Recent Advances in Molecular Mechanisms of Nucleoside Antivirals
Abstract
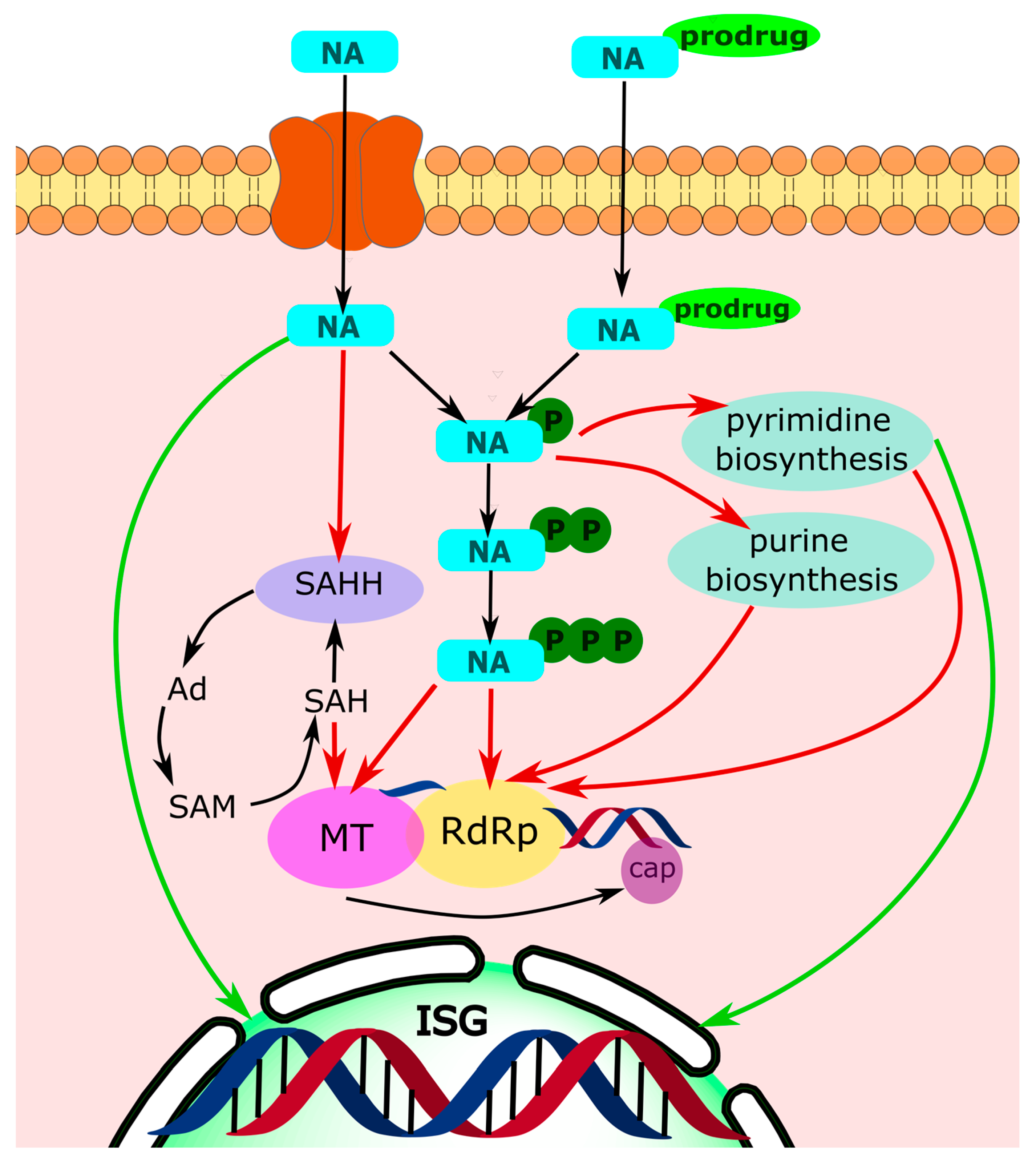
1. Introduction
2. Polymerase Targeting
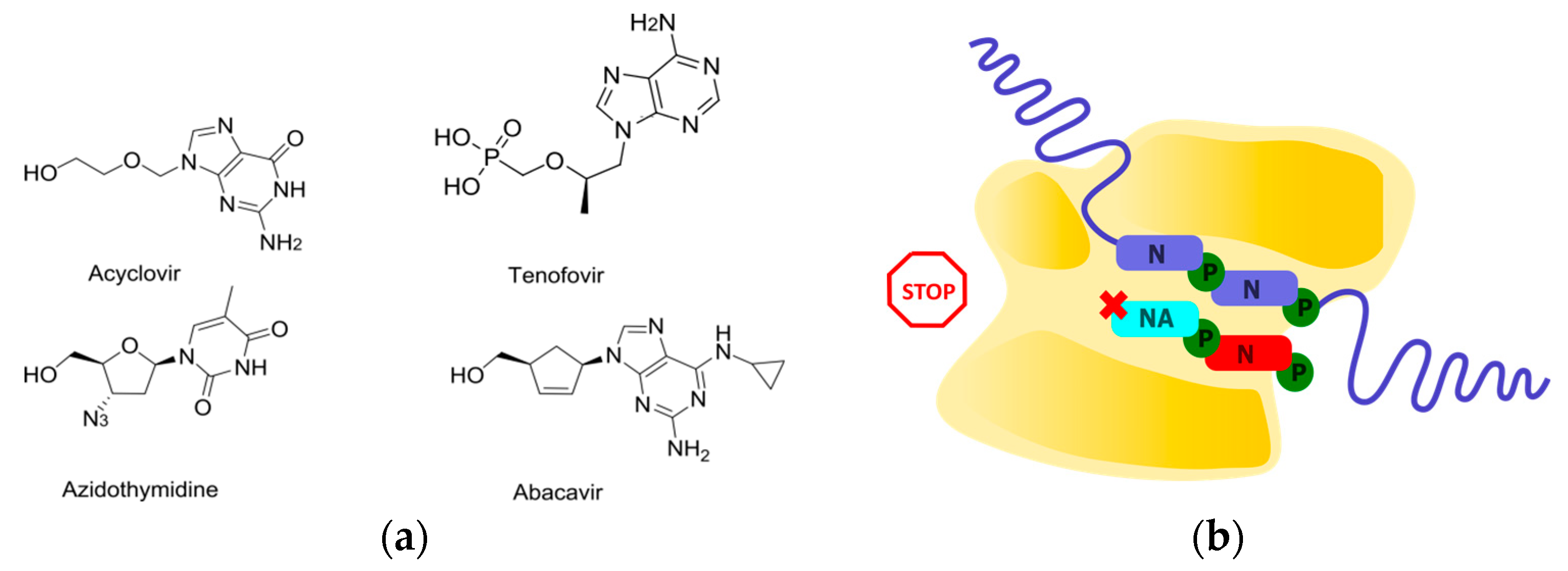
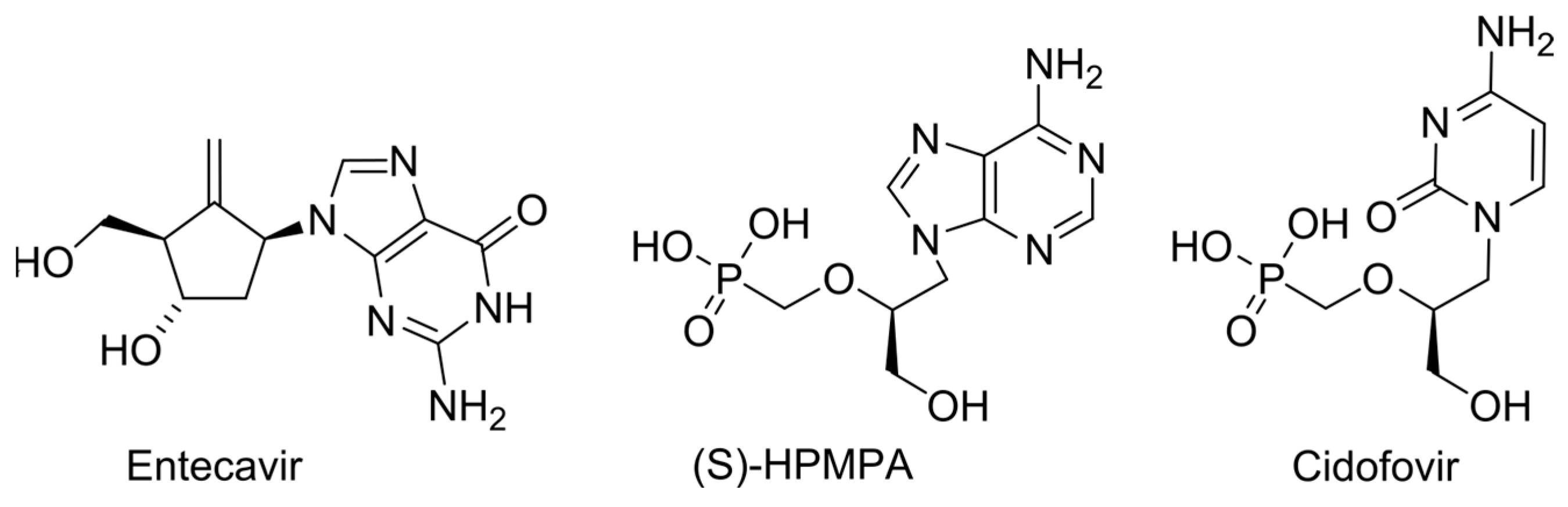
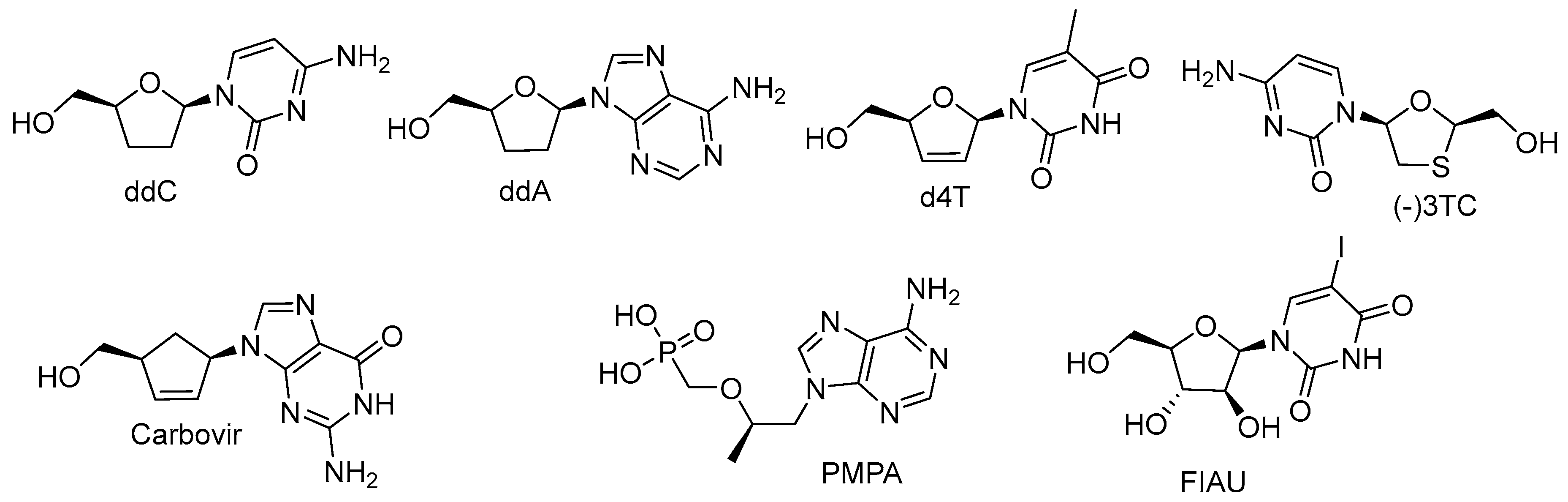
2.1. Obligate Termination
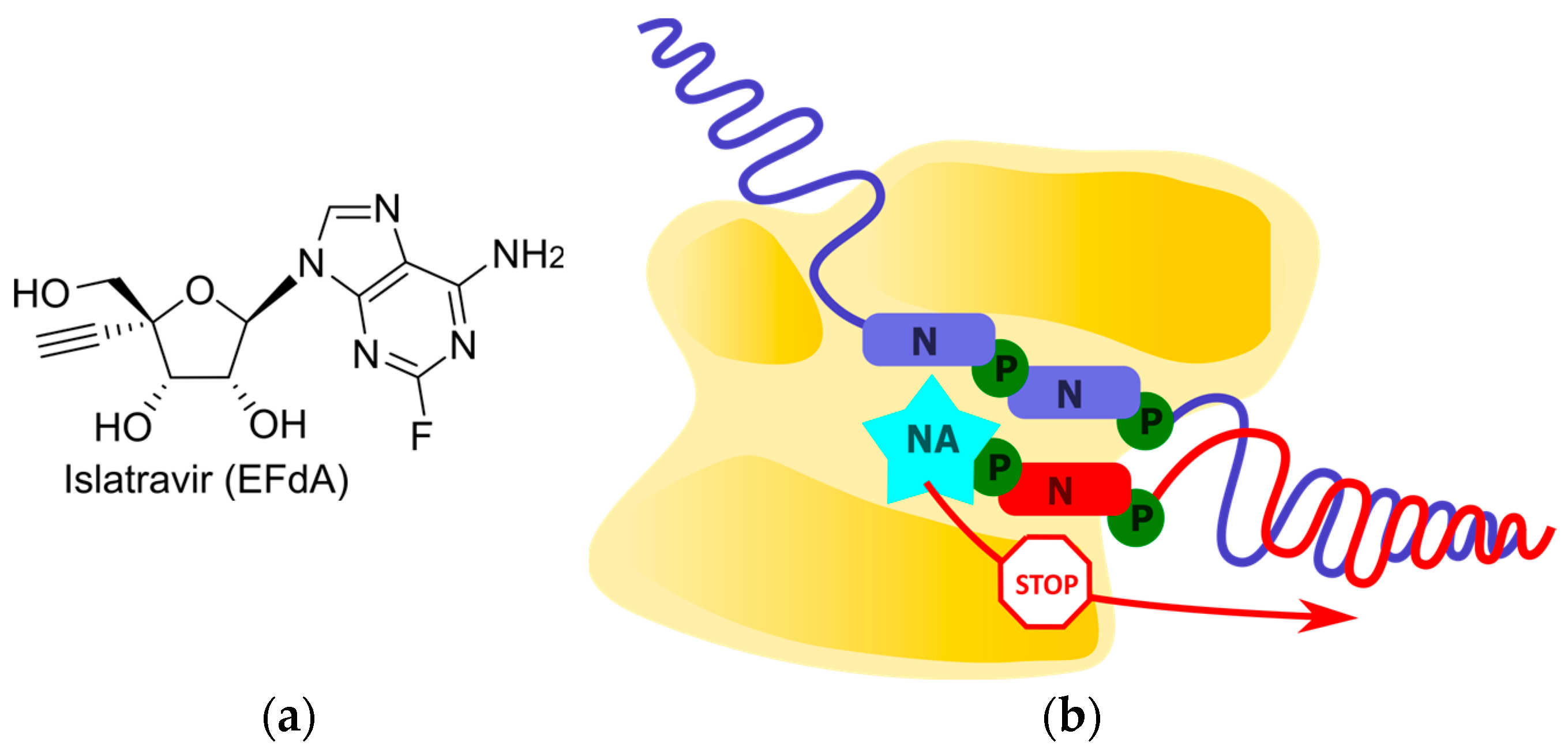
2.2. Non-Obligate Termination
2.2.1. Translocation Inhibition
2.2.2. Arrest of Chain Elongation through the Disruption of H-Bond Network between Polymerase and Incoming NTP
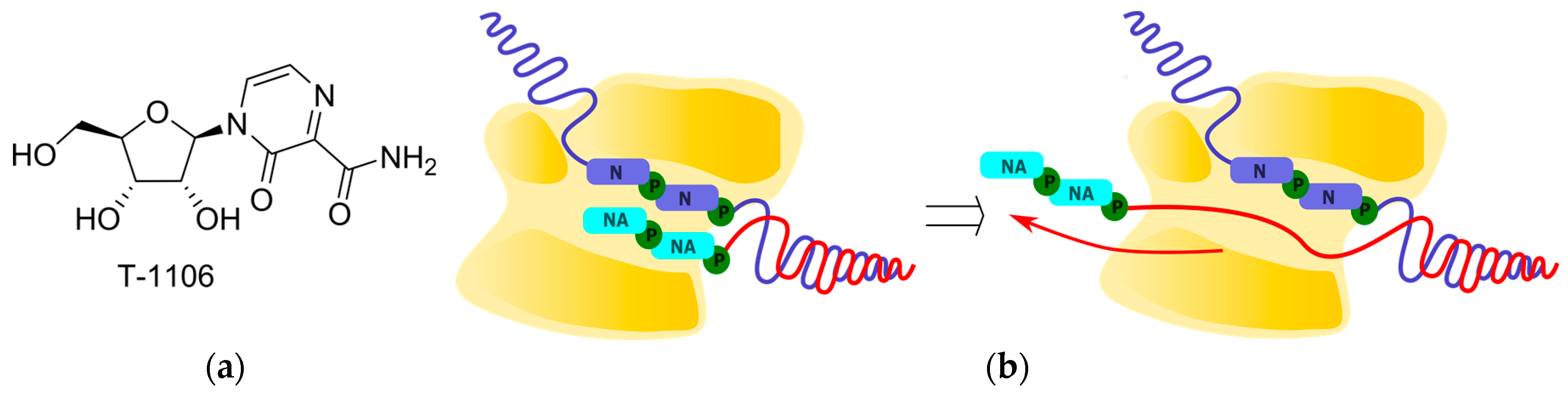
2.2.3. Polymerase Backtracking
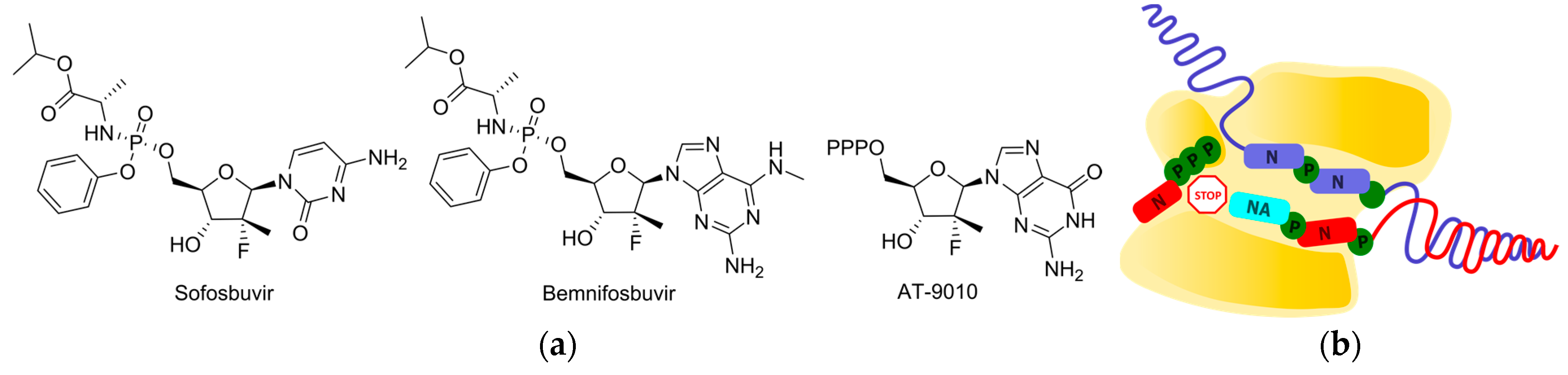
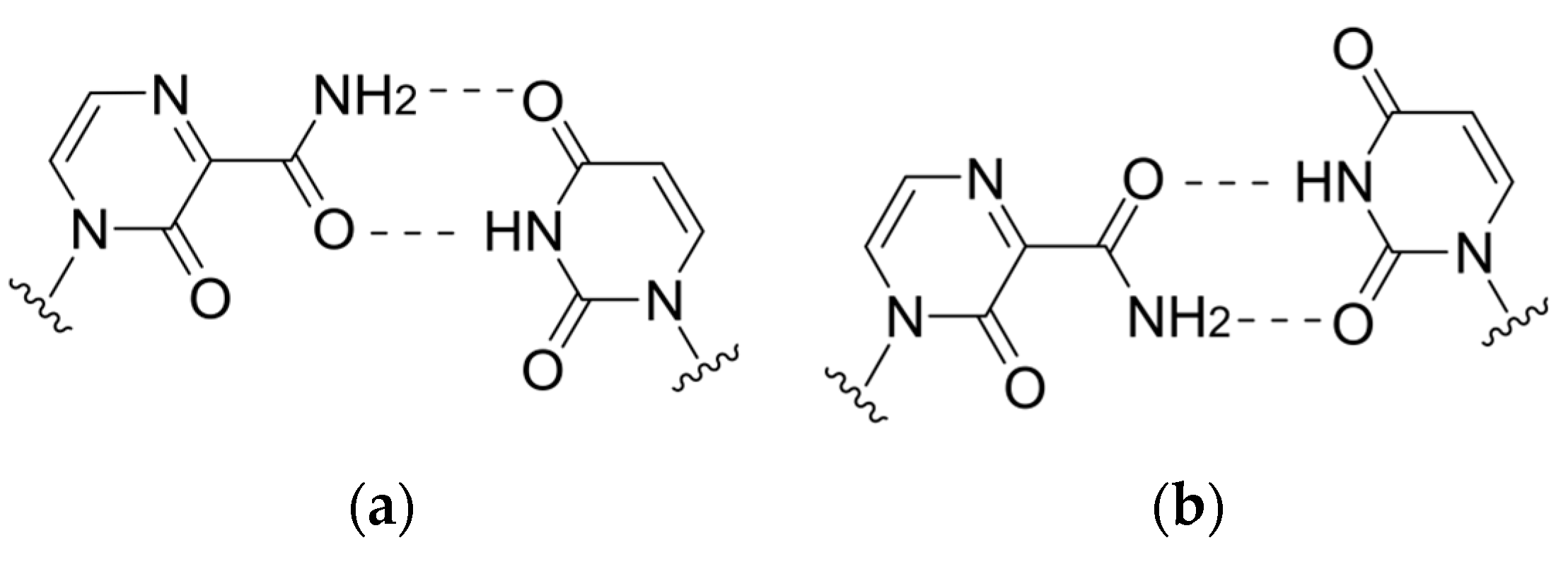
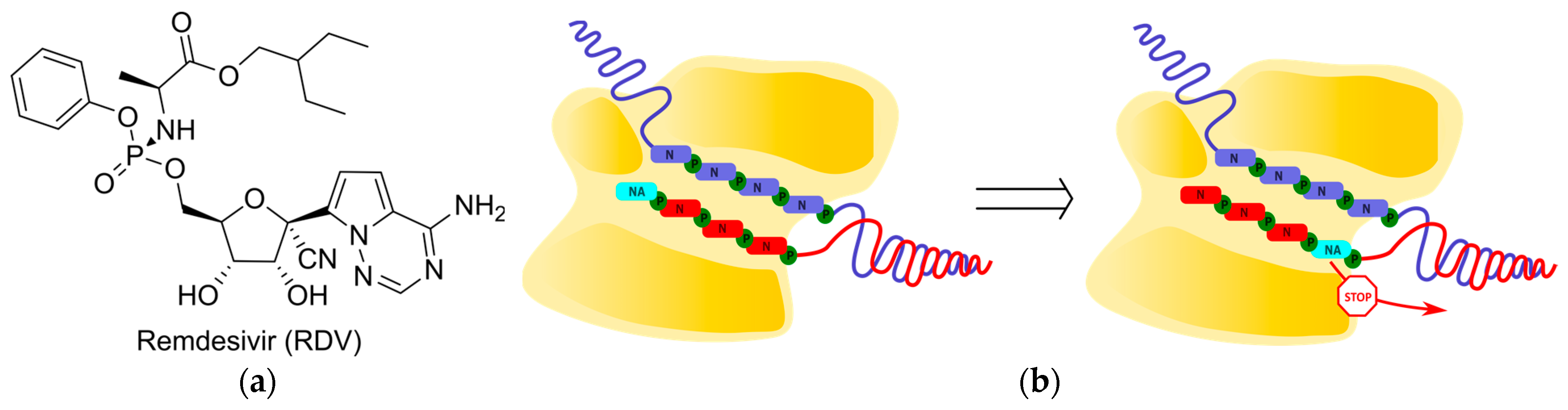
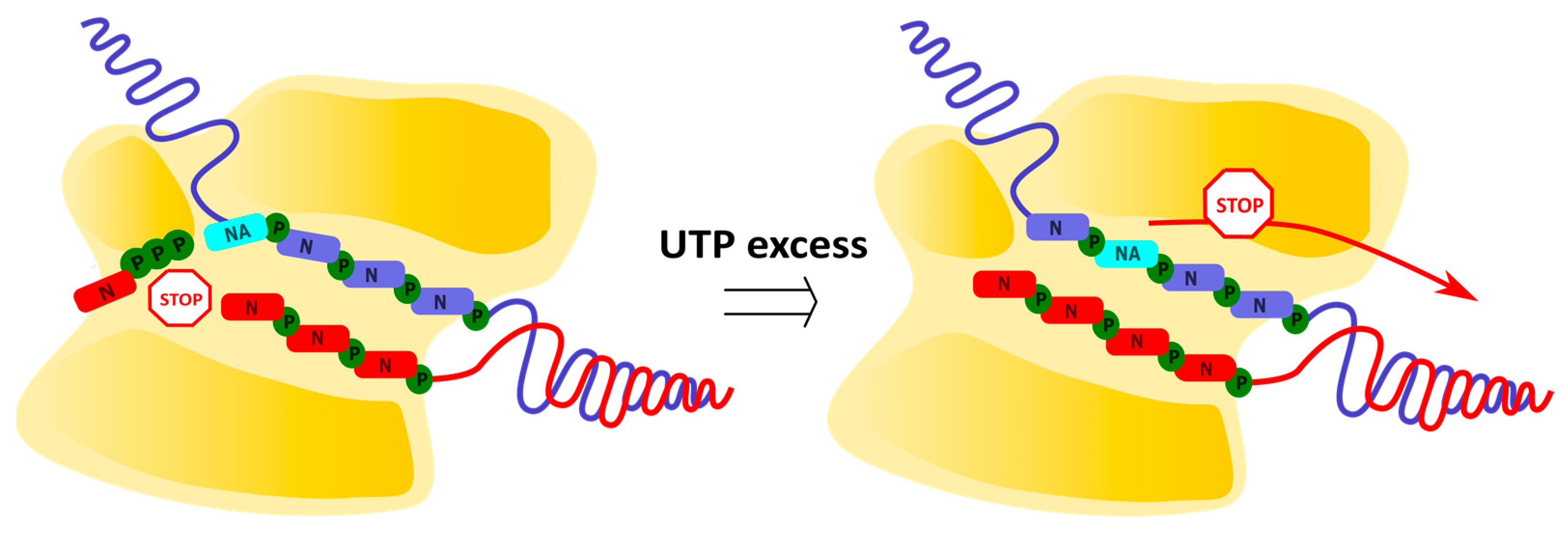
2.3. Delayed Termination
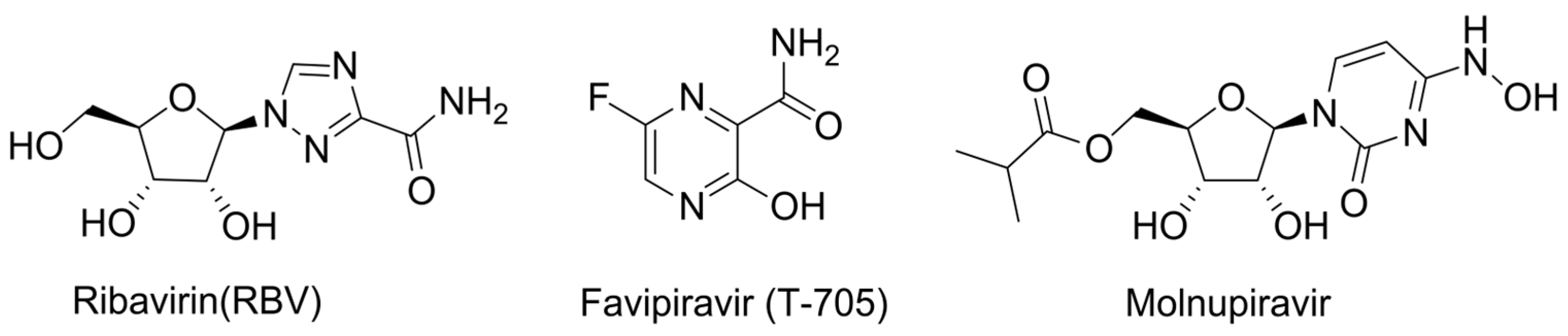
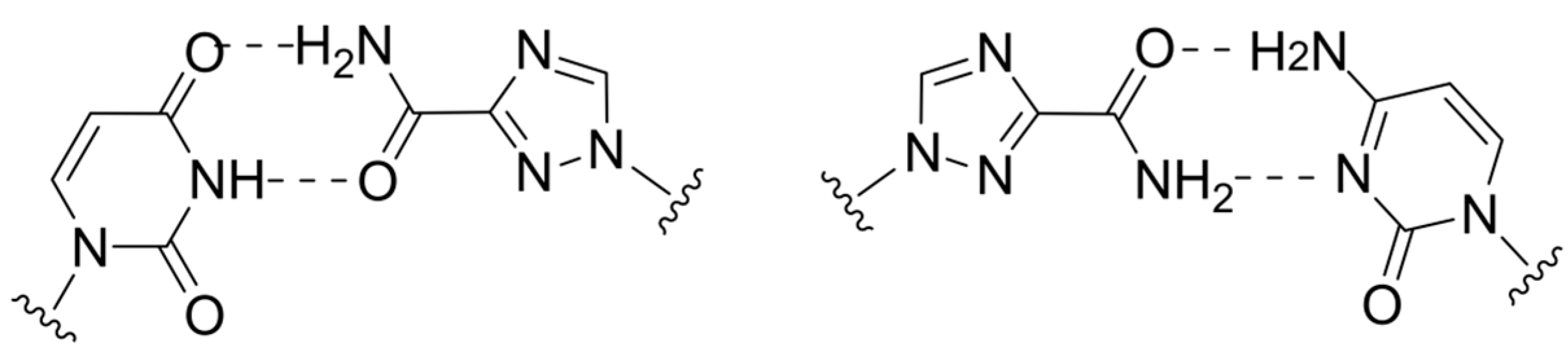
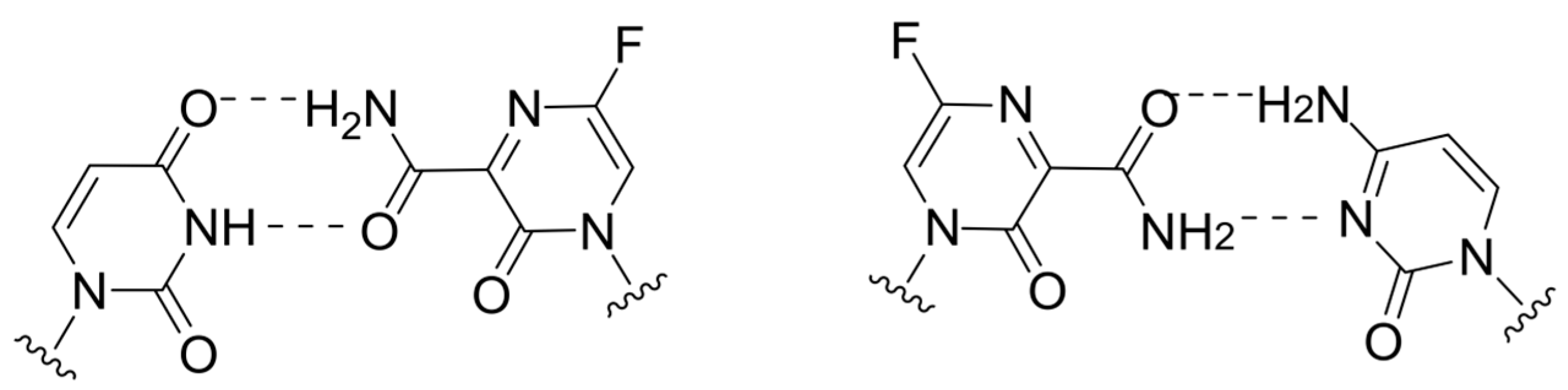
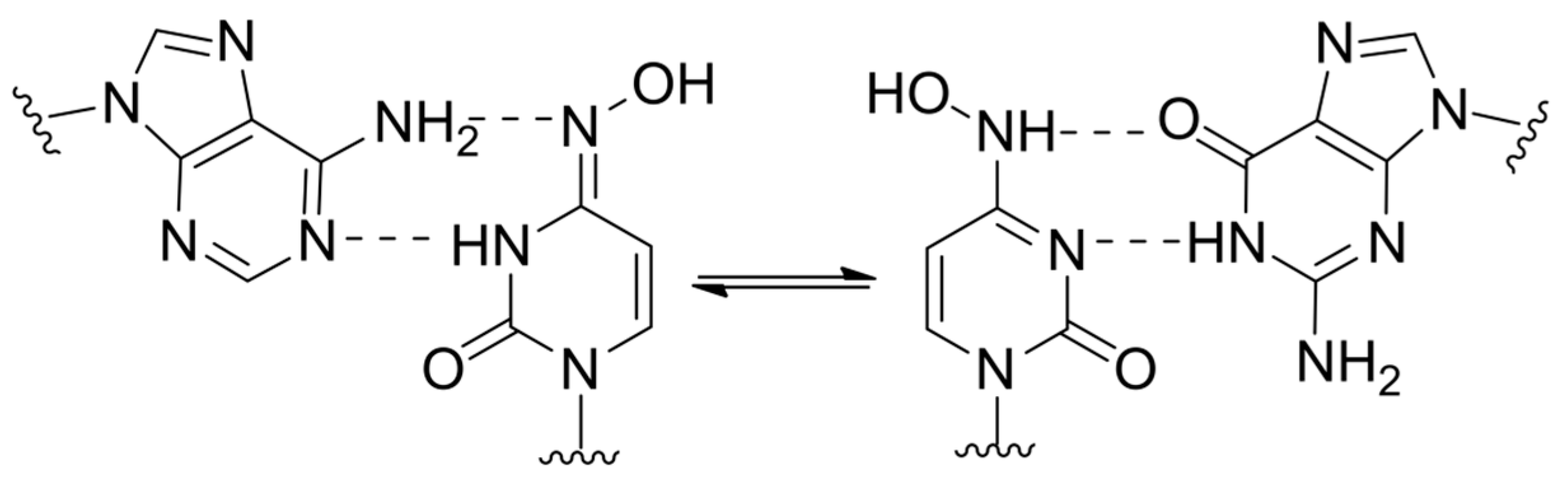
2.4. Lethal Mutagenesis
2.5. RdRp Inhibition without NA Incorporation
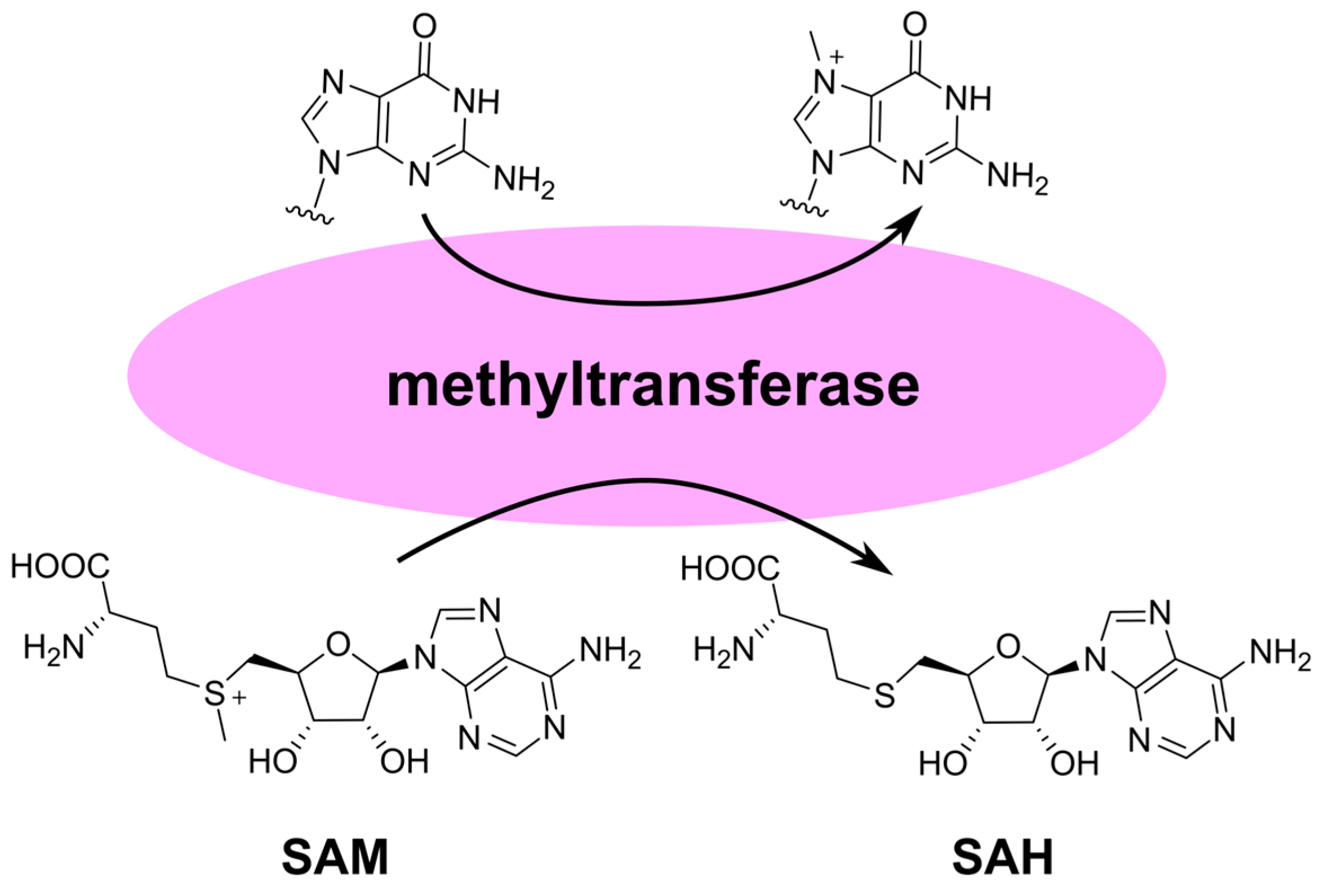
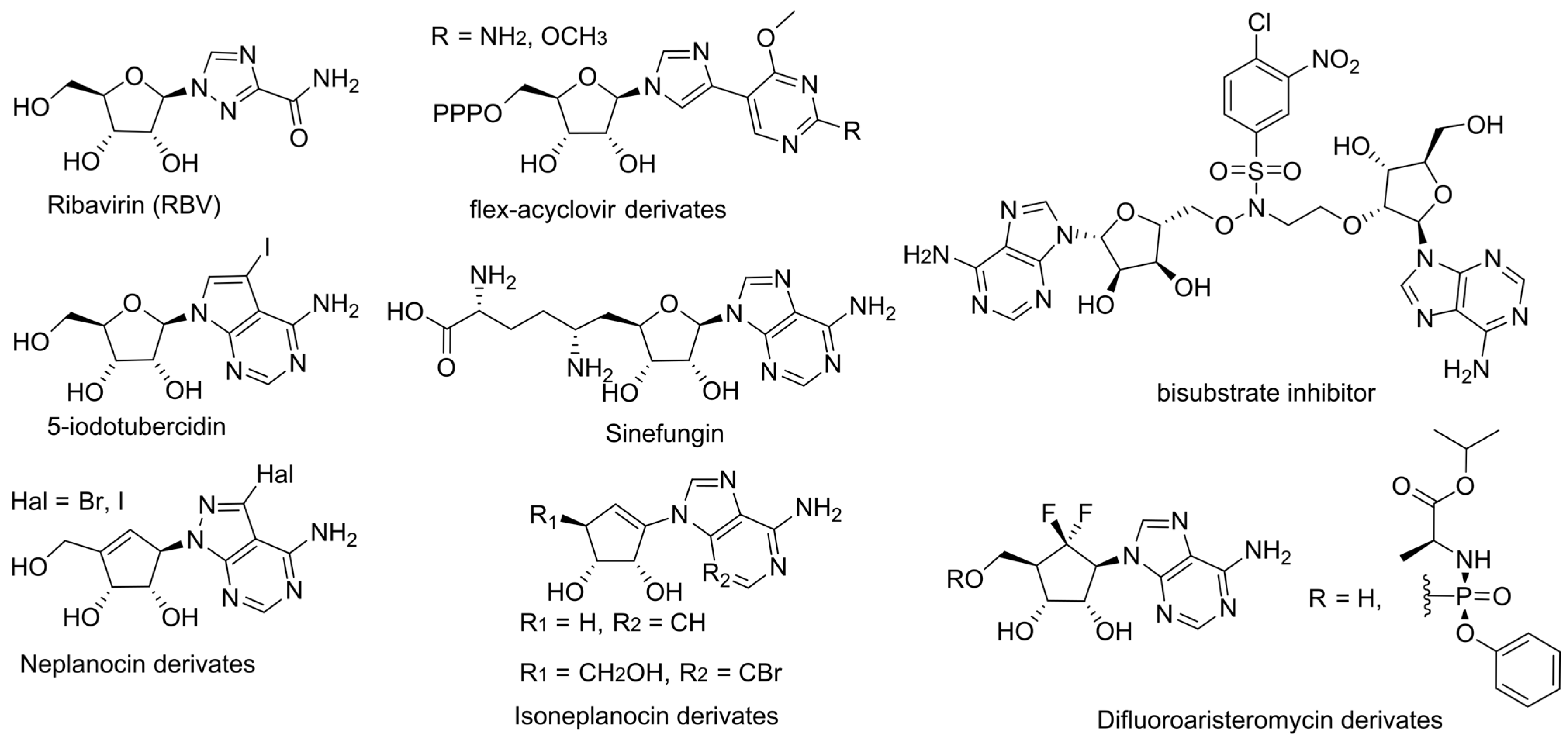
3. Inhibition of Viral Genome Methylation
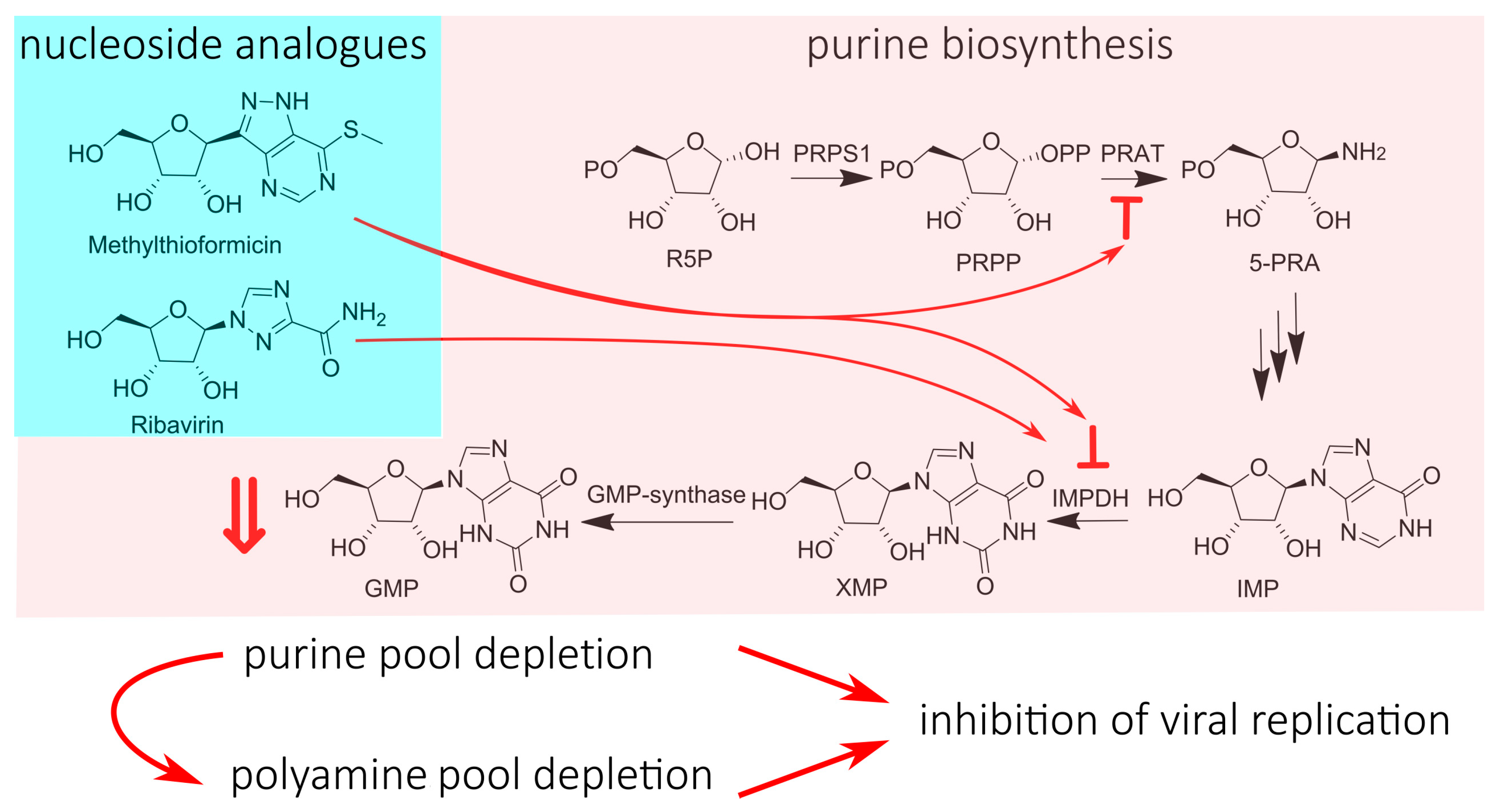
4. Nucleotide Pool Depletion
5. Immunomodulation
6. Prodrugs
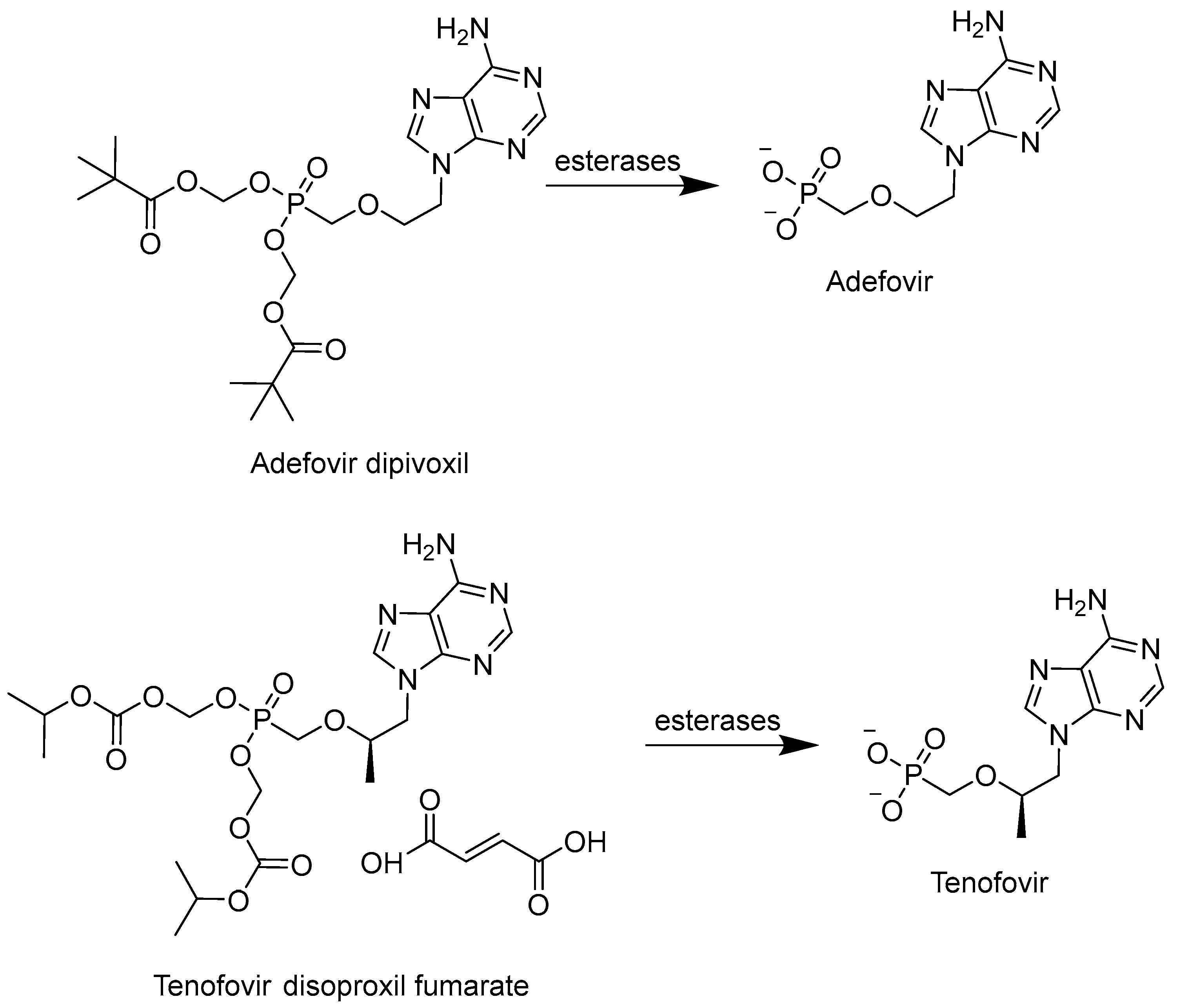
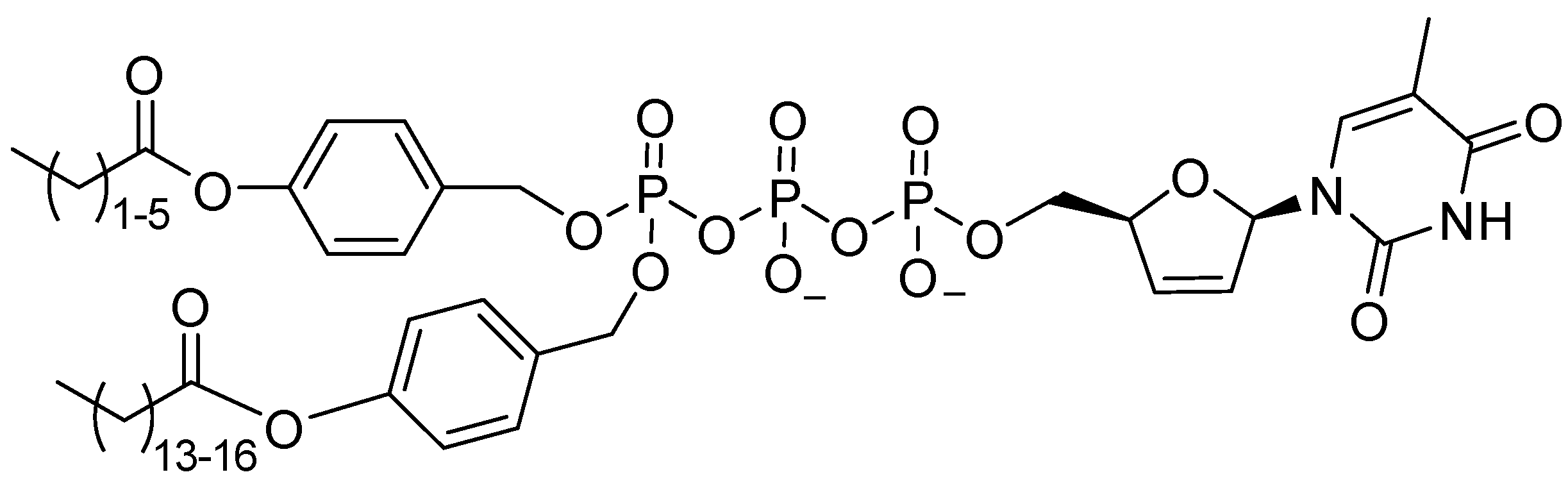
6.1. Phosphonates and POM/POC-Masked Prodrugs
6.2. McGuigan’s ProTides
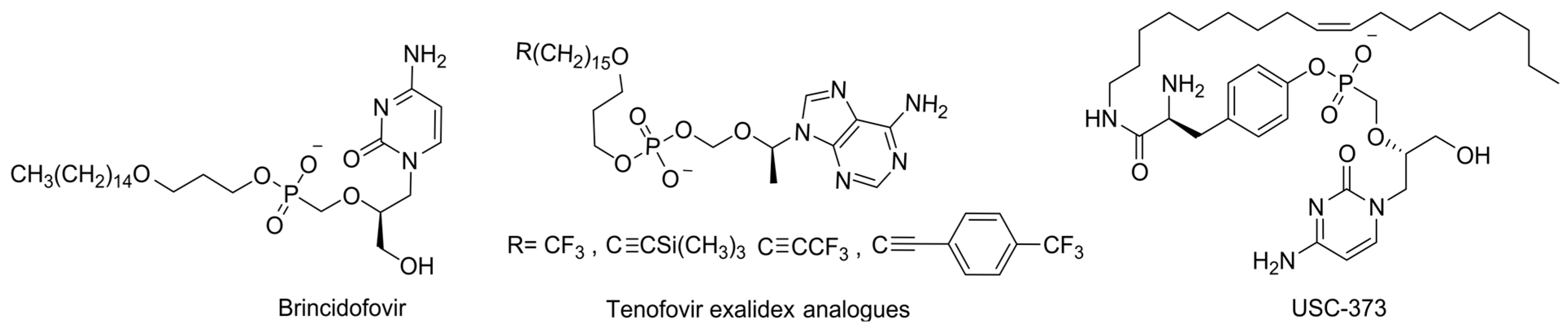
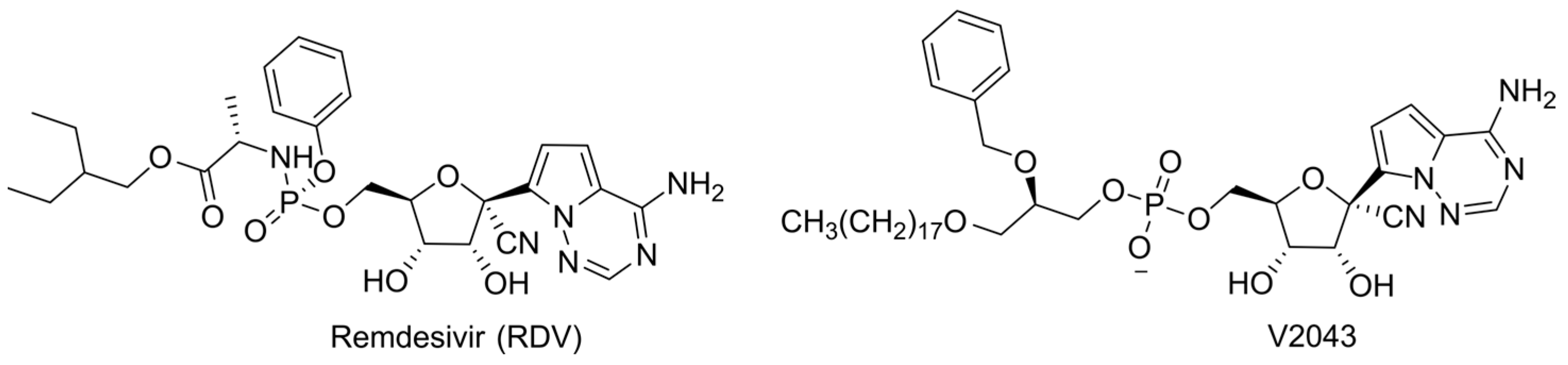
6.3. Lipid-Masked Prodrugs
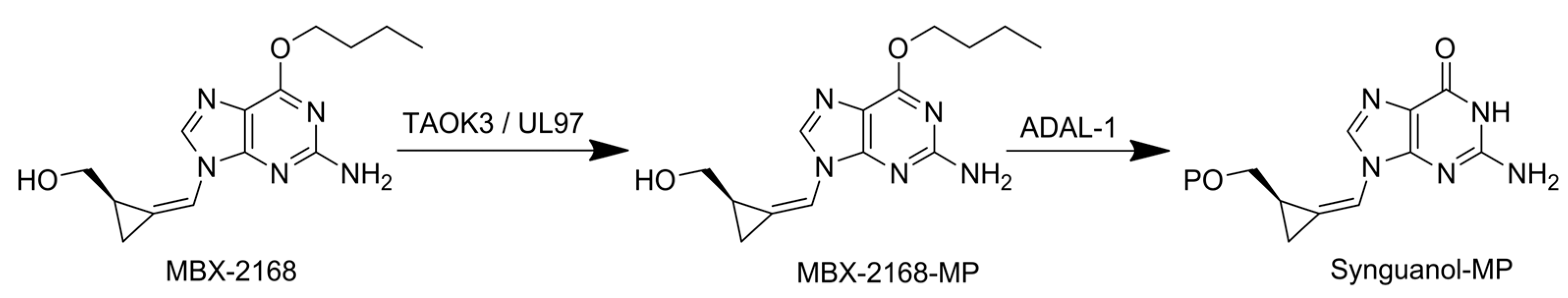
6.4. 6-Modified Purine Analogues
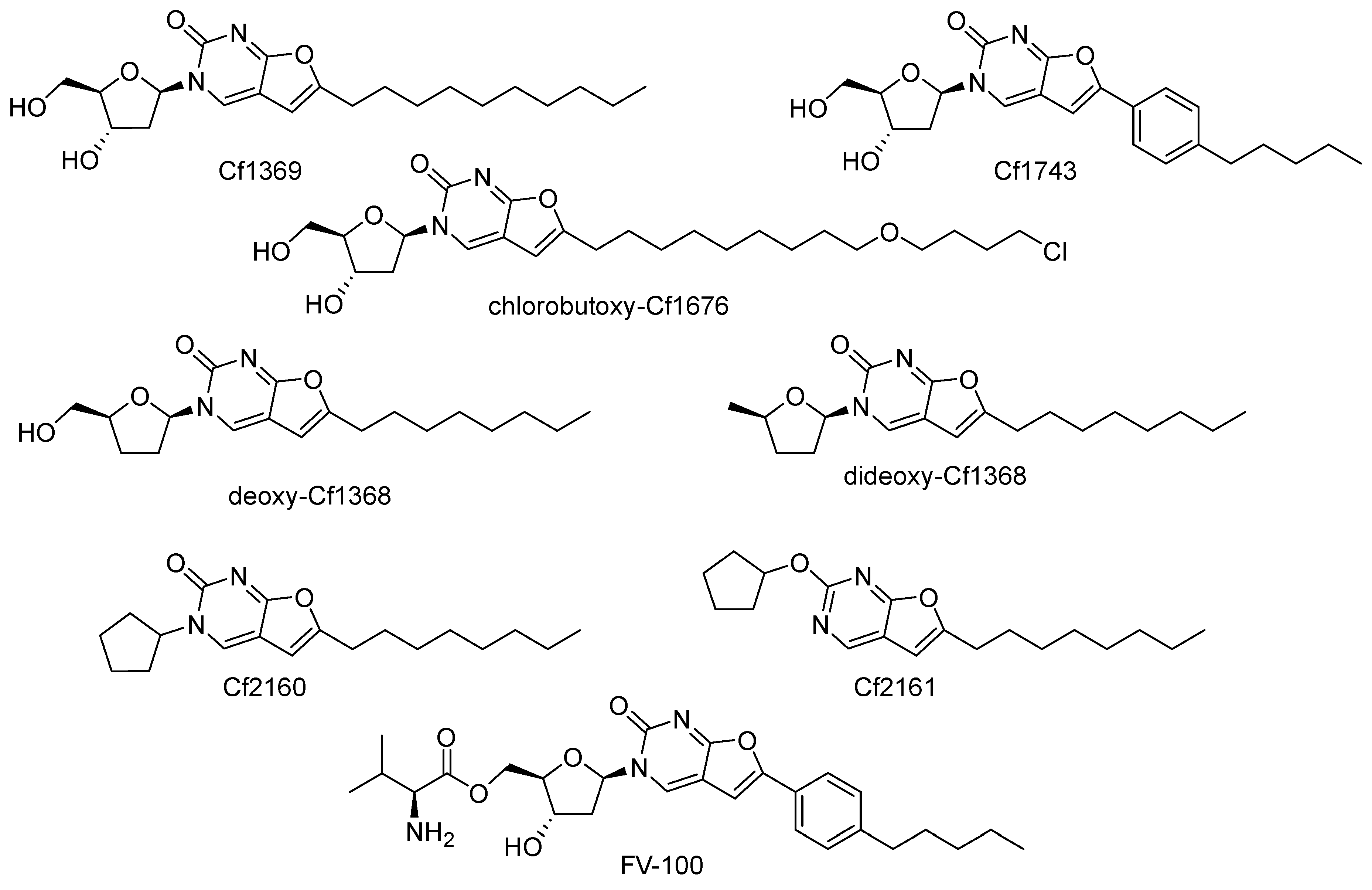
7. Furo[2,3-d]pyrimidin-2-one Compounds (Bicyclic Nucleoside Analogues, BCNAs)
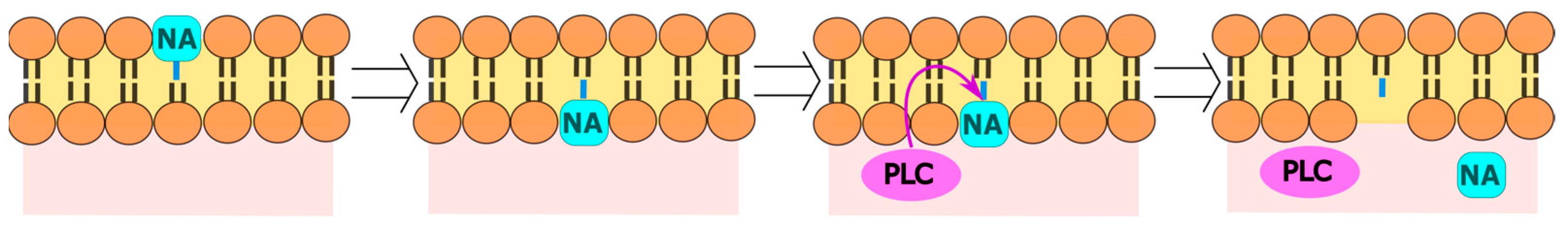
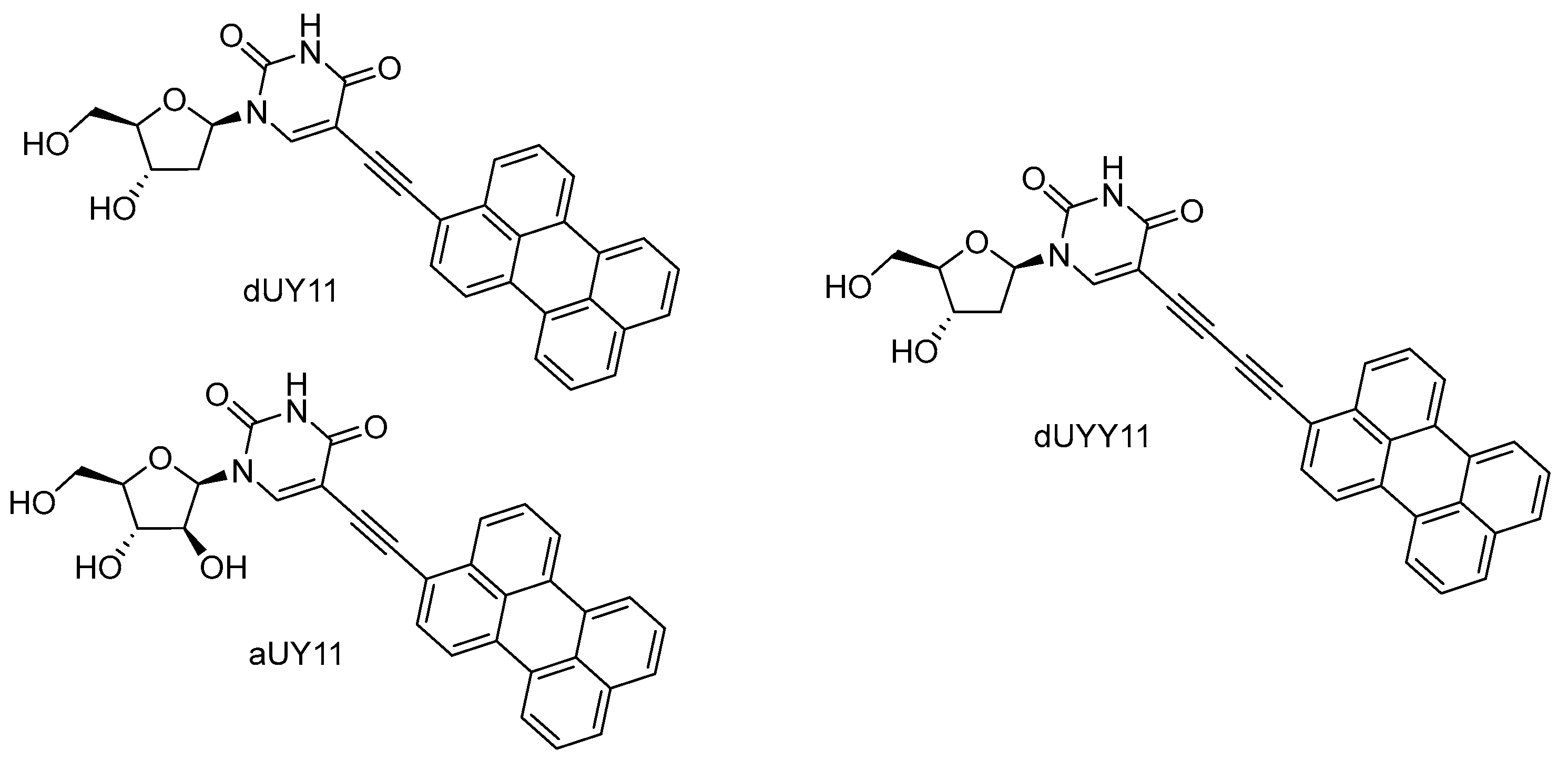
8. Perylene Nucleoside Derivatives Exerting Antiviral Activity through a Non-Nucleoside Mechanism
9. Adverse Effects
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chowell, G.; Echevarría-Zuno, S.; Viboud, C.; Simonsen, L.; Tamerius, J.; Miller, M.A.; Borja-Aburto, V.H. Characterizing the epidemiology of the 2009 influenza A/H1N1 pandemic in Mexico. PLoS Med. 2011, 8, e1000436. [Google Scholar] [CrossRef]
- Cenciarelli, O.; Pietropaoli, S.; Malizia, A.; Carestia, M.; D’Amico, F.; Sassolini, A.; Di Giovanni, D.; Rea, S.; Gabbarini, V.; Tamburrini, A.; et al. Ebola virus disease 2013–2014 outbreak in West Africa: An analysis of the epidemic spread and response. Int. J. Microbiol. 2015, 2015, 769121. [Google Scholar] [CrossRef] [PubMed]
- Mackay, I.M.; Arden, K.E. MERS coronavirus: Diagnostics, epidemiology and transmission. Virol. J. 2015, 12, 222. [Google Scholar] [CrossRef] [PubMed]
- Contini, C.; Di Nuzzo, M.; Barp, N.; Bonazza, A.; De Giorgio, R.; Tognon, M.; Rubino, S. The novel zoonotic COVID-19 pandemic: An expected global health concern. J. Infect. Dev. Ctries. 2020, 14, 254–264. [Google Scholar] [CrossRef]
- Walsh, D.; Mohr, I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011, 9, 860–875. [Google Scholar] [CrossRef] [PubMed]
- Nijhuis, M.; Van Maarseveen, N.M.; Boucher, C.A.B. Antiviral resistance and impact on viral replication capacity: Evolution of viruses under antiviral pressure occurs in three phases. In Antiviral Strategies; Kräusslich, H.-G., Bartenschlager, R., Eds.; Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2009; Volume 189, pp. 299–320. ISBN 978-3-540-79085-3. [Google Scholar] [CrossRef]
- Mercorelli, B.; Palù, G.; Loregian, A. Drug repurposing for viral infectious diseases: How far are we? Tr. Microbiol. 2018, 26, 865–876. [Google Scholar] [CrossRef]
- Jordheim, L.P.; Durantel, D.; Zoulim, F.; Dumontet, C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013, 12, 447–464. [Google Scholar] [CrossRef]
- De Clercq, E.; Li, G. Approved antiviral drugs over the Past 50 Years. Clin. Microbiol. Rev. 2016, 29, 695–747. [Google Scholar] [CrossRef]
- Ismail, M.M.F.; Ayoup, M.S. Review on fluorinated nucleoside/non-nucleoside FDA-approved antiviral drugs. RSC Adv. 2022, 12, 31032–31045. [Google Scholar] [CrossRef]
- Lowe, P.T.; O’Hagan, D. 4′-Fluoro-nucleosides and nucleotides: From nucleocidin to an emerging class of therapeutics. Chem. Soc. Rev. 2023, 52, 248–276. [Google Scholar] [CrossRef]
- Chang, J. 4′-Modified nucleosides for antiviral drug discovery: Achievements and perspectives. Acc. Chem. Res. 2022, 55, 565–578. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fan, P.-H.; Lin, G.-M.; Chang, W.-C.; Liu, H. Recent progress in unusual carbohydrate-containing natural products biosynthesis. In Comprehensive Natural Products III; Elsevier: Amsterdam, The Netherlands, 2020; Volume 2, pp. 336–392. ISBN 978-0-08-102691-5. [Google Scholar] [CrossRef]
- Yan, Y.-C.; Zhang, H.; Hu, K.; Zhou, S.-M.; Chen, Q.; Qu, R.-Y.; Yang, G.-F. A mini-review on synthesis and antiviral activity of natural product oxetanocin A derivatives. Bioorg. Med. Chem. 2022, 72, 116968. [Google Scholar] [CrossRef] [PubMed]
- De Jonghe, S.; Herdewijn, P. An overview of marketed nucleoside and nucleotide analogs. Curr. Protoc. 2022, 2, e376. [Google Scholar] [CrossRef] [PubMed]
- Eyer, L.; Valdés, J.J.; Gil, V.A.; Nencka, R.; Hřebabecký, H.; Šála, M.; Salát, J.; Černý, J.; Palus, M.; De Clercq, E.; et al. Nucleoside inhibitors of tick-borne encephalitis virus. Antimicrob. Agents Chemother. 2015, 59, 5483–5493. [Google Scholar] [CrossRef]
- Eyer, L.; Šmídková, M.; Nencka, R.; Neča, J.; Kastl, T.; Palus, M.; De Clercq, E.; Růžek, D. Structure-activity relationships of nucleoside analogues for inhibition of tick-borne encephalitis virus. Antivir. Res. 2016, 133, 119–129. [Google Scholar] [CrossRef]
- Eyer, L.; Nencka, R.; Huvarová, I.; Palus, M.; Joao Alves, M.; Gould, E.A.; De Clercq, E.; Růžek, D. Nucleoside inhibitors of Zika virus. J. Infect. Dis. 2016, 214, 707–711. [Google Scholar] [CrossRef]
- De Clercq, E. C-Nucleosides to be revisited: Miniperspective. J. Med. Chem. 2016, 59, 2301–2311. [Google Scholar] [CrossRef]
- Eyer, L.; Nencka, R.; De Clercq, E.; Seley-Radtke, K.; Růžek, D. Nucleoside analogs as a rich source of antiviral agents active against arthropod-borne flaviviruses. Antivir. Chem. Chemother. 2018, 26, 204020661876129. [Google Scholar] [CrossRef]
- Seley-Radtke, K.L.; Yates, M.K. The evolution of nucleoside analogue antivirals: A review for chemists and non-chemists. Part 1: Early structural modifications to the nucleoside scaffold. Antivir. Res. 2018, 154, 66–86. [Google Scholar] [CrossRef]
- Yates, M.K.; Seley-Radtke, K.L. The evolution of antiviral nucleoside analogues: A review for chemists and non-chemists. Part II: Complex modifications to the nucleoside scaffold. Antivir. Res. 2019, 162, 5–21. [Google Scholar] [CrossRef]
- De Clercq, E. New nucleoside analogues for the treatment of hemorrhagic fever virus infections. Chem. Asian J. 2019, 14, 3962–3968. [Google Scholar] [CrossRef]
- Pruijssers, A.J.; Denison, M.R. Nucleoside analogues for the treatment of coronavirus infections. Curr. Opin. Virol. 2019, 35, 57–62. [Google Scholar] [CrossRef]
- Ami, E.; Ohrui, H. Intriguing antiviral modified nucleosides: A retrospective view into the future treatment of COVID-19. ACS Med. Chem. Lett. 2021, 12, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Kataev, V.E.; Garifullin, B.F. Antiviral nucleoside analogs. Chem. Heterocycl. Compd. 2021, 57, 326–341. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, D.; Vijayakumar, B.G.; Kannan, T. Advances in nucleoside and nucleotide analogues in tackling human immunodeficiency virus and hepatitis virus infections. ChemMedChem 2021, 16, 1403–1419. [Google Scholar] [CrossRef]
- Borbone, N.; Piccialli, G.; Roviello, G.N.; Oliviero, G. Nucleoside analogs and nucleoside precursors as drugs in the fight against SARS-CoV-2 and other coronaviruses. Molecules 2021, 26, 986. [Google Scholar] [CrossRef] [PubMed]
- Zenchenko, A.A.; Drenichev, M.S.; Il’icheva, I.A.; Mikhailov, S.N. Antiviral and antimicrobial nucleoside derivatives: Structural features and mechanisms of action. Mol. Biol. 2021, 55, 786–812. [Google Scholar] [CrossRef] [PubMed]
- Geraghty, R.; Aliota, M.; Bonnac, L. Broad-spectrum antiviral strategies and nucleoside analogues. Viruses 2021, 13, 667. [Google Scholar] [CrossRef]
- Stevaert, A.; Groaz, E.; Naesens, L. Nucleoside analogs for management of respiratory virus infections: Mechanism of action and clinical efficacy. Curr. Opin. Virol. 2022, 57, 101279. [Google Scholar] [CrossRef]
- Roy, V.; Agrofoglio, L.A. Nucleosides and emerging viruses: A new story. Drug Discov. Today 2022, 27, 1945–1953. [Google Scholar] [CrossRef]
- Abdullah Al Awadh, A. Nucleotide and nucleoside-based drugs: Past, present, and future. Saudi J. Biol. Sci. 2022, 29, 103481. [Google Scholar] [CrossRef]
- Hadj Hassine, I.; Ben M’hadheb, M.; Menéndez-Arias, L. Lethal mutagenesis of RNA viruses and approved drugs with antiviral mutagenic activity. Viruses 2022, 14, 841. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, Z. Bench-to-bedside: Innovation of small molecule anti-SARS-CoV-2 drugs in China. Eur. J. Med. Chem. 2023, 257, 115503. [Google Scholar] [CrossRef] [PubMed]
- Maria, C.; Rauter, A.P. Nucleoside analogues: N-glycosylation methodologies, synthesis of antiviral and antitumor drugs and potential against drug-resistant bacteria and Alzheimer’s disease. Carbohydr. Res. 2023, 532, 108889. [Google Scholar] [CrossRef] [PubMed]
- Ojeda-Porras, A.C.; Roy, V.; Agrofoglio, L.A. Chemical approaches to carbocyclic nucleosides. Chem. Rec. 2022, 22, e202100307. [Google Scholar] [CrossRef] [PubMed]
- Rajapaksha, D.G.; Mondal, S.; Wang, J.W.; Meanwell, M.W. A guide for the synthesis of key nucleoside scaffolds in drug discovery. Med. Chem. Res. 2023, 32, 1315–1333. [Google Scholar] [CrossRef]
- Kausar, S.; Said Khan, F.; Ishaq Mujeeb Ur Rehman, M.; Akram, M.; Riaz, M.; Rasool, G.; Hamid Khan, A.; Saleem, I.; Shamim, S.; Malik, A. A review: Mechanism of action of antiviral drugs. Int. J. Immunopathol. Pharmacol. 2021, 35, 205873842110026. [Google Scholar] [CrossRef]
- Wright, N.J.; Lee, S.-Y. Toward a molecular basis of cellular nucleoside transport in humans. Chem. Rev. 2021, 121, 5336–5358. [Google Scholar] [CrossRef]
- Gong, P.; Peersen, O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 2010, 107, 22505–22510. [Google Scholar] [CrossRef]
- Shannon, A.; Canard, B. Kill or corrupt: Mechanisms of action and drug-resistance of nucleotide analogues against SARS-CoV-2. Antivir. Res. 2023, 210, 105501. [Google Scholar] [CrossRef]
- Esposito, F.; Corona, A.; Tramontano, E. HIV-1 reverse transcriptase still remains a new drug target: Structure, function, classical inhibitors, and new inhibitors with innovative mechanisms of actions. Mol. Biol. Int. 2012, 2012, 586401. [Google Scholar] [CrossRef]
- Shaw, T.A.; Ablenas, C.J.; Desrochers, G.F.; Powdrill, M.H.; Bilodeau, D.A.; Vincent-Rocan, J.-F.; Niu, M.; Monette, A.; Mouland, A.J.; Beauchemin, A.M.; et al. A bifunctional nucleoside probe for the inhibition of the human immunodeficiency virus-type 1 reverse transcriptase. Bioconjug. Chem. 2020, 31, 1537–1544. [Google Scholar] [CrossRef]
- Markowitz, M.; Sarafianos, S.G. 4′-Ethynyl-2-fluoro-2′-deoxyadenosine, MK-8591: A novel HIV-1 reverse transcriptase translocation inhibitor. Curr. Opin. HIV AIDS 2018, 13, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Michailidis, E.; Marchand, B.; Kodama, E.N.; Singh, K.; Matsuoka, M.; Kirby, K.A.; Ryan, E.M.; Sawani, A.M.; Nagy, E.; Ashida, N.; et al. Mechanism of inhibition of HIV-1 reverse transcriptase by 4′-ethynyl-2-fluoro-2′-deoxyadenosine triphosphate, a translocation-defective reverse transcriptase inhibitor. J. Biol. Chem. 2009, 284, 35681–35691. [Google Scholar] [CrossRef] [PubMed]
- Kirby, K.A.; Singh, K.; Michailidis, E.; Marchand, B.; Kodama, E.N.; Ashida, N.; Mitsuya, H.; Parniak, M.A.; Sarafianos, S.G. The sugar ring conformation of 4′-ethynyl-2-fluoro-2′-deoxyadenosine and its recognition by the polymerase active site of HIV reverse transcriptase. Cell. Mol. Biol. 2011, 57, 40–46. [Google Scholar] [PubMed]
- Wu, V.H.; Smith, R.A.; Masoum, S.; Raugi, D.N.; Ba, S.; Seydi, M.; Grobler, J.A.; Gottlieb, G.S. MK-8591 (4′-ethynyl-2-fluoro-2′-deoxyadenosine) exhibits potent activity against HIV-2 isolates and drug-resistant HIV-2 mutants in culture. Antimicrob. Agents Chemother. 2017, 61, e00744-17. [Google Scholar] [CrossRef]
- Diamond, T.L.; Ngo, W.; Xu, M.; Goh, S.L.; Rodriguez, S.; Lai, M.-T.; Asante-Appiah, E.; Grobler, J.A. Islatravir has a high barrier to resistance and exhibits a differentiated resistance profile from approved nucleoside reverse transcriptase inhibitors (NRTIs). Antimicrob. Agents Chemother. 2022, 66, e00133-22. [Google Scholar] [CrossRef]
- Han, J.; Kiss, L.; Mei, H.; Remete, A.M.; Ponikvar-Svet, M.; Sedgwick, D.M.; Roman, R.; Fustero, S.; Moriwaki, H.; Soloshonok, V.A. Chemical aspects of human and environmental overload with fluorine. Chem. Rev. 2021, 121, 4678–4742. [Google Scholar] [CrossRef]
- Glyn, R.J.; Pattison, G. Effects of replacing oxygenated functionality with fluorine on lipophilicity. J. Med. Chem. 2021, 64, 10246–10259. [Google Scholar] [CrossRef]
- Appleby, T.C.; Perry, J.K.; Murakami, E.; Barauskas, O.; Feng, J.; Cho, A.; Fox, D.; Wetmore, D.R.; McGrath, M.E.; Ray, A.S.; et al. Structural basis for RNA replication by the hepatitis C virus polymerase. Science 2015, 347, 771–775. [Google Scholar] [CrossRef]
- Shannon, A.; Fattorini, V.; Sama, B.; Selisko, B.; Feracci, M.; Falcou, C.; Gauffre, P.; El Kazzi, P.; Delpal, A.; Decroly, E.; et al. A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase. Nat. Commun. 2022, 13, 621. [Google Scholar] [CrossRef] [PubMed]
- Keating, G.M. Sofosbuvir: A review of its use in patients with chronic hepatitis C. Drugs 2014, 74, 1127–1146. [Google Scholar] [CrossRef]
- Good, S.S.; Westover, J.; Jung, K.H.; Zhou, X.-J.; Moussa, A.; La Colla, P.; Collu, G.; Canard, B.; Sommadossi, J.-P. AT-527, a double prodrug of a guanosine nucleotide analog, is a potent inhibitor of SARS-CoV-2 in vitro and a promising oral antiviral for treatment of COVID-19. Antimicrob. Agents Chemother. 2021, 65, e02479-20. [Google Scholar] [CrossRef] [PubMed]
- Deval, J.; Powdrill, M.H.; D’Abramo, C.M.; Cellai, L.; Götte, M. Pyrophosphorolytic excision of nonobligate chain terminators by hepatitis C virus NS5B polymerase. Antimicrob. Agents Chemother. 2007, 51, 2920–2928. [Google Scholar] [CrossRef] [PubMed]
- Boehr, A.K.; Arnold, J.J.; Oh, H.S.; Cameron, C.E.; Boehr, D.D. 2′-C-Methylated nucleotides terminate virus RNA synthesis by preventing active site closure of the viral RNA-dependent RNA polymerase. J. Biol. Chem. 2019, 294, 16897–16907. [Google Scholar] [CrossRef]
- Kouba, T.; Dubankova, A.; Drncova, P.; Donati, E.; Vidossich, P.; Speranzini, V.; Pflug, A.; Huchting, J.; Meier, C.; De Vivo, M.; et al. Direct observation of backtracking by influenza A and B polymerases upon consecutive incorporation of the nucleoside analog T1106. Cell Rep. 2023, 42, 111901. [Google Scholar] [CrossRef]
- Bouvet, M.; Imbert, I.; Subissi, L.; Gluais, L.; Canard, B.; Decroly, E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein Nsp10/Nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. USA 2012, 109, 9372–9377. [Google Scholar] [CrossRef]
- Boyer, P.L.; Julias, J.G.; Marquez, V.E.; Hughes, S.H. Fixed conformation nucleoside analogs effectively inhibit excision-proficient HIV-1 reverse transcriptases. J. Mol. Biol. 2005, 345, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Seifer, M.; Hamatake, R.K.; Colonno, R.J.; Standring, D.N. In vitro inhibition of hepadnavirus polymerases by the triphosphates of BMS-200475 and lobucavir. Antimicrob. Agents Chemother. 1998, 42, 3200–3208. [Google Scholar] [CrossRef] [PubMed]
- Tchesnokov, E.P.; Obikhod, A.; Schinazi, R.F.; Gótte, M. Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. J. Biol. Chem. 2008, 283, 34218–34228. [Google Scholar] [CrossRef]
- Magee, W.C.; Evans, D.H. The antiviral activity and mechanism of action of (S)-[3-hydroxy-2-(phosphonomethoxy)propyl] (HPMP) nucleosides. Antivir. Res. 2012, 96, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Magee, W.C.; Aldern, K.A.; Hostetler, K.Y.; Evans, D.H. Cidofovir and (S)-9-[3-hydroxy-(2-phosphonomethoxy)propyl]adenine are highly effective inhibitors of vaccinia virus DNA polymerase when incorporated into the template strand. Antimicrob. Agents Chemother. 2008, 52, 586–597. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shi, Y.; Reiss, K.; Maschietto, F.; Lolis, E.; Konigsberg, W.H.; Lisi, G.P.; Batista, V.S. Structural insights into binding of remdesivir triphosphate within the replication–transcription complex of SARS-CoV-2. Biochemistry 2022, 61, 1966–1973. [Google Scholar] [CrossRef]
- Tchesnokov, E.P.; Gordon, C.J.; Woolner, E.; Kocinkova, D.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2020, 295, 16156–16165. [Google Scholar] [CrossRef]
- Luo, X.; Wang, X.; Yao, Y.; Gao, X.; Zhang, L. Unveiling the “template-dependent” inhibition on the viral transcription of SARS-CoV-2. J. Phys. Chem. Lett. 2022, 13, 7197–7205. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.J.; Lee, H.W.; Tchesnokov, E.P.; Perry, J.K.; Feng, J.Y.; Bilello, J.P.; Porter, D.P.; Götte, M. Efficient incorporation and template-dependent polymerase inhibition are major determinants for the broad-spectrum antiviral activity of remdesivir. J. Biol. Chem. 2022, 298, 101529. [Google Scholar] [CrossRef]
- Radoshitzky, S.R.; Iversen, P.; Lu, X.; Zou, J.; Kaptein, S.J.F.; Stuthman, K.S.; Van Tongeren, S.A.; Steffens, J.; Gong, R.; Truong, H.; et al. Expanded profiling of remdesivir as a broad-spectrum antiviral and low potential for interaction with other medications in vitro. Sci. Rep. 2023, 13, 3131. [Google Scholar] [CrossRef]
- Swanstrom, R.; Schinazi, R.F. Lethal mutagenesis as an antiviral strategy. Science 2022, 375, 497–498. [Google Scholar] [CrossRef]
- Graci, J.D.; Cameron, C.E. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 2006, 16, 37–48. [Google Scholar] [CrossRef]
- Goldhill, D.H.; Yan, A.; Frise, R.; Zhou, J.; Shelley, J.; Gallego Cortés, A.; Miah, S.; Akinbami, O.; Galiano, M.; Zambon, M.; et al. Favipiravir-resistant influenza A virus shows potential for transmission. PLoS Pathog. 2021, 17, e1008937. [Google Scholar] [CrossRef]
- Kabinger, F.; Stiller, C.; Schmitzová, J.; Dienemann, C.; Kokic, G.; Hillen, H.S.; Höbartner, C.; Cramer, P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021, 28, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Hill, C.S.; Sarkar, S.; Tse, L.V.; Woodburn, B.M.D.; Schinazi, R.F.; Sheahan, T.P.; Baric, R.S.; Heise, M.T.; Swanstrom, R. β-d-N4-Hydroxycytidine inhibits SARS-CoV-2 through lethal mutagenesis but is also mutagenic to mammalian cells. J. Infect. Dis. 2021, 224, 415–419. [Google Scholar] [CrossRef]
- Lehmann, K.C.; Gulyaeva, A.; Zevenhoven-Dobbe, J.C.; Janssen, G.M.C.; Ruben, M.; Overkleeft, H.S.; van Veelen, P.A.; Samborskiy, D.V.; Kravchenko, A.A.; Leontovich, A.M.; et al. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015, 43, 8416–8434. [Google Scholar] [CrossRef]
- Matyugina, E.; Petushkov, I.; Surzhikov, S.; Kezin, V.; Maslova, A.; Ivanova, O.; Smirnova, O.; Kirillov, I.; Fedyakina, I.; Kulbachinskiy, A.; et al. Nucleoside analogs that inhibit SARS-CoV-2 replication by blocking interaction of virus polymerase with RNA. Int. J. Mol. Sci. 2023, 24, 3361. [Google Scholar] [CrossRef] [PubMed]
- Onwubiko, N.O.; Diaz, S.A.; Krečmerová, M.; Nasheuer, H.P. Alkoxylalkyl esters of nucleotide analogs inhibit polyomavirus DNA replication and large T antigen activities. Antimicrob. Agents Chemother. 2021, 65, e01641-20. [Google Scholar] [CrossRef]
- Ramdhan, P.; Li, C. Targeting viral methyltransferases: An approach to antiviral treatment for ssRNA viruses. Viruses 2022, 14, 379. [Google Scholar] [CrossRef]
- Banerjee, A.K. 5′-Terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol. Rev. 1980, 44, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Bougie, I.; Bisaillon, M. The broad spectrum antiviral nucleoside ribavirin as a substrate for a viral RNA capping enzyme. J. Biol. Chem. 2004, 279, 22124–22130. [Google Scholar] [CrossRef]
- Goswami, B.B.; Borek, E.; Sharma, O.K.; Fujitaki, J.; Smith, R.A. The broad spectrum antiviral agent ribavirin inhibits capping of mRNA. Biochem. Biophys. Res. Commun. 1979, 89, 830–836. [Google Scholar] [CrossRef]
- Kentsis, A.; Topisirovic, I.; Culjkovic, B.; Shao, L.; Borden, K.L.B. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA Cap. Proc. Natl. Acad. Sci. USA 2004, 101, 18105–18110. [Google Scholar] [CrossRef] [PubMed]
- Scheidel, L.M.; Stollar, V. Mutations that confer resistance to mycophenolic acid and ribavirin on Sindbis virus map to the nonstructural protein nsP1. Virology 1991, 181, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Thames, J.E.; Waters, C.D.; Valle, C.; Bassetto, M.; Aouadi, W.; Martin, B.; Selisko, B.; Falat, A.; Coutard, B.; Brancale, A.; et al. Synthesis and biological evaluation of novel flexible nucleoside analogues that inhibit flavivirus replication in vitro. Bioorg. Med. Chem. 2020, 28, 115713. [Google Scholar] [CrossRef] [PubMed]
- Mahalapbutr, P.; Kongtaworn, N.; Rungrotmongkol, T. Structural insight into the recognition of S-adenosyl-l-homocysteine and sinefungin in SARS-CoV-2 Nsp16/Nsp10 RNA cap 2′-O-methyltransferase. Comput. Struct. Biotechnol. J. 2020, 18, 2757–2765. [Google Scholar] [CrossRef]
- Zhang, J.; Zheng, Y.G. SAM/SAH analogs as versatile tools for SAM-dependent methyltransferases. ACS Chem. Biol. 2016, 11, 583–597. [Google Scholar] [CrossRef]
- Ahmed-Belkacem, R.; Sutto-Ortiz, P.; Guiraud, M.; Canard, B.; Vasseur, J.-J.; Decroly, E.; Debart, F. Synthesis of adenine dinucleosides SAM analogs as specific inhibitors of SARS-CoV nsp14 RNA cap guanine-N7-methyltransferase. Eur. J. Med. Chem. 2020, 201, 112557. [Google Scholar] [CrossRef] [PubMed]
- Mudgal, R.; Mahajan, S.; Tomar, S. Inhibition of Chikungunya virus by an adenosine analog targeting the SAM-dependent nsP1 Methyltransferase. FEBS Lett. 2020, 594, 678–694. [Google Scholar] [CrossRef]
- Yoon, J.; Kim, G.; Jarhad, D.B.; Kim, H.-R.; Shin, Y.-S.; Qu, S.; Sahu, P.K.; Kim, H.O.; Lee, H.W.; Wang, S.B.; et al. Design, synthesis, and anti-RNA virus activity of 6′-fluorinated-aristeromycin analogues. J. Med. Chem. 2019, 62, 6346–6362. [Google Scholar] [CrossRef]
- Toyama, M.; Watashi, K.; Ikeda, M.; Yamashita, A.; Okamoto, M.; Moriishi, K.; Muramatsu, M.; Wakita, T.; Sharon, A.; Baba, M. Novel neplanocin A derivatives as selective inhibitors of hepatitis B virus with a unique mechanism of action. Antimicrob. Agents Chemother. 2022, 66, e02073-21. [Google Scholar] [CrossRef]
- Liu, C.; Coleman, R.; Archer, A.; Hussein, I.; Bowlin, T.L.; Chen, Q.; Schneller, S.W. Enantiomeric 4′-truncated 3-deaza-1′,6′-isoneplanocins: Synthesis and antiviral properties including Ebola. Bioorg. Med. Chem. Lett. 2019, 29, 2480–2482. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, B.; Brecher, M.; Banavali, N.; Jones, S.A.; Li, Z.; Zhang, J.; Nag, D.; Kramer, L.D.; Ghosh, A.K.; et al. S-Adenosyl-homocysteine is a weakly bound inhibitor for a flaviviral methyltransferase. PLoS ONE 2013, 8, e76900. [Google Scholar] [CrossRef] [PubMed]
- Tate, P.M.; Mastrodomenico, V.; Mounce, B.C. Ribavirin induces polyamine depletion via nucleotide depletion to limit virus replication. Cell Rep. 2019, 28, 2620–2633.e4. [Google Scholar] [CrossRef] [PubMed]
- Mounce, B.C.; Cesaro, T.; Moratorio, G.; Hooikaas, P.J.; Yakovleva, A.; Werneke, S.W.; Smith, E.C.; Poirier, E.Z.; Simon-Loriere, E.; Prot, M.; et al. Inhibition of polyamine biosynthesis is a broad-spectrum strategy against RNA viruses. J. Virol. 2016, 90, 9683–9692. [Google Scholar] [CrossRef] [PubMed]
- Müller, W.E.G.; Maidhof, A.; Taschner, H.; Zahn, R.K. Virazole (1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide; a cytostatic agent. Biochem. Pharmacol. 1977, 26, 1071–1075. [Google Scholar] [CrossRef]
- Takizawa, N.; Takada, H.; Umekita, M.; Igarashi, M.; Takahashi, Y. Anti-influenza virus activity of methylthio-formycin distinct from that of T-705. Front. Microbiol. 2022, 13, 802671. [Google Scholar] [CrossRef] [PubMed]
- Cartenì-Farina, M.; Cacciapuoti, G.; Porcelli, M.; Ragione, F.D.; Lancieri, M.; Geraci, G.; Zappia, V. Studies on the metabolic effects of methylthioformycin. Biochim. Biophys. Acta 1984, 805, 158–164. [Google Scholar] [CrossRef]
- McCluskey, G.D.; Mohamady, S.; Taylor, S.D.; Bearne, S.L. Exploring the potent inhibition of CTP synthase by gemcitabine-5′-triphosphate. ChemBioChem 2016, 17, 2240–2249. [Google Scholar] [CrossRef]
- Lee, K.; Kim, D.-E.; Jang, K.-S.; Kim, S.-J.; Cho, S.; Kim, C. Gemcitabine, a broad-spectrum antiviral drug, suppresses enterovirus infections through innate immunity induced by the inhibition of pyrimidine biosynthesis and nucleotide depletion. Oncotarget 2017, 8, 115315–115325. [Google Scholar] [CrossRef]
- Song, J.-H.; Kim, S.-R.; Heo, E.-Y.; Lee, J.-Y.; Kim, D.; Cho, S.; Chang, S.-Y.; Yoon, B.-I.; Seong, J.; Ko, H.-J. Antiviral activity of gemcitabine against human rhinovirus in vitro and in vivo. Antivir. Res. 2017, 145, 6–13. [Google Scholar] [CrossRef]
- Stein, D.S.; Moore, K.H.P. Phosphorylation of nucleoside analog antiretrovirals: A review for clinicians. Pharmacotherapy 2001, 21, 11–34. [Google Scholar] [CrossRef]
- Yeo, K.L.; Chen, Y.-L.; Xu, H.Y.; Dong, H.; Wang, Q.-Y.; Yokokawa, F.; Shi, P.-Y. Synergistic suppression of dengue virus replication using a combination of nucleoside analogs and nucleoside synthesis inhibitors. Antimicrob. Agents Chemother. 2015, 59, 2086–2093. [Google Scholar] [CrossRef] [PubMed]
- Samuel, C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Hultgren, C.; Milich, D.R.; Weiland, O.; Sällberg, M. The antiviral compound ribavirin modulates the T helper (Th)1/Th2 subset balance in hepatitis B and C virus-specific immune responses. J. Gen. Virol. 1998, 79, 2381–2391. [Google Scholar] [CrossRef]
- Murata, K.; Asano, M.; Matsumoto, A.; Sugiyama, M.; Nishida, N.; Tanaka, E.; Inoue, T.; Sakamoto, M.; Enomoto, N.; Shirasaki, T.; et al. Induction of IFN-λ3 as an additional effect of nucleotide, not nucleoside, analogues: A new potential target for HBV infection. Gut 2018, 67, 362–371. [Google Scholar] [CrossRef]
- Li, Y.; Li, P.; Li, Y.; Zhang, R.; Yu, P.; Ma, Z.; Kainov, D.E.; De Man, R.A.; Peppelenbosch, M.P.; Pan, Q. Drug screening identified gemcitabine inhibiting hepatitis E virus by inducing interferon-like response via activation of STAT1 phosphorylation. Antivir. Res. 2020, 184, 104967. [Google Scholar] [CrossRef]
- Bonnac, L.F.; Dreis, C.D.; Geraghty, R.J. Structure activity relationship, 6-modified purine riboside analogues to activate HSTING, stimulator of interferon genes. Bioorg. Med. Chem. Lett. 2020, 30, 126819. [Google Scholar] [CrossRef]
- Galegov, G.A. Phosphazide (nikavir) is a highly effective drug for the treatment of HIV/AIDS infection. Probl. Virol. 2017, 62, 5–11. [Google Scholar] [CrossRef]
- Krečmerová, M.; Majer, P.; Rais, R.; Slusher, B.S. Phosphonates and phosphonate prodrugs in medicinal chemistry: Past successes and future prospects. Front. Chem. 2022, 10, 889737. [Google Scholar] [CrossRef]
- Singh, U.S.; Konreddy, A.K.; Kothapalli, Y.; Liu, D.; Lloyd, M.G.; Annavarapu, V.; White, C.A.; Bartlett, M.G.; Moffat, J.F.; Chu, C.K. Prodrug strategies for the development of β-l-5-((E)-2-bromovinyl)-1-((2S,4S)-2-(hydroxymethyl)-1,3-(dioxolane-4-yl))uracil (l-BHDU) against varicella zoster virus (VZV). J. Med. Chem. 2023, 66, 7038–7053. [Google Scholar] [CrossRef]
- Wiemer, A.J.; Wiemer, D.F. Prodrugs of phosphonates and phosphates: Crossing the membrane barrier. In Phosphorus Chemistry I; Montchamp, J.-L., Ed.; Topics in Current Chemistry; Springer: Cham, Switzerland, 2014; Volume 360, pp. 115–160. ISBN 978-3-319-15472-5. [Google Scholar] [CrossRef]
- Serpi, M.; Pertusati, F. An overview of ProTide technology and its implications to drug discovery. Expert Opin. Drug Discov. 2021, 16, 1149–1161. [Google Scholar] [CrossRef]
- McGuigan, C.; Pathirana, R.N.; Balzarini, J.; De Clercq, E. Intracellular delivery of bioactive AZT nucleotides by aryl phosphate derivatives of AZT. J. Med. Chem. 1993, 36, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Pertusati, F.; Serafini, S.; Albadry, N.; Snoeck, R.; Andrei, G. Phosphonoamidate prodrugs of C5-substituted pyrimidine acyclic nucleosides for antiviral therapy. Antivir. Res. 2017, 143, 262–268. [Google Scholar] [CrossRef]
- Jones, S.A.; Murakami, E.; Delaney, W.; Furman, P.; Hu, J. Noncompetitive inhibition of hepatitis B virus reverse transcriptase protein priming and DNA synthesis by the nucleoside analog clevudine. Antimicrob. Agents Chemother. 2013, 57, 4181–4189. [Google Scholar] [CrossRef] [PubMed]
- Squires, K.E.; Mayers, D.L.; Bluemling, G.R.; Kolykhalov, A.A.; Guthrie, D.B.; Reddy, P.; Mitchell, D.G.; Saindane, M.T.; Sticher, Z.M.; Edpuganti, V.; et al. ATI-2173, a novel liver-targeted non-chain-terminating nucleotide for hepatitis B virus cure regimens. Antimicrob. Agents Chemother. 2020, 64, e00836-20. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, S.; Li, J.; Wang, W.; Zhu, H.-J. Cell-dependent activation of ProTide prodrugs and its implications in antiviral studies. ACS Pharmacol. Transl. Sci. 2023, 6. [Google Scholar] [CrossRef]
- Bhatia, H.K.; Singh, H.; Grewal, N.; Natt, N.K. Sofosbuvir: A novel treatment option for chronic hepatitis C infection. J. Pharmacol. Pharmacother. 2014, 5, 278–284. [Google Scholar] [CrossRef]
- Childs-Kean, L.M.; Egelund, E.F.; Jourjy, J. Tenofovir alafenamide for the treatment of chronic hepatitis B monoinfection. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2018, 38, 1051–1057. [Google Scholar] [CrossRef]
- Schooley, R.T.; Carlin, A.F.; Beadle, J.R.; Valiaeva, N.; Zhang, X.-Q.; Clark, A.E.; McMillan, R.E.; Leibel, S.L.; McVicar, R.N.; Xie, J.; et al. Rethinking remdesivir: Synthesis, antiviral activity, and pharmacokinetics of oral lipid prodrugs. Antimicrob. Agents Chemother. 2021, 65, e01155-21. [Google Scholar] [CrossRef]
- Hostetler, K.Y. Alkoxyalkyl prodrugs of acyclic nucleoside phosphonates enhance oral antiviral activity and reduce toxicity: Current state of the art. Antivir. Res. 2009, 82, A84–A98. [Google Scholar] [CrossRef]
- Tippin, T.K.; Morrison, M.E.; Brundage, T.M.; Momméja-Marin, H. Brincidofovir is not a substrate for the human organic anion transporter 1: A mechanistic explanation for the lack of nephrotoxicity observed in clinical studies. Ther. Drug Monit. 2016, 38, 777–786. [Google Scholar] [CrossRef]
- Lo, M.K.; Shrivastava-Ranjan, P.; Chatterjee, P.; Flint, M.; Beadle, J.R.; Valiaeva, N.; Murphy, J.; Schooley, R.T.; Hostetler, K.Y.; Montgomery, J.M.; et al. Broad-spectrum in vitro antiviral activity of ODBG-P-RVn: An orally-available, lipid-modified monophosphate prodrug of remdesivir parent nucleoside (GS-441524). Microbiol. Spectr. 2021, 9, e01537-21. [Google Scholar] [CrossRef] [PubMed]
- Carlin, A.F.; Beadle, J.R.; Clark, A.E.; Gully, K.L.; Moreira, F.R.; Baric, R.S.; Graham, R.L.; Valiaeva, N.; Leibel, S.L.; Bray, W.; et al. 1-O-Octadecyl-2-O-benzyl-sn-glyceryl-3-phospho-GS-441524 (V2043). Evaluation of oral V2043 in a mouse model of SARS-CoV-2 infection and synthesis and antiviral evaluation of additional phospholipid esters with enhanced anti-SARS-CoV-2 activity. J. Med. Chem. 2023, 66, 5802–5819. [Google Scholar] [CrossRef] [PubMed]
- Hostetler, K.Y.; Beadle, J.R.; Trahan, J.; Aldern, K.A.; Owens, G.; Schriewer, J.; Melman, L.; Buller, R.M. Oral 1-O-octadecyl-2-O-benzyl-sn-glycero-3-cidofovir targets the lung and is effective against a lethal respiratory challenge with ectromelia virus in mice. Antivir. Res. 2007, 73, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Toti, K.S.; Pribut, N.; D’Erasmo, M.; Dasari, M.; Sharma, S.K.; Bartsch, P.W.; Burton, S.L.; Gold, H.B.; Bushnev, A.; Derdeyn, C.A.; et al. Expanding the toolbox of metabolically stable lipid prodrug strategies. Front. Pharmacol. 2023, 13, 1083284. [Google Scholar] [CrossRef]
- Lloyd, M.G.; Liu, D.; Lyu, J.; Fan, J.; Overhulse, J.M.; Kashemirov, B.A.; Prichard, M.N.; McKenna, C.E.; Moffat, J.F. An acyclic phosphonate prodrug of HPMPC is effective against VZV in skin organ culture and mice. Antivir. Res. 2022, 199, 105275. [Google Scholar] [CrossRef]
- Toth, K.; Spencer, J.F.; Ying, B.; Tollefson, A.E.; Hartline, C.B.; Richard, E.T.; Fan, J.; Lyu, J.; Kashemirov, B.A.; Harteg, C.; et al. USC-087 protects Syrian hamsters against lethal challenge with human species C adenoviruses. Antivir. Res. 2018, 153, 1–9. [Google Scholar] [CrossRef]
- Weinschenk, L.; Schols, D.; Balzarini, J.; Meier, C. Nucleoside diphosphate prodrugs: Nonsymmetric diPPro-nucleotides. J. Med. Chem. 2015, 58, 6114–6130. [Google Scholar] [CrossRef]
- Zhao, C.; Jia, X.; Schols, D.; Balzarini, J.; Meier, C. γ-Non-symmetrically dimasked triPPPro prodrugs as potential antiviral agents against HIV. ChemMedChem 2021, 16, 499–512. [Google Scholar] [CrossRef]
- Daluge, S.M.; Good, S.S.; Faletto, M.B.; Miller, W.H.; St Clair, M.H.; Boone, L.R.; Tisdale, M.; Parry, N.R.; Reardon, J.E.; Dornsife, R.E.; et al. 1592U89, a novel carbocyclic nucleoside analog with potent, selective anti-human immunodeficiency virus activity. Antimicrob. Agents Chemother. 1997, 41, 1082–1093. [Google Scholar] [CrossRef]
- Vollmer, K.J.; Burns, A.C.; Sauer, H.E.; Williams, J.D.; Komazin-Meredith, G.; Cardinale, S.; Butler, M.; Aron, Z.; Hussein, I.; Busch, M.G.; et al. Activation of 6-alkoxy-substituted methylenecyclopropane nucleoside analogs requires enzymatic modification by adenosine deaminase-like protein 1. Antimicrob. Agents Chemother. 2019, 63, e01301-19. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liang, C.; Zou, L.; Yin, Y.; Wang, J.; Chen, D.; Lan, W. Advance of structural modification of nucleosides scaffold. Eur. J. Med. Chem. 2021, 214, 113233. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, S.; Zhu, Y.; Pan, Y.; Zhang, L.; Yang, Z. Overcoming the delivery barrier of oligonucleotide drugs and enhancing nucleoside drug efficiency: The use of nucleolipids. Med. Res. Rev. 2020, 40, 1178–1199. [Google Scholar] [CrossRef] [PubMed]
- McGuigan, C.; Yarnold, C.J.; Jones, G.; Velázquez, S.; Barucki, H.; Brancale, A.; Andrei, G.; Snoeck, R.; De Clercq, E.; Balzarini, J. Potent and selective inhibition of varicella-zoster virus (VZV) by nucleoside analogues with an unusual bicyclic base. J. Med. Chem. 1999, 42, 4479–4484. [Google Scholar] [CrossRef] [PubMed]
- McGuigan, C.; Barucki, H.; Blewett, S.; Carangio, A.; Erichsen, J.T.; Andrei, G.; Snoeck, R.; De Clercq, E.; Balzarini, J. Highly potent and selective inhibition of varicella-zoster virus by bicyclic furopyrimidine nucleosides bearing an aryl side chain. J. Med. Chem. 2000, 43, 4993–4997. [Google Scholar] [CrossRef]
- Balzarini, J.; McGuigan, C. Chemotherapy of varicella-zoster virus by a novel class of highly specific anti-VZV bicyclic pyrimidine nucleosides. Biochim. Biophys. Acta 2002, 1587, 287–295. [Google Scholar] [CrossRef]
- McGuigan, C.; Brancale, A.; Andrei, G.; Snoeck, R.; De Clercq, E.; Balzarini, J. Novel bicyclic furanopyrimidines with dual anti-VZV and -HCMV activity. Bioorg. Med. Chem. Lett. 2003, 13, 4511–4513. [Google Scholar] [CrossRef]
- McGuigan, C.; Pathirana, R.N.; Snoeck, R.; Andrei, G.; De Clercq, E.; Balzarini, J. Discovery of a new family of inhibitors of human cytomegalovirus (HCMV) based upon lipophilic alkyl furano pyrimidine dideoxy nucleosides: Action via a novel non-nucleosidic mechanism. J. Med. Chem. 2004, 47, 1847–1851. [Google Scholar] [CrossRef]
- Bidet, O.; McGuigan, C.; Snoeck, R.; Andrei, G.; De Clercq, E.; Balzarini, J. Non-Nucleoside Structures Retain Full Anti-HCMV Potency of the Dideoxy Furanopyrimidine Family. Antivir. Chem. Chemother. 2004, 15, 329–332. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E. Nucleoside analogues exerting antiviral activity through a non-nucleoside mechanism. Nucleosides Nucleotides Nucleic Acids 2004, 23, 457–470. [Google Scholar] [CrossRef]
- McGuigan, C.; Pathirana, R.N.; Migliore, M.; Adak, R.; Luoni, G.; Jones, A.T.; Diez-Torrubia, A.; Camarasa, M.-J.; Velazquez, S.; Henson, G.; et al. Preclinical development of bicyclic nucleoside analogues as potent and selective inhibitors of varicella zoster virus. J. Antimicrob. Chemother. 2007, 60, 1316–1330. [Google Scholar] [CrossRef] [PubMed]
- Pentikis, H.S.; Matson, M.; Atiee, G.; Boehlecke, B.; Hutchins, J.T.; Patti, J.M.; Henson, G.W.; Morris, A. Pharmacokinetics and safety of FV-100, a novel oral anti-herpes zoster nucleoside analogue, Administered in Single and Multiple Doses to Healthy Young Adult and Elderly Adult Volunteers. Antimicrob. Agents Chemother. 2011, 55, 2847–2854. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tyring, S.K.; Lee, P.; Hill, G.T.; Silverfield, J.C.; Moore, A.Y.; Matkovits, T.; Sullivan-Bolyai, J. FV-100 versus valacyclovir for the prevention of post-herpetic neuralgia and the treatment of acute herpes zoster-associated pain: A randomized-controlled trial. J. Med. Virol. 2017, 89, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E. FV-100 for the treatment of varicella-virus (VZV) infections: Quo vadis? Viruses 2022, 14, 770. [Google Scholar] [CrossRef]
- St. Vincent, M.R.; Colpitts, C.C.; Ustinov, A.V.; Muqadas, M.; Joyce, M.A.; Barsby, N.L.; Epand, R.F.; Epand, R.M.; Khramyshev, S.A.; Valueva, O.A.; et al. Rigid amphipathic fusion inhibitors, small molecule antiviral compounds against enveloped viruses. Proc. Natl. Acad. Sci. USA 2010, 107, 17339–17344. [Google Scholar] [CrossRef]
- Colpitts, C.C.; Ustinov, A.V.; Epand, R.F.; Epand, R.M.; Korshun, V.A.; Schang, L.M. 5-(Perylen-3-yl)ethynyl-arabino-uridine (aUY11), an arabino-based rigid amphipathic fusion inhibitor, targets virion envelope lipids to inhibit fusion of influenza virus, hepatitis C virus, and other enveloped viruses. J. Virol. 2013, 87, 3640–3654. [Google Scholar] [CrossRef]
- Speerstra, S.; Chistov, A.A.; Proskurin, G.V.; Aralov, A.V.; Ulashchik, E.A.; Streshnev, P.P.; Shmanai, V.V.; Korshun, V.A.; Schang, L.M. Antivirals acting on viral envelopes via biophysical mechanisms of action. Antivir. Res. 2018, 149, 164–173. [Google Scholar] [CrossRef]
- Orlov, A.A.; Chistov, A.A.; Kozlovskaya, L.I.; Ustinov, A.V.; Korshun, V.A.; Karganova, G.G.; Osolodkin, D.I. Rigid amphipathic nucleosides suppress reproduction of the tick-borne encephalitis virus. MedChemComm 2016, 7, 495–499. [Google Scholar] [CrossRef]
- Hakobyan, A.; Galindo, I.; Nañez, A.; Arabyan, E.; Karalyan, Z.; Chistov, A.A.; Streshnev, P.P.; Korshun, V.A.; Alonso, C.; Zakaryan, H. Rigid amphipathic fusion inhibitors demonstrate antiviral activity against African swine fever virus. J. Gen. Virol. 2018, 99, 148–156. [Google Scholar] [CrossRef]
- Chistov, A.A.; Orlov, A.A.; Streshnev, P.P.; Slesarchuk, N.A.; Aparin, I.O.; Rathi, B.; Brylev, V.A.; Kutyakov, S.V.; Mikhura, I.V.; Ustinov, A.V.; et al. Compounds based on 5-(perylen-3-ylethynyl)uracil scaffold: High activity against tick-borne encephalitis virus and non-specific activity against enterovirus A. Eur. J. Med. Chem. 2019, 171, 93–103. [Google Scholar] [CrossRef]
- Aralov, A.V.; Proskurin, G.V.; Orlov, A.A.; Kozlovskaya, L.I.; Chistov, A.A.; Kutyakov, S.V.; Karganova, G.G.; Palyulin, V.A.; Osolodkin, D.I.; Korshun, V.A. Perylenyltriazoles inhibit reproduction of enveloped viruses. Eur. J. Med. Chem. 2017, 138, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Slesarchuk, N.A.; Khvatov, E.V.; Chistov, A.A.; Proskurin, G.V.; Nikitin, T.D.; Lazarevich, A.I.; Ulanovskaya, A.A.; Ulashchik, E.A.; Orlov, A.A.; Jegorov, A.V.; et al. Simplistic perylene-related compounds as inhibitors of tick-borne encephalitis virus reproduction. Bioorg. Med. Chem. Lett. 2020, 30, 127100. [Google Scholar] [CrossRef] [PubMed]
- Shtro, A.A.; Garshinina, A.V.; Alferova, V.A.; Kamzeeva, P.N.; Volok, V.P.; Kolpakova, E.S.; Nikitin, T.D.; Chistov, A.A.; Belyaev, E.S.; Korshun, V.A.; et al. Cationic perylene antivirals with aqueous solubility for studies in vivo. Pharmaceuticals 2022, 15, 1178. [Google Scholar] [CrossRef]
- Vigant, F.; Hollmann, A.; Lee, J.; Santos, N.C.; Jung, M.E.; Lee, B. The rigid amphipathic fusion inhibitor dUY11 acts through photosensitization of viruses. J. Virol. 2014, 88, 1849–1853. [Google Scholar] [CrossRef] [PubMed]
- Chistov, A.A.; Chumakov, S.P.; Mikhnovets, I.E.; Nikitin, T.D.; Slesarchuk, N.A.; Uvarova, V.I.; Rubekina, A.A.; Nikolaeva, Y.V.; Radchenko, E.V.; Khvatov, E.V.; et al. 5-(Perylen-3-ylethynyl)uracil as an antiviral scaffold: Potent suppression of enveloped virus reproduction by 3-methyl derivatives in vitro. Antivir. Res. 2023, 209, 105508. [Google Scholar] [CrossRef]
- Straková, P.; Bednář, P.; Kotouček, J.; Holoubek, J.; Fořtová, A.; Svoboda, P.; Štefánik, M.; Huvarová, I.; Šimečková, P.; Mašek, J.; et al. Antiviral activity of singlet oxygen-photogenerating perylene compounds against SARS-CoV-2: Interaction with the viral envelope and photodynamic virion inactivation. Virus Res. 2023, 334, 199158. [Google Scholar] [CrossRef]
- Johnson, A.A.; Ray, A.S.; Hanes, J.; Suo, Z.; Colacino, J.M.; Anderson, K.S.; Johnson, K.A. Toxicity of antiviral nucleoside analogs and the human mitochondrial DNA polymerase. J. Biol. Chem. 2001, 276, 40847–40857. [Google Scholar] [CrossRef]
- Arnold, J.J.; Sharma, S.D.; Feng, J.Y.; Ray, A.S.; Smidansky, E.D.; Kireeva, M.L.; Cho, A.; Perry, J.; Vela, J.E.; Park, Y.; et al. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog. 2012, 8, e1003030. [Google Scholar] [CrossRef]
- Feng, J.Y.; Xu, Y.; Barauskas, O.; Perry, J.K.; Ahmadyar, S.; Stepan, G.; Yu, H.; Babusis, D.; Park, Y.; McCutcheon, K.; et al. Role of mitochondrial RNA polymerase in the toxicity of nucleotide inhibitors of hepatitis C virus. Antimicrob. Agents Chemother. 2016, 60, 806–817. [Google Scholar] [CrossRef]
- Feng, J.Y. Addressing the selectivity and toxicity of antiviral nucleosides. Antivir. Chem. Chemother. 2018, 26, 204020661875852. [Google Scholar] [CrossRef]
- Lynx, M.D.; McKee, E.E. 3′-Azido-3′-deoxythymidine (AZT) is a competitive inhibitor of thymidine phosphorylation in isolated rat heart and liver mitochondria. Biochem. Pharmacol. 2006, 72, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Susan-Resiga, D.; Bentley, A.T.; Lynx, M.D.; LaClair, D.D.; McKee, E.E. Zidovudine inhibits thymidine phosphorylation in the isolated perfused rat heart. Antimicrob. Agents Chemother. 2007, 51, 1142–1149. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, Y.; Barauskas, O.; Kim, C.; Babusis, D.; Murakami, E.; Kornyeyev, D.; Lee, G.; Stepan, G.; Perron, M.; Bannister, R.; et al. Off-target in vitro profiling demonstrates that remdesivir is a highly selective antiviral agent. Antimicrob. Agents Chemother. 2021, 65, e02237-20. [Google Scholar] [CrossRef] [PubMed]
- Bjork, J.A.; Wallace, K.B. Remdesivir; molecular and functional measures of mitochondrial safety. Toxicol. Appl. Pharmacol. 2021, 433, 115783. [Google Scholar] [CrossRef]
- Ostrov, D.A.; Grant, B.J.; Pompeu, Y.A.; Sidney, J.; Harndahl, M.; Southwood, S.; Oseroff, C.; Lu, S.; Jakoncic, J.; De Oliveira, C.A.F.; et al. Drug hypersensitivity caused by alteration of the MHC-presented self-peptide repertoire. Proc. Natl. Acad. Sci. USA 2012, 109, 9959–9964. [Google Scholar] [CrossRef]
- Norcross, M.A.; Luo, S.; Lu, L.; Boyne, M.T.; Gomarteli, M.; Rennels, A.D.; Woodcock, J.; Margulies, D.H.; McMurtrey, C.; Vernon, S.; et al. Abacavir induces loading of novel self-peptides into HLA-B*57:01 an autoimmune model for HLA-associated drug hypersensitivity. AIDS 2012, 26, F21–F29. [Google Scholar] [CrossRef]
- Illing, P.T.; Vivian, J.P.; Dudek, N.L.; Kostenko, L.; Chen, Z.; Bharadwaj, M.; Miles, J.J.; Kjer-Nielsen, L.; Gras, S.; Williamson, N.A.; et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 2012, 486, 554–558. [Google Scholar] [CrossRef]
- Thaljeh, L.F.; Rothschild, J.A.; Naderi, M.; Coghill, L.M.; Brown, J.M.; Brylinski, M. Hinge region in DNA packaging terminase PUL15 of herpes simplex virus: A potential allosteric target for antiviral drugs. Biomolecules 2019, 9, 603. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamzeeva, P.N.; Aralov, A.V.; Alferova, V.A.; Korshun, V.A. Recent Advances in Molecular Mechanisms of Nucleoside Antivirals. Curr. Issues Mol. Biol. 2023, 45, 6851-6879. https://doi.org/10.3390/cimb45080433
Kamzeeva PN, Aralov AV, Alferova VA, Korshun VA. Recent Advances in Molecular Mechanisms of Nucleoside Antivirals. Current Issues in Molecular Biology. 2023; 45(8):6851-6879. https://doi.org/10.3390/cimb45080433
Chicago/Turabian StyleKamzeeva, Polina N., Andrey V. Aralov, Vera A. Alferova, and Vladimir A. Korshun. 2023. "Recent Advances in Molecular Mechanisms of Nucleoside Antivirals" Current Issues in Molecular Biology 45, no. 8: 6851-6879. https://doi.org/10.3390/cimb45080433
APA StyleKamzeeva, P. N., Aralov, A. V., Alferova, V. A., & Korshun, V. A. (2023). Recent Advances in Molecular Mechanisms of Nucleoside Antivirals. Current Issues in Molecular Biology, 45(8), 6851-6879. https://doi.org/10.3390/cimb45080433







