Establishment of a Novel Anti-CD44 Variant 10 Monoclonal Antibody C44Mab-18 for Immunohistochemical Analysis against Oral Squamous Cell Carcinomas
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Production of Hybridoma Cells
2.3. Enzyme–Linked Immunosorbent Assay (ELISA)
2.4. Flow Cytometry
2.5. Determination of Apparent Dissociation Constant (KD) via Flow Cytometry
2.6. Western Blot Analysis
2.7. Immunohistochemical Analysis of Formalin-Fixed Paraffin-Embedded (FFPE) Tissues
3. Results
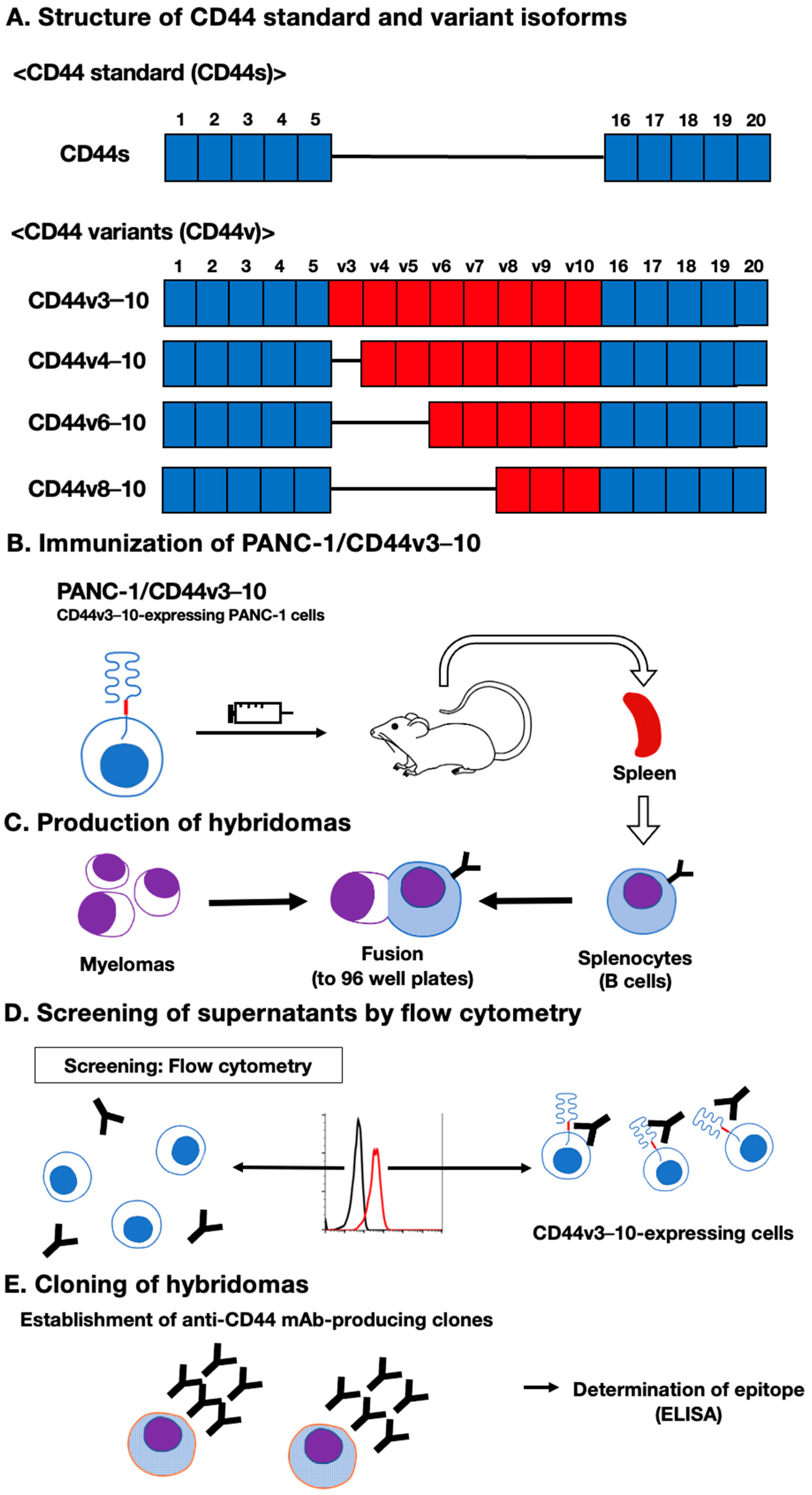
3.1. Establishment of an Anti-CD44 mAbs via Immunization of PANC-1/CD44v3–10 Cells
3.2. Flow Cytometric Analysis of C44Mab-18- to CD44-Expressing Cells
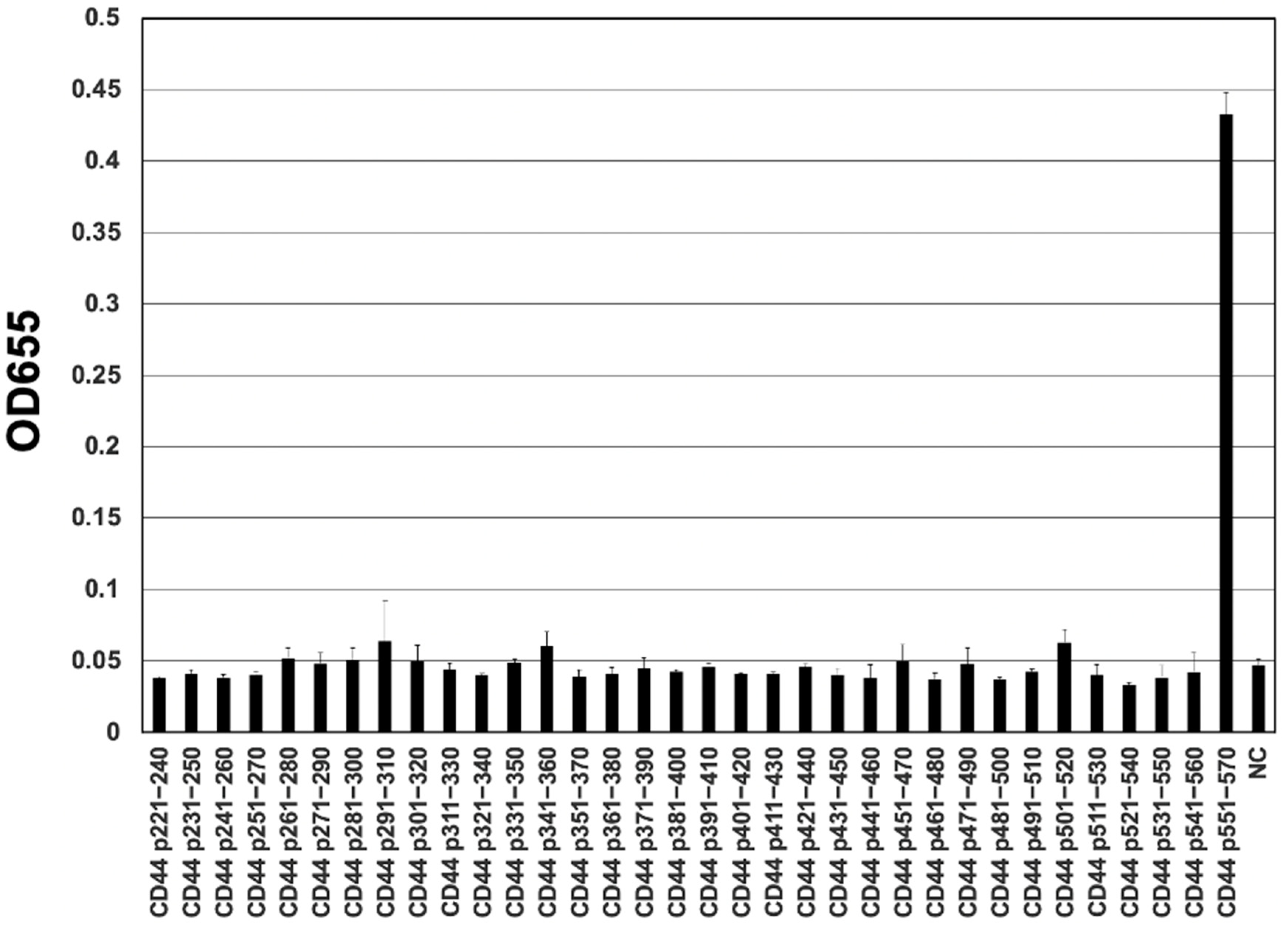
3.3. Epitope Mapping of C44Mab-18 by ELISA
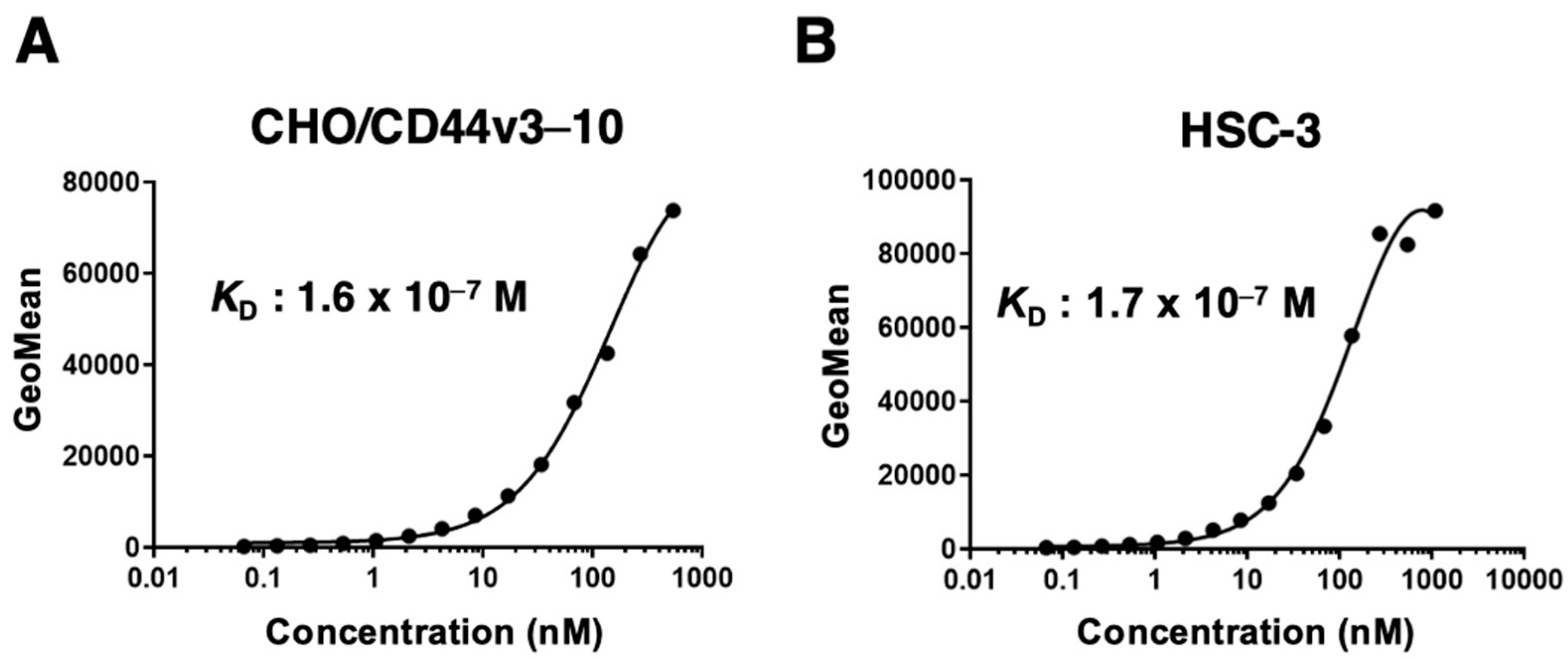
3.4. Determination of the Apparent Binding Affinity of C44Mab-18 via Flow Cytometry
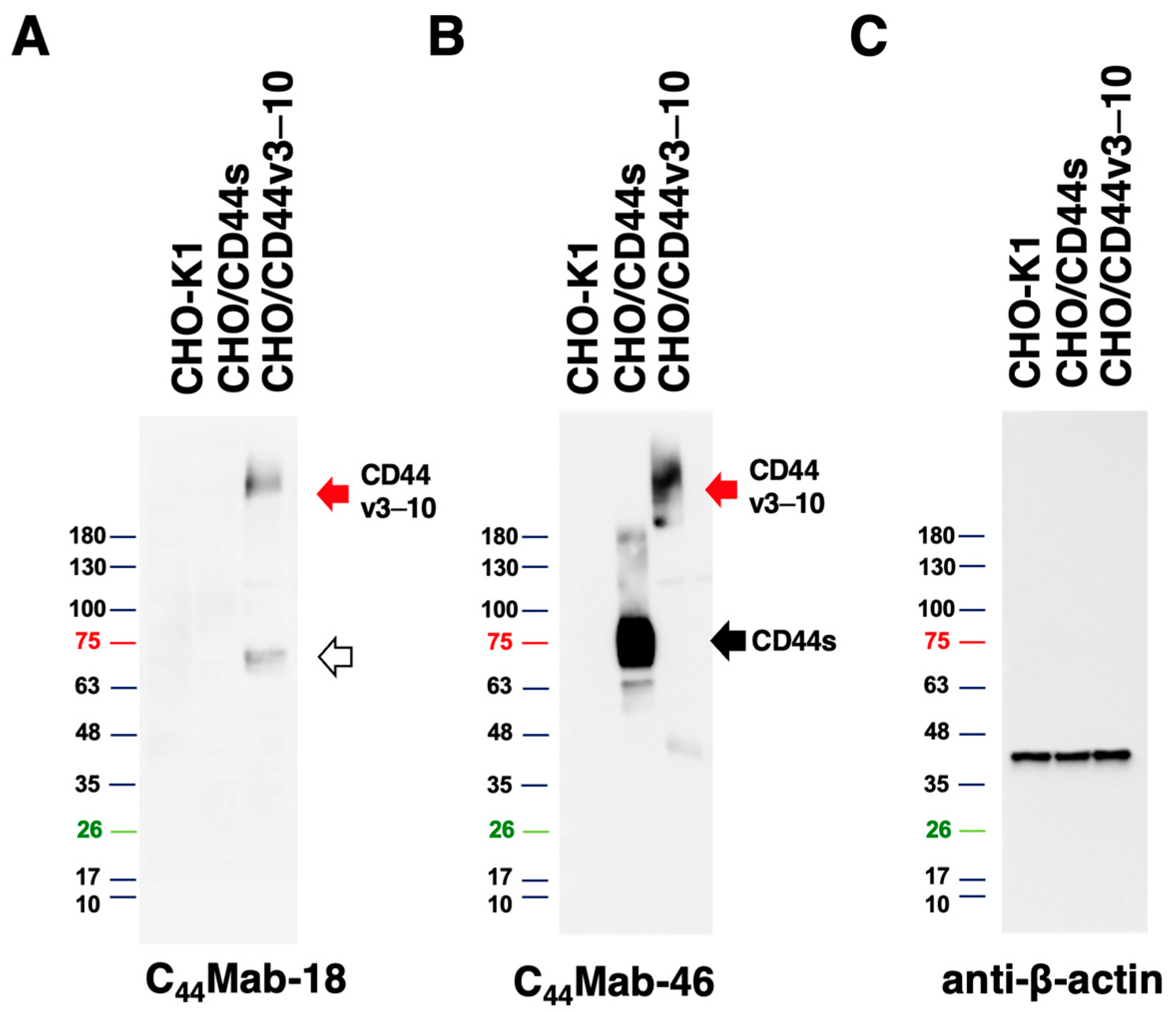
3.5. Western Blot Analysis
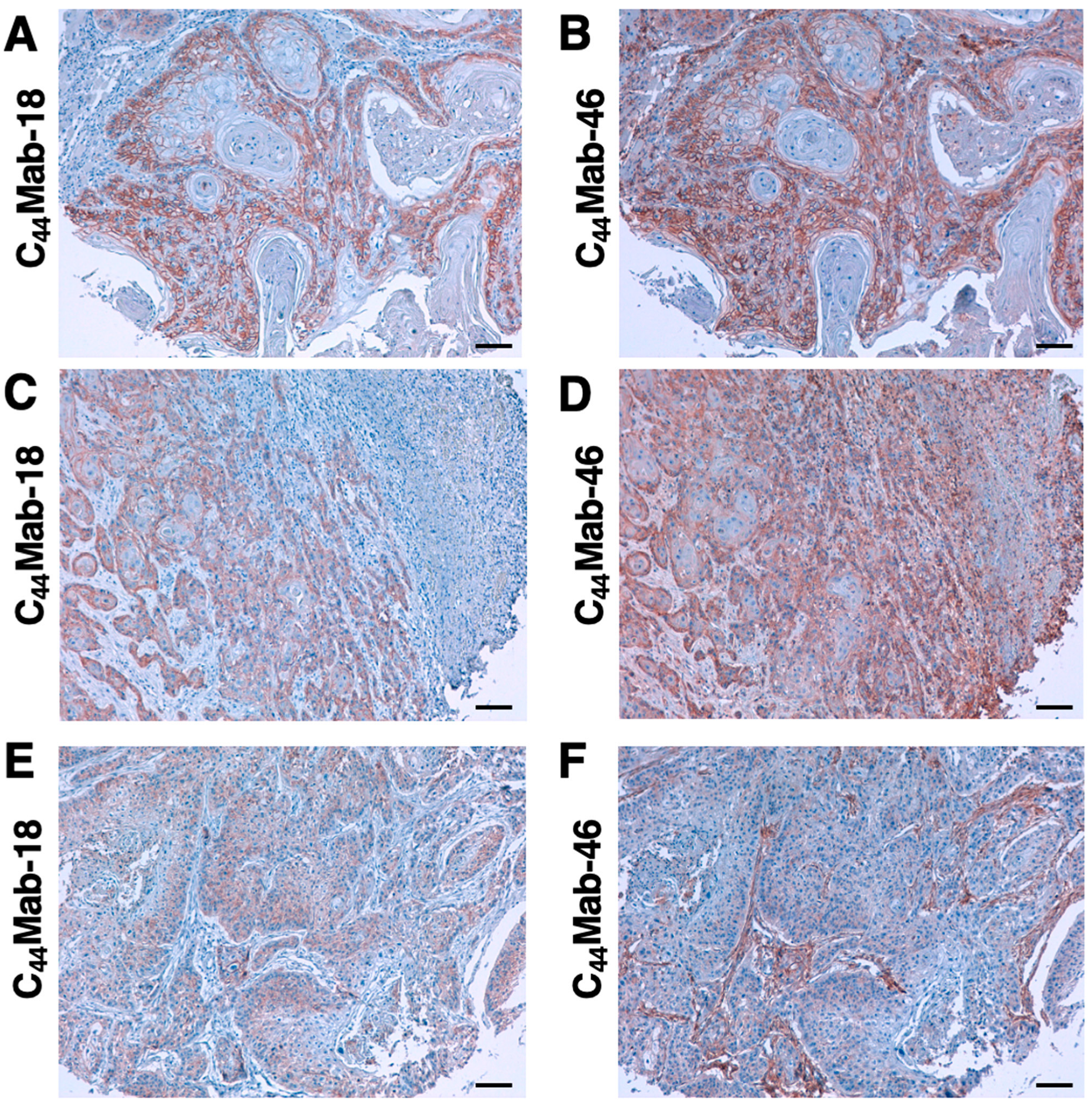
3.6. Immunohistochemical Analysis Using C44Mab-18 against Tumor Tissues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mody, M.D.; Rocco, J.W.; Yom, S.S.; Haddad, R.I.; Saba, N.F. Head and neck cancer. Lancet 2021, 398, 2289–2299. [Google Scholar] [CrossRef]
- Xing, D.T.; Khor, R.; Gan, H.; Wada, M.; Ermongkonchai, T.; Ng, S.P. Recent Research on Combination of Radiotherapy with Targeted Therapy or Immunotherapy in Head and Neck Squamous Cell Carcinoma: A Review for Radiation Oncologists. Cancers 2021, 13, 5716. [Google Scholar] [CrossRef]
- Muzaffar, J.; Bari, S.; Kirtane, K.; Chung, C.H. Recent Advances and Future Directions in Clinical Management of Head and Neck Squamous Cell Carcinoma. Cancers 2021, 13, 338. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Prim. 2020, 6, 92. [Google Scholar] [CrossRef]
- Ludwig, N.; Szczepanski, M.J.; Gluszko, A.; Szafarowski, T.; Azambuja, J.H.; Dolg, L.; Gellrich, N.C.; Kampmann, A.; Whiteside, T.L.; Zimmerer, R.M. CD44(+) tumor cells promote early angiogenesis in head and neck squamous cell carcinoma. Cancer Lett. 2019, 467, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Slevin, M.; Krupinski, J.; Gaffney, J.; Matou, S.; West, D.; Delisser, H.; Savani, R.C.; Kumar, S. Hyaluronan-mediated angiogenesis in vascular disease: Uncovering RHAMM and CD44 receptor signaling pathways. Matrix Biol. 2007, 26, 58–68. [Google Scholar] [CrossRef]
- Valastyan, S.; Weinberg, R.A. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef] [PubMed]
- de Visser, K.E.; Joyce, J.A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef]
- Zöller, M. CD44, Hyaluronan, the Hematopoietic Stem Cell, and Leukemia-Initiating Cells. Front. Immunol. 2015, 6, 235. [Google Scholar] [CrossRef] [PubMed]
- Hassn Mesrati, M.; Syafruddin, S.E.; Mohtar, M.A.; Syahir, A. CD44: A Multifunctional Mediator of Cancer Progression. Biomolecules 2021, 11, 1850. [Google Scholar] [CrossRef] [PubMed]
- Zöller, M. CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer 2011, 11, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Prince, M.E.; Sivanandan, R.; Kaczorowski, A.; Wolf, G.T.; Kaplan, M.J.; Dalerba, P.; Weissman, I.L.; Clarke, M.F.; Ailles, L.E. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. USA 2007, 104, 973–978. [Google Scholar] [CrossRef]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.J.; Divi, V.; Owen, J.H.; Bradford, C.R.; Carey, T.E.; Papagerakis, S.; Prince, M.E. Metastatic potential of cancer stem cells in head and neck squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 2010, 136, 1260–1266. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, T.; Nagano, O.; Yae, T.; Tamada, M.; Motohara, T.; Oshima, H.; Oshima, M.; Ikeda, T.; Asaba, R.; Yagi, H.; et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell 2011, 19, 387–400. [Google Scholar] [CrossRef]
- Kagami, T.; Yamade, M.; Suzuki, T.; Uotani, T.; Tani, S.; Hamaya, Y.; Iwaizumi, M.; Osawa, S.; Sugimoto, K.; Baba, S.; et al. High expression level of CD44v8-10 in cancer stem-like cells is associated with poor prognosis in esophageal squamous cell carcinoma patients treated with chemoradiotherapy. Oncotarget 2018, 9, 34876–34888. [Google Scholar] [CrossRef]
- Hagiwara, M.; Kikuchi, E.; Tanaka, N.; Kosaka, T.; Mikami, S.; Saya, H.; Oya, M. Variant isoforms of CD44 involves acquisition of chemoresistance to cisplatin and has potential as a novel indicator for identifying a cisplatin-resistant population in urothelial cancer. BMC Cancer 2018, 18, 113. [Google Scholar] [CrossRef]
- Yamada, S.; Itai, S.; Nakamura, T.; Yanaka, M.; Kaneko, M.K.; Kato, Y. Detection of high CD44 expression in oral cancers using the novel monoclonal antibody, C(44)Mab-5. Biochem. Biophys. Rep. 2018, 14, 64–68. [Google Scholar] [CrossRef]
- Goto, N.; Suzuki, H.; Tanaka, T.; Asano, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Anti-CD44 Monoclonal Antibody for Multiple Applications against Esophageal Squamous Cell Carcinomas. Int. J. Mol. Sci. 2022, 23, 5535. [Google Scholar] [CrossRef] [PubMed]
- Takei, J.; Asano, T.; Suzuki, H.; Kaneko, M.K.; Kato, Y. Epitope Mapping of the Anti-CD44 Monoclonal Antibody (C44Mab-46) Using Alanine-Scanning Mutagenesis and Surface Plasmon Resonance. Monoclon. Antib. Immunodiagn. Immunother. 2021, 40, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Kaneko, M.K.; Takei, J.; Tateyama, N.; Kato, Y. Epitope Mapping of the Anti-CD44 Monoclonal Antibody (C44Mab-46) Using the REMAP Method. Monoclon. Antib. Immunodiagn. Immunother. 2021, 40, 156–161. [Google Scholar] [CrossRef]
- Asano, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Epitope Mapping System: RIEDL Insertion for Epitope Mapping Method. Monoclon. Antib. Immunodiagn. Immunother. 2021, 40, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Takei, J.; Kaneko, M.K.; Ohishi, T.; Hosono, H.; Nakamura, T.; Yanaka, M.; Sano, M.; Asano, T.; Sayama, Y.; Kawada, M.; et al. A defucosylated antiCD44 monoclonal antibody 5mG2af exerts antitumor effects in mouse xenograft models of oral squamous cell carcinoma. Oncol. Rep. 2020, 44, 1949–1960. [Google Scholar] [CrossRef]
- Suzuki, H.; Kitamura, K.; Goto, N.; Ishikawa, K.; Ouchida, T.; Tanaka, T.; Kaneko, M.K.; Kato, Y. A Novel Anti-CD44 Variant 3 Monoclonal Antibody C(44)Mab-6 Was Established for Multiple Applications. Int. J. Mol. Sci. 2023, 24, 8411. [Google Scholar] [CrossRef]
- Suzuki, H.; Tanaka, T.; Goto, N.; Kaneko, M.K.; Kato, Y. Development of a Novel Anti-CD44 Variant 4 Monoclonal Antibody C44Mab-108 for Immunohistochemistry. Curr. Issues Mol. Biol. 2023, 45, 1875–1888. [Google Scholar] [CrossRef]
- Kudo, Y.; Suzuki, H.; Tanaka, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Anti-CD44 variant 5 Monoclonal Antibody C44Mab-3 for Multiple Applications against Pancreatic Carcinomas. Antibodies 2023, 12, 31. [Google Scholar] [CrossRef]
- Ejima, R.; Suzuki, H.; Tanaka, T.; Asano, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Anti-CD44 Variant 6 Monoclonal Antibody C(44)Mab-9 for Multiple Applications against Colorectal Carcinomas. Int. J. Mol. Sci. 2023, 24, 4007. [Google Scholar] [CrossRef]
- Suzuki, H.; Ozawa, K.; Tanaka, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Anti-CD44 Variant 7/8 Monoclonal Antibody, C44Mab-34, for Multiple Applications against Oral Carcinomas. Biomedicines 2023, 11, 1099. [Google Scholar] [CrossRef]
- Tawara, M.; Suzuki, H.; Goto, N.; Tanaka, T.; Kaneko, M.K.; Kato, Y. A Novel Anti-CD44 Variant 9 Monoclonal Antibody C44Mab-1 was Developed for Immunohistochemical Analyses Against Colorectal Cancers. Curr. Issues Mol. Biol. 2023, 45, 3658–3673. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Kaneko, M.K.; Kuno, A.; Uchiyama, N.; Amano, K.; Chiba, Y.; Hasegawa, Y.; Hirabayashi, J.; Narimatsu, H.; Mishima, K.; et al. Inhibition of tumor cell-induced platelet aggregation using a novel anti-podoplanin antibody reacting with its platelet-aggregation-stimulating domain. Biochem. Biophys. Res. Commun. 2006, 349, 1301–1307. [Google Scholar] [CrossRef] [PubMed]
- Heider, K.H.; Sproll, M.; Susani, S.; Patzelt, E.; Beaumier, P.; Ostermann, E.; Ahorn, H.; Adolf, G.R. Characterization of a high-affinity monoclonal antibody specific for CD44v6 as candidate for immunotherapy of squamous cell carcinomas. Cancer Immunol. Immunother. 1996, 43, 245–253. [Google Scholar] [CrossRef]
- Heider, K.H.; Mulder, J.W.; Ostermann, E.; Susani, S.; Patzelt, E.; Pals, S.T.; Adolf, G.R. Splice variants of the cell surface glycoprotein CD44 associated with metastatic tumour cells are expressed in normal tissues of humans and cynomolgus monkeys. Eur. J. Cancer 1995, 31a, 2385–2391. [Google Scholar] [CrossRef] [PubMed]
- Gansauge, F.; Gansauge, S.; Zobywalski, A.; Scharnweber, C.; Link, K.H.; Nussler, A.K.; Beger, H.G. Differential expression of CD44 splice variants in human pancreatic adenocarcinoma and in normal pancreas. Cancer Res. 1995, 55, 5499–5503. [Google Scholar]
- Beham-Schmid, C.; Heider, K.H.; Hoefler, G.; Zatloukal, K. Expression of CD44 splice variant v10 in Hodgkin's disease is associated with aggressive behaviour and high risk of relapse. J. Pathol. 1998, 186, 383–389. [Google Scholar] [CrossRef]
- Mishra, M.N.; Chandavarkar, V.; Sharma, R.; Bhargava, D. Structure, function and role of CD44 in neoplasia. J. Oral Maxillofac. Pathol. 2019, 23, 267–272. [Google Scholar] [CrossRef]
- Hirata, K.; Suzuki, H.; Imaeda, H.; Matsuzaki, J.; Tsugawa, H.; Nagano, O.; Asakura, K.; Saya, H.; Hibi, T. CD44 variant 9 expression in primary early gastric cancer as a predictive marker for recurrence. Br. J. Cancer 2013, 109, 379–386. [Google Scholar] [CrossRef]
- Shitara, K.; Doi, T.; Nagano, O.; Imamura, C.K.; Ozeki, T.; Ishii, Y.; Tsuchihashi, K.; Takahashi, S.; Nakajima, T.E.; Hironaka, S.; et al. Dose-escalation study for the targeting of CD44v(+) cancer stem cells by sulfasalazine in patients with advanced gastric cancer (EPOC1205). Gastric Cancer 2017, 20, 341–349. [Google Scholar] [CrossRef]
- Erb, U.; Megaptche, A.P.; Gu, X.; Büchler, M.W.; Zöller, M. CD44 standard and CD44v10 isoform expression on leukemia cells distinctly influences niche embedding of hematopoietic stem cells. J. Hematol. Oncol. 2014, 7, 29. [Google Scholar] [CrossRef]
- Holm, F.; Hellqvist, E.; Mason, C.N.; Ali, S.A.; Delos-Santos, N.; Barrett, C.L.; Chun, H.J.; Minden, M.D.; Moore, R.A.; Marra, M.A.; et al. Reversion to an embryonic alternative splicing program enhances leukemia stem cell self-renewal. Proc. Natl. Acad. Sci. USA 2015, 112, 15444–15449. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Huang, H.; Tang, Y.; Li, Q.; Wang, J.; Li, D.; Zhong, Z.; Zou, P.; You, Y.; Cao, Y.; et al. CD44v6 chimeric antigen receptor T cell specificity towards AML with FLT3 or DNMT3A mutations. Clin. Transl. Med. 2022, 12, e1043. [Google Scholar] [CrossRef] [PubMed]
- Verel, I.; Heider, K.H.; Siegmund, M.; Ostermann, E.; Patzelt, E.; Sproll, M.; Snow, G.B.; Adolf, G.R.; van Dongen, G.A. Tumor targeting properties of monoclonal antibodies with different affinity for target antigen CD44V6 in nude mice bearing head-and-neck cancer xenografts. Int. J. Cancer 2002, 99, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Suzuki, H.; Ohishi, T.; Asano, T.; Tanaka, T.; Yanaka, M.; Nakamura, T.; Yoshikawa, T.; Kawada, M.; Kaneko, M.K.; et al. Antitumor activities of a defucosylated anti-EpCAM monoclonal antibody in colorectal carcinoma xenograft models. Int. J. Mol. Med. 2023, 51, 18. [Google Scholar] [CrossRef]
- Kaneko, M.K.; Ohishi, T.; Kawada, M.; Kato, Y. A cancer-specific anti-podocalyxin monoclonal antibody (60-mG(2a)-f) exerts antitumor effects in mouse xenograft models of pancreatic carcinoma. Biochem. Biophys. Rep. 2020, 24, 100826. [Google Scholar] [CrossRef]
- Kato, Y.; Kaneko, M.K. A cancer-specific monoclonal antibody recognizes the aberrantly glycosylated podoplanin. Sci. Rep. 2014, 4, 5924. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Kaneko, M.K.; Kato, Y. Roles of Podoplanin in Malignant Progression of Tumor. Cells 2022, 11, 575. [Google Scholar] [CrossRef]
- Ishikawa, A.; Waseda, M.; Ishii, T.; Kaneko, M.K.; Kato, Y.; Kaneko, S. Improved anti-solid tumor response by humanized anti-podoplanin chimeric antigen receptor transduced human cytotoxic T cells in an animal model. Genes Cells 2022, 27, 549–558. [Google Scholar] [CrossRef]
- Chalise, L.; Kato, A.; Ohno, M.; Maeda, S.; Yamamichi, A.; Kuramitsu, S.; Shiina, S.; Takahashi, H.; Ozone, S.; Yamaguchi, J.; et al. Efficacy of cancer-specific anti-podoplanin CAR-T cells and oncolytic herpes virus G47Δ combination therapy against glioblastoma. Mol. Ther.-Oncolytics 2022, 26, 265–274. [Google Scholar] [CrossRef]
- Shiina, S.; Ohno, M.; Ohka, F.; Kuramitsu, S.; Yamamichi, A.; Kato, A.; Motomura, K.; Tanahashi, K.; Yamamoto, T.; Watanabe, R.; et al. CAR T Cells Targeting Podoplanin Reduce Orthotopic Glioblastomas in Mouse Brains. Cancer Immunol. Res. 2016, 4, 259–268. [Google Scholar] [CrossRef]

| No. | Age | Sex | Organ /Anatomic Site | Pathology Diagnosis | TNM | C44Mab-18 | C44Mab-46 |
|---|---|---|---|---|---|---|---|
| 1 | 78 | M | Tongue | SCC of tongue | T2N0M0 | + | + |
| 2 | 40 | M | Tongue | SCC of tongue | T2N0M0 | + | ++ |
| 3 | 75 | F | Tongue | SCC of tongue | T2N0M0 | - | + |
| 4 | 35 | F | Tongue | SCC of tongue | T2N0M0 | ++ | ++ |
| 5 | 61 | M | Tongue | SCC of tongue | T2N0M0 | ++ | +++ |
| 6 | 41 | F | Tongue | SCC of tongue | T2N0M0 | + | + |
| 7 | 64 | M | Tongue | SCC of right tongue | T2N2M0 | ++ | ++ |
| 8 | 76 | M | Tongue | SCC of tongue | T1N0M0 | ++ | ++ |
| 9 | 50 | F | Tongue | SCC of tongue | T2N0M0 | ++ | ++ |
| 10 | 44 | M | Tongue | SCC of tongue | T2N1M0 | ++ | +++ |
| 11 | 53 | F | Tongue | SCC of tongue | T1N0M0 | + | ++ |
| 12 | 46 | F | Tongue | SCC of tongue | T2N0M0 | ++ | + |
| 13 | 50 | M | Tongue | SCC of root of tongue | T3N1M0 | ++ | + |
| 14 | 36 | F | Tongue | SCC of tongue | T1N0M0 | ++ | +++ |
| 15 | 63 | F | Tongue | SCC of tongue | T1N0M0 | + | + |
| 16 | 46 | M | Tongue | SCC of tongue | T2N0M0 | + | - |
| 17 | 58 | M | Tongue | SCC of tongue | T2N0M0 | + | + |
| 18 | 64 | M | Lip | SCC of lower lip | T1N0M0 | + | +++ |
| 19 | 57 | M | Lip | SCC of lower lip | T2N0M0 | + | +++ |
| 20 | 61 | M | Lip | SCC of lower lip | T1N0M0 | + | ++ |
| 21 | 60 | M | Gum | SCC of gum | T3N0M0 | ++ | + |
| 22 | 60 | M | Gum | SCC of gum | T1N0M0 | +++ | +++ |
| 23 | 69 | M | Gum | SCC of upper gum | T3N0M0 | ++ | ++ |
| 24 | 53 | M | Bucca cavioris | SCC of bucca cavioris | T2N0M0 | ++ | + |
| 25 | 55 | M | Bucca cavioris | SCC of bucca cavioris | T1N0M0 | +++ | + |
| 26 | 58 | M | Tongue | SCC of base of tongue | T1N0M0 | ++ | ++ |
| 27 | 63 | M | Oral cavity | SCC | T1N0M0 | +++ | ++ |
| 28 | 48 | F | Tongue | SCC of tongue | T1N0M0 | + | + |
| 29 | 80 | M | Lip | SCC of lower lip | T1N0M0 | +++ | +++ |
| 30 | 77 | M | Tongue | SCC of base of tongue | T2N0M0 | ++ | ++ |
| 31 | 59 | M | Tongue | SCC of tongue | T2N0M0 | + | - |
| 32 | 77 | F | Tongue | SCC of tongue | T1N0M0 | + | ++ |
| 33 | 56 | M | Tongue | SCC of root of tongue | T2N1M0 | + | + |
| 34 | 60 | M | Tongue | SCC of tongue | T2N1M0 | ++ | ++ |
| 35 | 62 | M | Tongue | SCC of tongue | T2N0M0 | + | ++ |
| 36 | 67 | F | Tongue | SCC of tongue | T2N0M0 | - | ++ |
| 37 | 47 | F | Tongue | SCC of tongue | T2N0M0 | +++ | +++ |
| 38 | 37 | M | Tongue | SCC of tongue | T2N1M0 | - | - |
| 39 | 55 | F | Tongue | SCC of tongue | T2N0M0 | + | + |
| 40 | 56 | F | Bucca cavioris | SCC of bucca cavioris | T2N0M0 | + | + |
| 41 | 49 | M | Bucca cavioris | SCC of bucca cavioris | T1N0M0 | - | - |
| 42 | 45 | M | Bucca cavioris | SCC of bucca cavioris | T2N0M0 | - | - |
| 43 | 42 | M | Bucca cavioris | SCC of bucca cavioris | T3N0M0 | +++ | ++ |
| 44 | 44 | M | Jaw | SCC of right drop jaw | T1N0M0 | + | +++ |
| 45 | 40 | F | Tongue | SCC of base of tongue | T2N0M0 | - | ++ |
| 46 | 49 | M | Bucca cavioris | SCC of bucca cavioris | T1N0M0 | ++ | +++ |
| 47 | 56 | F | Tongue | SCC of base of tongue | T2N0M0 | - | + |
| 48 | 42 | M | Bucca cavioris | SCC a of bucca cavioris | T3N0M0 | +++ | +++ |
| 49 | 87 | F | Face | SCC a of left face | T2N0M0 | - | + |
| 50 | 50 | M | Gum | SCC of gum | T2N0M0 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishikawa, K.; Suzuki, H.; Kaneko, M.K.; Kato, Y. Establishment of a Novel Anti-CD44 Variant 10 Monoclonal Antibody C44Mab-18 for Immunohistochemical Analysis against Oral Squamous Cell Carcinomas. Curr. Issues Mol. Biol. 2023, 45, 5248-5262. https://doi.org/10.3390/cimb45070333
Ishikawa K, Suzuki H, Kaneko MK, Kato Y. Establishment of a Novel Anti-CD44 Variant 10 Monoclonal Antibody C44Mab-18 for Immunohistochemical Analysis against Oral Squamous Cell Carcinomas. Current Issues in Molecular Biology. 2023; 45(7):5248-5262. https://doi.org/10.3390/cimb45070333
Chicago/Turabian StyleIshikawa, Kenichiro, Hiroyuki Suzuki, Mika K. Kaneko, and Yukinari Kato. 2023. "Establishment of a Novel Anti-CD44 Variant 10 Monoclonal Antibody C44Mab-18 for Immunohistochemical Analysis against Oral Squamous Cell Carcinomas" Current Issues in Molecular Biology 45, no. 7: 5248-5262. https://doi.org/10.3390/cimb45070333
APA StyleIshikawa, K., Suzuki, H., Kaneko, M. K., & Kato, Y. (2023). Establishment of a Novel Anti-CD44 Variant 10 Monoclonal Antibody C44Mab-18 for Immunohistochemical Analysis against Oral Squamous Cell Carcinomas. Current Issues in Molecular Biology, 45(7), 5248-5262. https://doi.org/10.3390/cimb45070333








