H.; Muramatsu, M. Genomic binding-site cloning reveals an estrogen-responsive gene that encodes a RING finger protein. Proc. Natl. Acad. Sci. USA 1993, 90, 11117–11121. [Google Scholar] [CrossRef]
- Tecalco-Cruz, A.C.; Abraham-Juárez, M.J.; Solleiro-Villavicencio, H.; Ramírez-Jarquín, J.O. TRIM25: A central factor in breast cancer. World J. Clin. Oncol. 2021, 12, 646–655. [Google Scholar] [CrossRef]
- Urano, T.; Saito, T.; Tsukui, T.; Fujita, M.; Hosoi, T.; Muramatsu, M.; Ouchi, Y.; Inoue, S. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature 2002, 417, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.Y.; Wen, Y.Y.; Lin, Y.l.; Pham, L.; Su, C.H.; Yang, H.; Chen, J.; Lee, M.H. Roles for negative cell regulator 14-3-3σ in control of MDM2 activities. Oncogene 2007, 26, 7355–7362. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Teh, A.H.; Yap, B.K. Identification of peptide binding sequence of TRIM25 on 14-3-3σ by bioinformatics and biophysical techniques. J. Biomol. Struct. Dyn. 2023, 41, 13260–13270. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Cui, H.; Zhang, H. Overexpression of TRIM25 in lung cancer regulates tumor cell progression. Technol. Cancer Res. Treat. 2016, 15, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Elabd, S.; Hammer, S.; Solozobova, V.; Yan, H.; Bartel, F.; Inoue, S.; Henrich, T.; Wittbrodt, J.; Loosli, F.; et al. TRIM25 has a dual function in the p53/Mdm2 circuit. Oncogene 2015, 34, 5729–5738. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.I.; Suzuki, T.; Tanaka, T.; Fujimura, T.; Takahashi, S.; Urano, T.; Ikeda, K.; Inoue, S. TRIM25 enhances cell growth and cell survival by modulating p53 signals via interaction with G3BP2 in prostate cancer. Oncogene 2018, 37, 2165–2180. [Google Scholar] [CrossRef]
- Wen, J.T.; Chen, X.; Liu, X.; Xie, B.M.; Chen, J.W.; Qin, H.L.; Zhao, Y. Small nucleolar RNA and C/D Box 15B regulate the TRIM25/P53 complex to promote the development of endometrial cancer. J. Oncol. 2022, 2022, 7762708. [Google Scholar] [CrossRef]
- He, Y.-m.; Zhou, X.-m.; Jiang, S.-y.; Zhang, Z.-b.; Cao, B.-y.; Liu, J.-b.; Zeng, Y.-y.; Zhao, J.; Mao, X.-l. TRIM25 activates AKT/mTOR by inhibiting PTEN via K63-linked polyubiquitination in non-small cell lung cancer. Acta Pharmacol. Sin. 2022, 43, 681–691. [Google Scholar] [CrossRef]
- Yuan, P.; Zheng, A.; Tang, Q. Tripartite motif protein 25 is associated with epirubicin resistance in hepatocellular carcinoma cells via regulating PTEN/AKT pathway. Cell Biol. Int. 2020, 44, 1503–1513. [Google Scholar] [CrossRef]
- Lee, N.-R.; Kim, H.-I.; Choi, M.-S.; Yi, C.-M.; Inn, K.-S. Regulation of MDA5-MAVS antiviral signaling axis by TRIM25 through TRAF6-mediated NF-κB activation. Mol. Cells 2015, 38, 759–764. [Google Scholar] [CrossRef]
- Gack, M.U.; Kirchhofer, A.; Shin, Y.C.; Inn, K.-S.; Liang, C.; Cui, S.; Myong, S.; Ha, T.; Hopfner, K.-P.; Jung, J.U. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. USA 2008, 105, 16743. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, J.; He, B.; Zhang, X.; Li, X.; Kuang, E. RTN3 inhibits RIG-I-mediated antiviral responses by impairing TRIM25-mediated K63-linked polyubiquitination. eLife 2021, 10, e68958. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.S.; Zhao, Y.; Sun, Y.; Zhong, L.; Cheng, Y.; Zhang, Y.; Ning, K.; Tao, Q.; Wang, Y.; Ying, Y. The epigenetic modifier PBRM1 restricts the basal activity of the innate immune system by repressing retinoic acid-inducible gene-I-like receptor signalling and is a potential prognostic biomarker for colon cancer. J. Pathol. 2018, 244, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Xie, P. TRAF molecules in cell signaling and in human diseases. J. Mol. Signal. 2013, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, K.; Huang, Y.; Sun, M.; Tian, Q.; Zhang, S.; Qin, Y. TRIM25 promotes TNF-α–induced NF-κB activation through potentiating the K63-linked ubiquitination of TRAF2. J. Immunol. 2020, 204, 1499–1507. [Google Scholar] [CrossRef]
- Shi, J.-H.; Sun, S.-C. Tumor Necrosis Factor Receptor-associated Factor regulation of Nuclear Factor κB and Mitogen-Activated Protein Kinase pathways. Front. Immunol. 2018, 9, 1849. [Google Scholar] [CrossRef]
- Dainichi, T.; Matsumoto, R.; Mostafa, A.; Kabashima, K. Immune control by TRAF6-mediated pathways of epithelial cells in the EIME (Epithelial Immune Microenvironment). Front. Immunol. 2019, 10, 1107. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, X.; Cui, K.; Liu, C.; Wu, M.; Prochownik, E.V.; Li, Y. The MAP3K13-TRIM25-FBXW7α axis affects c-Myc protein stability and tumor development. Cell Death Differ. 2020, 27, 420–433. [Google Scholar] [CrossRef]
- Zhou, D.-d.; Yu, J.-j.; Hu, Z.-w.; Hua, F. TRIM25 enhances EGFR stability and signaling activity to promote lung cancer progression. Acta Pharm. Sin. 2019, 54, 1026–1035. [Google Scholar] [CrossRef]
- Hwang, Y.P.; Yun, H.J.; Choi, J.H.; Han, E.H.; Kim, H.G.; Song, G.Y.; Kwon, K.-i.; Jeong, T.C.; Jeong, H.G. Suppression of EGF-induced tumor cell migration and matrix metalloproteinase-9 expression by capsaicin via the inhibition of EGFR-mediated FAK/Akt, PKC/Raf/ERK, p38 MAPK, and AP-1 signaling. Mol. Nutr. Food Res. 2011, 55, 594–605. [Google Scholar] [CrossRef]
- Zimmer, S.; Kahl, P.; Buhl, T.M.; Steiner, S.; Wardelmann, E.; Merkelbach-Bruse, S.; Buettner, R.; Heukamp, L.C. Epidermal growth factor receptor mutations in non-small cell lung cancer influence downstream Akt, MAPK and Stat3 signaling. J. Cancer Res. Clin. Oncol. 2009, 135, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, M.; Betensky, R.A.; Batchelor, T.T.; Bernay, D.C.; Louis, D.N.; Nutt, C.L. Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: Correlation with EGFR status, tumor grade, and survival. J. Neuropathol. Exp. Neurol. 2006, 65, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tao, S.; Liao, L.; Li, Y.; Li, H.; Li, Z.; Lin, L.; Wan, X.; Yang, X.; Chen, L. TRIM25 promotes the cell survival and growth of hepatocellular carcinoma through targeting Keap1-Nrf2 pathway. Nat. Commun. 2020, 11, 348. [Google Scholar] [CrossRef]
- Ji, Y.; Li, F.; Zhang, H.; Yang, L.; Yi, Y.; Wang, L.; Chen, H.; Zhang, Y.; Yang, Z. Targeting TRIM40 signaling reduces esophagus cancer development: A mechanism involving in protection of oroxylin A. Int. Immunopharmacol. 2024, 137, 112362. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Buttari, B.; Panieri, E.; Profumo, E.; Saso, L. An overview of Nrf2 signaling pathway and its role in inflammation. Molecules 2020, 25, 5474. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.W.; Sikriwal, D.; Dong, X.; Guo, P.; Sun, X.; Dong, J.T. Oestrogen causes degradation of KLF5 by inducing the E3 ubiquitin ligase EFP in ER-positive breast cancer cells. Biochem. J. 2011, 437, 323–333. [Google Scholar] [CrossRef]
- Gao, Y.; Ding, Y.; Chen, H.; Chen, H.; Zhou, J. Targeting Krüppel-Like Factor 5 (KLF5) for cancer therapy. Curr. Top. Med. Chem. 2015, 15, 699–713. [Google Scholar] [CrossRef]
- Sun, N.; Xue, Y.; Dai, T.; Li, X.; Zheng, N. Tripartite motif containing 25 promotes proliferation and invasion of colorectal cancer cells through TGF-beta signaling. Biosci. Rep. 2017, 37, BSR20170805. [Google Scholar] [CrossRef]
- Czerwińska, P.; Mazurek, S.; Wiznerowicz, M. The complexity of TRIM28 contribution to cancer. J. Biomed. Sci. 2017, 24, 63. [Google Scholar] [CrossRef]
- Bunch, H.; Calderwood, S.K. TRIM28 as a novel transcriptional elongation factor. BMC Mol. Biol. 2015, 16, 14. [Google Scholar] [CrossRef]
- Urrutia, R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003, 4, 231. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Wang, Y.; Macfarlan, T.S. The role of KRAB-ZFPs in transposable element repression and mammalian evolution. Trends Genet. 2017, 33, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Iyengar, S.; Ivanov Alexey, V.; Jin Victor, X.; Rauscher Frank, J.; Farnham Peggy, J. Functional analysis of KAP1 genomic recruitment. Mol. Cell. Biol. 2011, 31, 1833–1847. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, A.A.; Kurka, T.; Jeggo, P.A. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 2011, 18, 831–839. [Google Scholar] [CrossRef]
- Venkov, C.D.; Link, A.J.; Jennings, J.L.; Plieth, D.; Inoue, T.; Nagai, K.; Xu, C.; Dimitrova, Y.N.; Rauscher, F.J., III; Neilson, E.G. A proximal activator of transcription in epithelial-mesenchymal transition. J. Clin. Investig. 2007, 117, 482–491. [Google Scholar] [CrossRef]
- Wang, C.; Ivanov, A.; Chen, L.; Fredericks, W.J.; Seto, E.; Rauscher, F.J., 3rd; Chen, J. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J. 2005, 24, 3279–3290. [Google Scholar] [CrossRef]
- Jin, J.O.; Lee, G.D.; Nam, S.H.; Lee, T.H.; Kang, D.H.; Yun, J.K.; Lee, P.C. Sequential ubiquitination of p53 by TRIM28, RLIM, and MDM2 in lung tumorigenesis. Cell Death Differ. 2021, 28, 1790–1803. [Google Scholar] [CrossRef]
- Saint-Germain, E.; Mignacca, L.; Vernier, M.; Bobbala, D.; Ilangumaran, S.; Ferbeyre, G. SOCS1 regulates senescence and ferroptosis by modulating the expression of p53 target genes. Aging 2017, 9, 2137–2162. [Google Scholar] [CrossRef]
- Li, J.; Lu, D.; Dou, H.; Liu, H.; Weaver, K.; Wang, W.; Li, J.; Yeh, E.T.H.; Williams, B.O.; Zheng, L.; et al. Desumoylase SENP6 maintains osteochondroprogenitor homeostasis by suppressing the p53 pathway. Nat. Commun. 2018, 9, 143. [Google Scholar] [CrossRef]
- Weon, J.L.; Potts, P.R. The MAGE protein family and cancer. Curr. Opin. Cell Biol. 2015, 37, 1–8. [Google Scholar] [CrossRef]
- Gao, X.; Li, Q.; Chen, G.; He, H.; Ma, Y. MAGEA3 promotes proliferation and suppresses apoptosis in cervical cancer cells by inhibiting the KAP1/p53 signaling pathway. Am. J. Transl. Res. 2020, 12, 3596–3612. [Google Scholar] [PubMed]
- Pineda, C.T.; Ramanathan, S.; Fon Tacer, K.; Weon, J.L.; Potts, M.B.; Ou, Y.H.; White, M.A.; Potts, P.R. Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell 2015, 160, 715–728. [Google Scholar] [CrossRef] [PubMed]
- Pineda, C.T.; Potts, P.R. Oncogenic MAGEA-TRIM28 ubiquitin ligase downregulates autophagy by ubiquitinating and degrading AMPK in cancer. Autophagy 2015, 11, 844–846. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Guo, C.; Zheng, Y.; Wang, Y.; Jin, Z.; Yin, Y. Post-transcriptional regulation of cancer/testis antigen MAGEC2 expression by TRIM28 in tumor cells. BMC Cancer 2018, 18, 971. [Google Scholar] [CrossRef]
- Liu, H.; Chen, H.; Deng, X.; Peng, Y.; Zeng, Q.; Song, Z.; He, W.; Zhang, L.; Xiao, T.; Gao, G.; et al. Knockdown of TRIM28 inhibits PDGF-BB-induced vascular smooth muscle cell proliferation and migration. Chem. Biol. Interact. 2019, 311, 108772. [Google Scholar] [CrossRef]
- Liang, M.; Sun, Z.; Chen, X.; Wang, L.; Wang, H.; Qin, L.; Zhao, W.; Geng, B. E3 ligase TRIM28 promotes anti-PD-1 resistance in non-small cell lung cancer by enhancing the recruitment of myeloid-derived suppressor cells. J. Exp. Clin. Cancer Res. 2023, 42, 275. [Google Scholar] [CrossRef]
- Park, H.H.; Kim, H.R.; Park, S.Y.; Hwang, S.M.; Hong, S.M.; Park, S.; Kang, H.C.; Morgan, M.J.; Cha, J.H.; Lee, D.; et al. RIPK3 activation induces TRIM28 derepression in cancer cells and enhances the anti-tumor microenvironment. Mol. Cancer 2021, 20, 107. [Google Scholar] [CrossRef]
- Kamitani, S.; Togi, S.; Ikeda, O.; Nakasuji, M.; Sakauchi, A.; Sekine, Y.; Muromoto, R.; Oritani, K.; Matsuda, T. Krüppel-associated box-associated protein 1 negatively regulates TNF-α-induced NF-κB transcriptional activity by influencing the interactions among STAT3, p300, and NF-κB/p65. J. Immunol. 2011, 187, 2476–2483. [Google Scholar] [CrossRef]
- Xu, F.; Li, X.; Yan, L.; Yuan, N.; Fang, Y.; Cao, Y.; Xu, L.; Zhang, X.; Xu, L.; Ge, C.; et al. Autophagy promotes the repair of radiation-induced DNA damage in bone marrow hematopoietic cells via enhanced STAT3 signaling. Radiat. Res. 2017, 187, 382–396. [Google Scholar] [CrossRef]
- Tsuruma, R.; Ohbayashi, N.; Kamitani, S.; Ikeda, O.; Sato, N.; Muromoto, R.; Sekine, Y.; Oritani, K.; Matsuda, T. Physical and functional interactions between STAT3 and KAP1. Oncogene 2008, 27, 3054–3059. [Google Scholar] [CrossRef]
- Zhao, C.; Zhao, Y.; Zhao, J.; Meng, G.; Huang, S.; Liu, Y.; Wang, S.; Qi, L. Acute myeloid leukemia cell-derived extracellular vesicles carrying microRNA-548ac regulate hematopoietic function via the TRIM28/STAT3 pathway. Cancer Gene Ther. 2022, 29, 918–929. [Google Scholar] [CrossRef] [PubMed]
- Kamitani, S.; Ohbayashi, N.; Ikeda, O.; Togi, S.; Muromoto, R.; Sekine, Y.; Ohta, K.; Ishiyama, H.; Matsuda, T. KAP1 regulates type I interferon/STAT1-mediated IRF-1 gene expression. Biochem. Biophys. Res. Commun. 2008, 370, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Narayan, V.; Pion, E.; Landré, V.; Müller, P.; Ball, K.L. Docking-dependent ubiquitination of the interferon regulatory factor-1 tumor suppressor protein by the ubiquitin ligase CHIP. J. Biol. Chem. 2011, 286, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wang, L.; Wang, C.; Shen, J.; Su, B.; Marisetty, A.L.; Fang, D.; Kassab, C.; Jeong, K.J.; Zhao, W.; et al. Verteporfin inhibits PD-L1 through autophagy and the STAT1-IRF1-TRIM28 signaling axis, exerting antitumor efficacy. Cancer Immunol. Res. 2020, 8, 952–965. [Google Scholar] [CrossRef]
- Liang, Q.; Deng, H.; Li, X.; Wu, X.; Tang, Q.; Chang, T.H.; Peng, H.; Rauscher, F.J., 3rd; Ozato, K.; Zhu, F. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J. Immunol. 2011, 187, 4754–4763. [Google Scholar] [CrossRef]
- Eames, H.L.; Saliba, D.G.; Krausgruber, T.; Lanfrancotti, A.; Ryzhakov, G.; Udalova, I.A. KAP1/TRIM28: An inhibitor of IRF5 function in inflammatory macrophages. Immunobiology 2012, 217, 1315–1324. [Google Scholar] [CrossRef]
- Sokolova, O.; Kähne, T.; Bryan, K.; Naumann, M. Interactome analysis of transforming growth factor-β-activated kinase 1 in Helicobacter pylori-infected cells revealed novel regulators tripartite motif 28 and CDC37. Oncotarget 2018, 9, 14366–14381. [Google Scholar] [CrossRef]
- Mukhopadhyay, H.; Lee, N.Y. Multifaceted roles of TAK1 signaling in cancer. Oncogene 2020, 39, 1402–1413. [Google Scholar] [CrossRef]
- Zonneville, J.; Wong, V.; Limoge, M.; Nikiforov, M.; Bakin, A.V. TAK1 signaling regulates p53 through a mechanism involving ribosomal stress. Sci. Rep. 2020, 10, 2517. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Huang, Y.; Dai, X.; Liu, Y.; Liu, Z.; Wang, Y.; Wang, N.; Zhang, P. Tripartite motif-containing 28 bridges endothelial inflammation and angiogenic activity by retaining expression of TNFR-1 and -2 and VEGFR2 in endothelial cells. FASEB J. 2017, 31, 2026–2036. [Google Scholar] [CrossRef]
- Kumar, J.; Kaur, G.; Ren, R.; Lu, Y.; Lin, K.; Li, J.; Huang, Y.; Patel, A.; Barton, M.C.; Macfarlan, T.; et al. KRAB domain of ZFP568 disrupts TRIM28-mediated abnormal interactions in cancer cells. NAR Cancer 2020, 2, zcaa007. [Google Scholar] [CrossRef]
- Li, J.; Xi, Y.; Li, W.; McCarthy, R.L.; Stratton, S.A.; Zou, W.; Li, W.; Dent, S.Y.; Jain, A.K.; Barton, M.C. TRIM28 interacts with EZH2 and SWI/SNF to activate genes that promote mammosphere formation. Oncogene 2017, 36, 2991–3001. [Google Scholar] [CrossRef]
- Iannetti, A.; Ledoux, A.C.; Tudhope, S.J.; Sellier, H.; Zhao, B.; Mowla, S.; Moore, A.; Hummerich, H.; Gewurz, B.E.; Cockell, S.J.; et al. Regulation of p53 and Rb links the alternative NF-κB pathway to EZH2 expression and cell senescence. PLoS Genet. 2014, 10, e1004642. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gou, H.; Yao, J.; Yi, K.; Jin, Z.; Matsuoka, M.; Zhao, T. The noncanonical role of EZH2 in cancer. Cancer Sci. 2021, 112, 1376–1382. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Tokita, K.; Wada, N.; Ito, K.; Yamauchi, C.; Ito, Y.; Ochiai, A. MEK–ERK pathway regulates EZH2 overexpression in association with aggressive breast cancer subtypes. Oncogene 2011, 30, 4118–4128. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Cai, J.; Wang, Q.; Wang, Y.; Liu, M.; Yang, J.; Zhou, J.; Kang, C.; Li, M.; Jiang, C. Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/β-catenin pathway by scaffolding EZH2. Clin. Cancer Res. 2018, 24, 684–695. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.T.; Li, Z.; Wu, Z.; Aau, M.; Guan, P.; Karuturi, R.K.; Liou, Y.C.; Yu, Q. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol. Cell 2011, 43, 798–810. [Google Scholar] [CrossRef]
- Lawrence, C.L.; Baldwin, A.S. Non-canonical EZH2 transcriptionally activates RelB in triple negative breast cancer. PLoS ONE 2016, 11, e0165005. [Google Scholar] [CrossRef]
- Su, X.; Wu, C.; Ye, X.; Zeng, M.; Zhang, Z.; Che, Y.; Zhang, Y.; Liu, L.; Lin, Y.; Yang, R. Embryonic lethality in mice lacking Trim59 due to impaired gastrulation development. Cell Death Dis. 2018, 9, 302. [Google Scholar] [CrossRef]
- Wang, M.; Chao, C.; Luo, G.; Wang, B.; Zhan, X.; Di, D.; Qian, Y.; Zhang, X. Prognostic significance of TRIM59 for cancer patient survival: A systematic review and meta-analysis. Medicine 2019, 98, e18024. [Google Scholar] [CrossRef]
- Guo, J.; Min, K.; Deng, L. Potential value of tripartite motif-containing 59 as a biomarker for predicting the prognosis of patients with lung cancer: A protocol for systematic review and meta-analysis. Medicine 2021, 100, e26868. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Ji, Z.; Wang, Y.; Li, J.; Cao, H.; Zhu, H.H.; Gao, W.Q. TRIM59 is up-regulated in gastric tumors, promoting ubiquitination and degradation of p53. Gastroenterology 2014, 147, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Xing, D.; Li, Z.; Shen, J.; Zhao, H.; Li, S. TRIM59 is upregulated and promotes cell proliferation and migration in human osteosarcoma. Mol. Med. Report. 2016, 13, 5200–5206. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dong, Y.; Zhao, L.; Su, L.; Diao, K.; Mi, X. TRIM59 overexpression correlates with poor prognosis and contributes to breast cancer progression through AKT signaling pathway. Mol. Carcinog. 2018, 57, 1792–1802. [Google Scholar] [CrossRef] [PubMed]
- Che, B.; Du, Y.; Yuan, R.; Xiao, H.; Zhang, W.; Shao, J.; Lu, H.; Yu, Y.; Xiang, M.; Hao, L.; et al. SLC35F2-SYVN1-TRIM59 axis critically regulates ferroptosis of pancreatic cancer cells by inhibiting endogenous p53. Oncogene. 2023, 42, 3260–3273. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Liu, H. TRIM59 knockdown blocks cisplatin resistance in A549/DDP cells through regulating PTEN/AKT/HK2. Gene 2020, 747, 144553. [Google Scholar] [CrossRef]
- Shen, H.; Zhang, J.; Zhang, Y.; Feng, Q.; Wang, H.; Li, G.; Jiang, W.; Li, X. Knockdown of tripartite motif 59 (TRIM59) inhibits proliferation in cholangiocarcinoma via the PI3K/AKT/mTOR signalling pathway. Gene 2019, 698, 50–60. [Google Scholar] [CrossRef]
- Li, R.; Weng, L.; Liu, B.; Zhu, L.; Zhang, X.; Tian, G.; Hu, L.; Li, Q.; Jiang, S.; Shang, M. TRIM59 predicts poor prognosis and promotes pancreatic cancer progression via the PI3K/AKT/mTOR-glycolysis signaling axis. J. Cell. Biochem. 2020, 121, 1986–1997. [Google Scholar] [CrossRef]
- Sang, Y.; Li, Y.; Song, L.; Alvarez, A.A.; Zhang, W.; Lv, D.; Tang, J.; Liu, F.; Chang, Z.; Hatakeyama, S.; et al. TRIM59 promotes gliomagenesis by inhibiting TC45 dephosphorylation of STAT3. Cancer Res. 2018, 78, 1792–1804. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, Z.; Zeng, J.; Zhang, S.; Chen, L.; Zhang, G.; Xu, W.; Song, L.; Guo, X. TRIM59 promotes gefitinib resistance in EGFR mutant lung adenocarcinoma cells. Life Sci. 2019, 224, 23–32. [Google Scholar] [CrossRef]
- Su, X.; Zhang, Q.; Yue, J.; Wang, Y.; Zhang, Y.; Yang, R. TRIM59 suppresses NO production by promoting the binding of PIAS1 and STAT1 in macrophages. Int. Immunopharmacol. 2020, 89, 107030. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lou, J.; Liu, H.; Liu, Y.; Xie, B.; Zhang, W.; Xie, J.; Pan, H.; Han, W. TRIM59 deficiency promotes M1 macrophage activation and inhibits colorectal cancer through the STAT1 signaling pathway. Sci. Rep. 2024, 14, 16081. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Yu, Y.; Dotti, G.; Shen, T.; Tan, X.; Savoldo, B.; Pass, A.K.; Chu, M.; Zhang, D.; Lu, X.; et al. PPM1A and PPM1B act as IKKβ phosphatases to terminate TNFα-induced IKKβ-NF-κB activation. Cell Signal. 2009, 21, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; An, H.; Jin, R.; Zou, M.; Guo, Y.; Su, P.F.; Liu, D.; Shyr, Y.; Yarbrough, W.G. PPM1A is a RelA phosphatase with tumor suppressor-like activity. Oncogene 2014, 33, 2918–2927. [Google Scholar] [CrossRef]
- Ying, H.; Ji, L.; Xu, Z.; Fan, X.; Tong, Y.; Liu, H.; Zhao, J.; Cai, X. TRIM59 promotes tumor growth in hepatocellular carcinoma and regulates the cell cycle by degradation of protein phosphatase 1B. Cancer Lett. 2020, 473, 13–24. [Google Scholar] [CrossRef]
- Wang, F.; Wang, H.; Sun, L.; Niu, C.; Xu, J. TRIM59 inhibits PPM1A through ubiquitination and activates TGF-β/Smad signaling to promote the invasion of ectopic endometrial stromal cells in endometriosis. Am. J. Physiol. Cell Physiol. 2020, 319, C392–C401. [Google Scholar] [CrossRef]
- Li, R.; Gong, Z.; Pan, C.; Xie, D.D.; Tang, J.Y.; Cui, M.; Xu, Y.F.; Yao, W.; Pang, Q.; Xu, Z.G.; et al. Metal-dependent protein phosphatase 1A functions as an extracellular signal-regulated kinase phosphatase. FEBS J. 2013, 280, 2700–2711. [Google Scholar] [CrossRef]
- Dvashi, Z.; Sar Shalom, H.; Shohat, M.; Ben-Meir, D.; Ferber, S.; Satchi-Fainaro, R.; Ashery-Padan, R.; Rosner, M.; Solomon, A.S.; Lavi, S. Protein phosphatase magnesium dependent 1A governs the wound healing-inflammation-angiogenesis cross talk on injury. Am. J. Pathol. 2014, 184, 2936–2950. [Google Scholar] [CrossRef]
- Schaaf, K.; Smith, S.R.; Duverger, A.; Wagner, F.; Wolschendorf, F.; Westfall, A.O.; Kutsch, O.; Sun, J. Mycobacterium tuberculosis exploits the PPM1A signaling pathway to block host macrophage apoptosis. Sci. Rep. 2017, 7, 42101. [Google Scholar] [CrossRef]
- Li, J.; Liu, N.; Tang, L.; Yan, B.; Chen, X.; Zhang, J.; Peng, C. The relationship between TRAF6 and tumors. Cancer Cell Int. 2020, 20, 429. [Google Scholar] [CrossRef]
- Han, T.; Guo, M.; Gan, M.; Yu, B.; Tian, X.; Wang, J.B. TRIM59 regulates autophagy through modulating both the transcription and the ubiquitination of BECN1. Autophagy 2018, 14, 2035–2048. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; He, L.; Zhou, Y. TRIM59 deficiency curtails breast cancer metastasis through SQSTM1-selective autophagic degradation of PDCD10. Autophagy 2019, 15, 747–749. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Chen, X.; Wang, L.; Qin, L.; Wang, H.; Sun, Z.; Zhao, W.; Geng, B. Cancer-derived exosomal TRIM59 regulates macrophage NLRP3 inflammasome activation to promote lung cancer progression. J. Exp. Clin. Cancer Res. 2020, 39, 176. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Zhu, Z.; Liu, S.; Hou, Y.; Tang, M.; Zhu, P.; Tian, Y.; Li, D.; Yan, D.; Zhu, X. TRIM59 protects mice from sepsis by regulating inflammation and phagocytosis in macrophages. Front. Immunol. 2020, 11, 263. [Google Scholar] [CrossRef]
- Kondo, T.; Watanabe, M.; Hatakeyama, S. TRIM59 interacts with ECSIT and negatively regulates NF-κB and IRF-3/7-mediated signal pathways. Biochem. Biophys. Res. Commun. 2012, 422, 501–507. [Google Scholar] [CrossRef]
- Mi Wi, S.; Park, J.; Shim, J.-H.; Chun, E.; Lee, K.-Y. Ubiquitination of ECSIT is crucial for the activation of p65/p50 NF-κBs in Toll-like receptor 4 signaling. Mol. Biol. Cell 2014, 26, 151–160. [Google Scholar] [CrossRef]
- Chen, G.; Chen, W.; Ye, M.; Tan, W.; Jia, B. TRIM59 knockdown inhibits cell proliferation by down-regulating the Wnt/beta-catenin signaling pathway in neuroblastoma. Biosci. Rep. 2019, 39, BSR20181277. [Google Scholar] [CrossRef]
- D’Amico, F.; Mukhopadhyay, R.; Ovaa, H.; Mulder, M.P.C. Targeting TRIM proteins: A quest towards drugging an emerging protein class. ChemBioChem 2021, 22, 2011–2031. [Google Scholar] [CrossRef]
- Eberhardt, W.; Haeussler, K.; Nasrullah, U.; Pfeilschifter, J. Multifaceted roles of TRIM proteins in colorectal carcinoma. Int. J. Mol. Sci. 2020, 21, 7532. [Google Scholar] [CrossRef]
- Tang, H.; Li, X.; Jiang, L.; Liu, Z.; Chen, L.; Chen, J.; Deng, M.; Zhou, F.; Zheng, X.; Liu, Z. RITA1 drives the growth of bladder cancer cells by recruiting TRIM25 to facilitate the proteasomal degradation of RBPJ. Cancer Sci. 2022, 113, 3071–3084. [Google Scholar] [CrossRef]
- Huang, N.; Sun, X.; Li, P.; Liu, X.; Zhang, X.; Chen, Q.; Xin, H. TRIM family contribute to tumorigenesis, cancer development, and drug resistance. Exp. Hematol. Oncol. 2022, 11, 75. [Google Scholar] [CrossRef]
- Sanchez, J.G.; Sparrer, K.M.J.; Chiang, C.; Reis, R.A.; Chiang, J.J.; Zurenski, M.A.; Wan, Y.; Gack, M.U.; Pornillos, O. TRIM25 binds RNA to modulate cellular anti-viral defense. J. Mol. Biol. 2018, 430, 5280–5293. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiang, D.C.; Yap, B.K. TRIM25, TRIM28 and TRIM59 and Their Protein Partners in Cancer Signaling Crosstalk: Potential Novel Therapeutic Targets for Cancer. Curr. Issues Mol. Biol. 2024, 46, 10745-10761. https://doi.org/10.3390/cimb46100638
Chiang DC, Yap BK. TRIM25, TRIM28 and TRIM59 and Their Protein Partners in Cancer Signaling Crosstalk: Potential Novel Therapeutic Targets for Cancer. Current Issues in Molecular Biology. 2024; 46(10):10745-10761. https://doi.org/10.3390/cimb46100638
Chicago/Turabian StyleChiang, De Chen, and Beow Keat Yap. 2024. "TRIM25, TRIM28 and TRIM59 and Their Protein Partners in Cancer Signaling Crosstalk: Potential Novel Therapeutic Targets for Cancer" Current Issues in Molecular Biology 46, no. 10: 10745-10761. https://doi.org/10.3390/cimb46100638
APA StyleChiang, D. C., & Yap, B. K. (2024). TRIM25, TRIM28 and TRIM59 and Their Protein Partners in Cancer Signaling Crosstalk: Potential Novel Therapeutic Targets for Cancer. Current Issues in Molecular Biology, 46(10), 10745-10761. https://doi.org/10.3390/cimb46100638





