De Novo Design of Anti-COVID Drugs Using Machine Learning-Based Equivariant Diffusion Model Targeting the Spike Protein
Abstract
1. Introduction
1.1. Diffusion Models for Molecules
1.2. Structure-Based Drug Design (SBDD)
2. Materials and Methods
2.1. Selection of Spike Protein
2.2. Domain Analysis and Selection of Binding Sites
2.3. Selection of Algorithm Model and Sampling Variables
2.4. ADME Properties and Toxicity Predictions
2.5. Molecular Docking
2.5.1. Protein Preparation and Ligand Preparation
2.5.2. Precision Docking
2.6. MD Simulation and Analysis
- Equilibration, Brownian Dynamics NVT, T = 10 K, small timesteps, and restraints on solute heavy atoms, 100 ps.
- Equilibration, NVT, T = 10 K, small timesteps, and restraints on solute heavy atoms, 12 ps.
- Equilibration, NVT and no restraints.
- Final simulation (production MD) for 100 ns with time step was set to 2 femto-seconds (fs) and temperature of 303 K. A cut-off short range method with a radius of 9.0 Å (this optimized value avoids overlapping of atoms). The Noose-Hover chain thermostat method was used with RESPA integrator. Simulation has a temperature increase of 10 K per time step after solvation of the binding pocket.
2.7. MM-GBSA Analysis
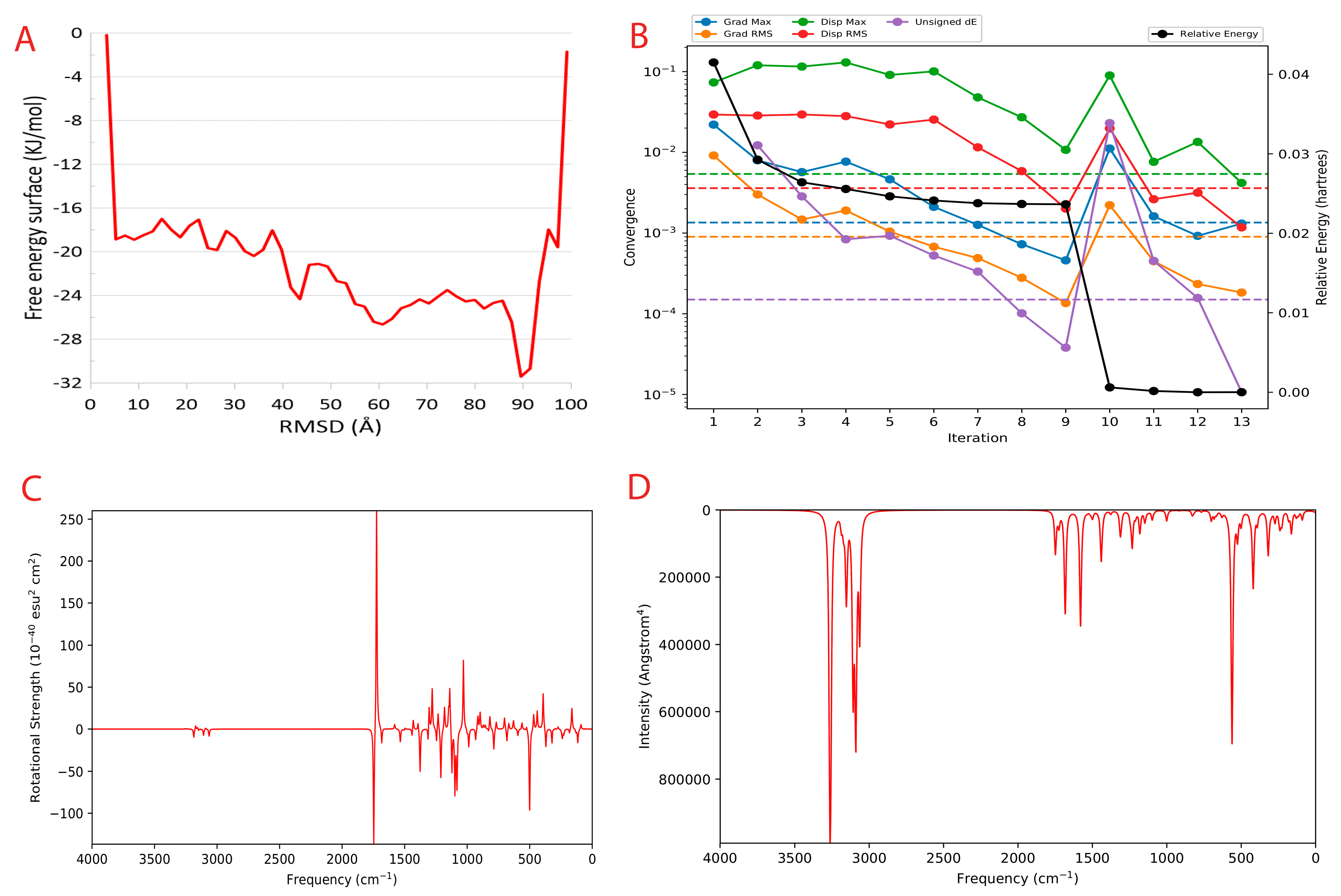
2.8. Metadynamics Simulation
2.9. Quantum Convergence and Single Point Energy Calculations
3. Results
3.1. Domain Analysis of Spike Protein
3.2. Novel Molecules Were Generated Using an Equivariant Diffusion Model
3.3. Molecular Docking and Interaction Pattern
3.4. CoECG-M1 Shows Potential Drug-like Ability via ADMET Predictions
- Lipinski Rule of 5 Violations = 0 (maximum is 4)
- Jorgensen Rule of 3 Violations = 0 (maximum is 3)
- % Human Oral Absorption in GI (+−20%) = 69 (<25% is poor)
- Qual. Model for Human Oral Absorption = Medium (>80% is high)
3.5. A Comprehensive Simulation Studies Provide Insights into Stability, Binding and Unbinding Energies of CoECG-M1
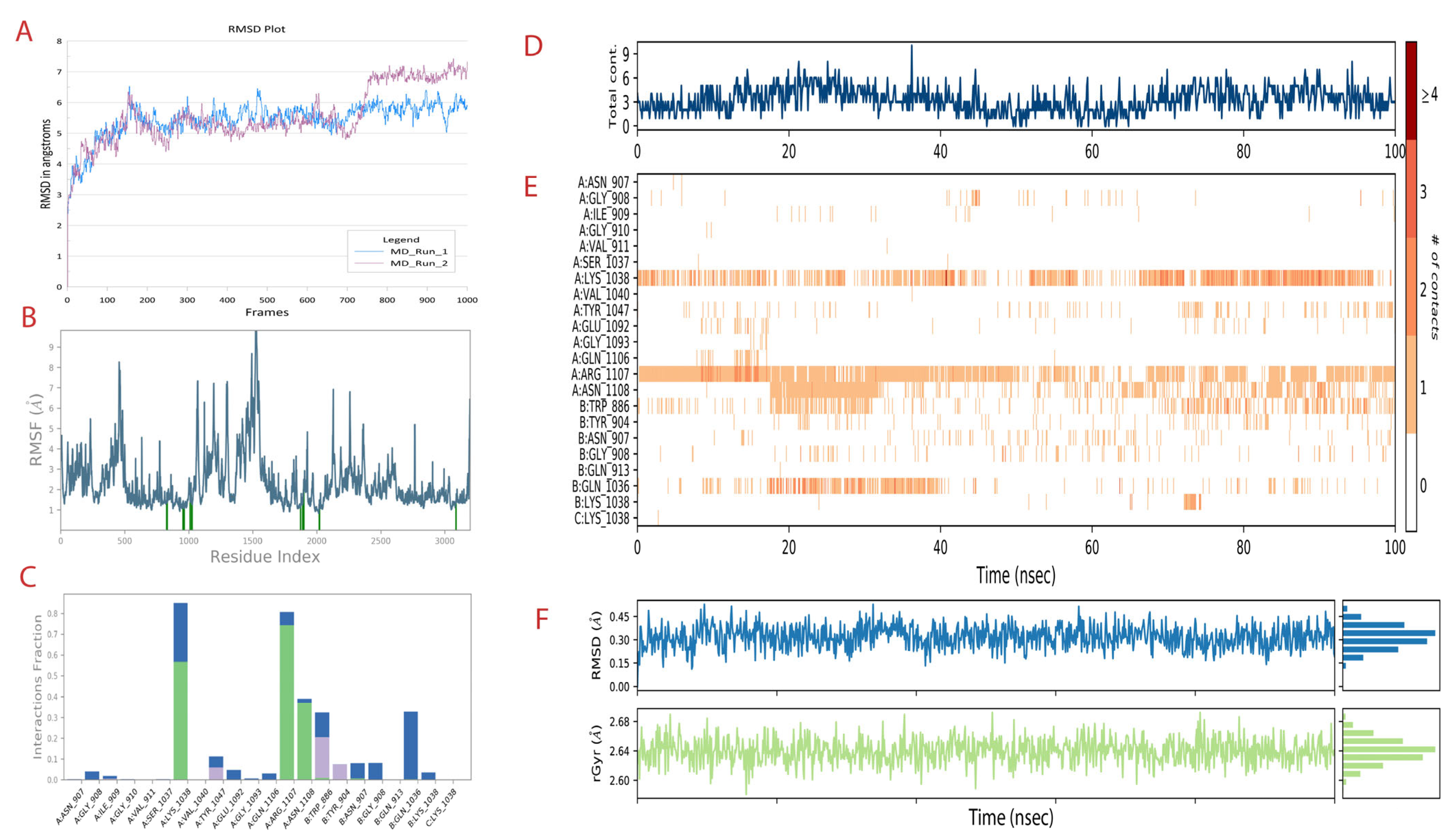
3.5.1. Molecular Dynamic Simulation for 100 ns
3.5.2. Molecular Mechanics with Generalized BORN and Surface area Solvation (MM/GBSA) Studies
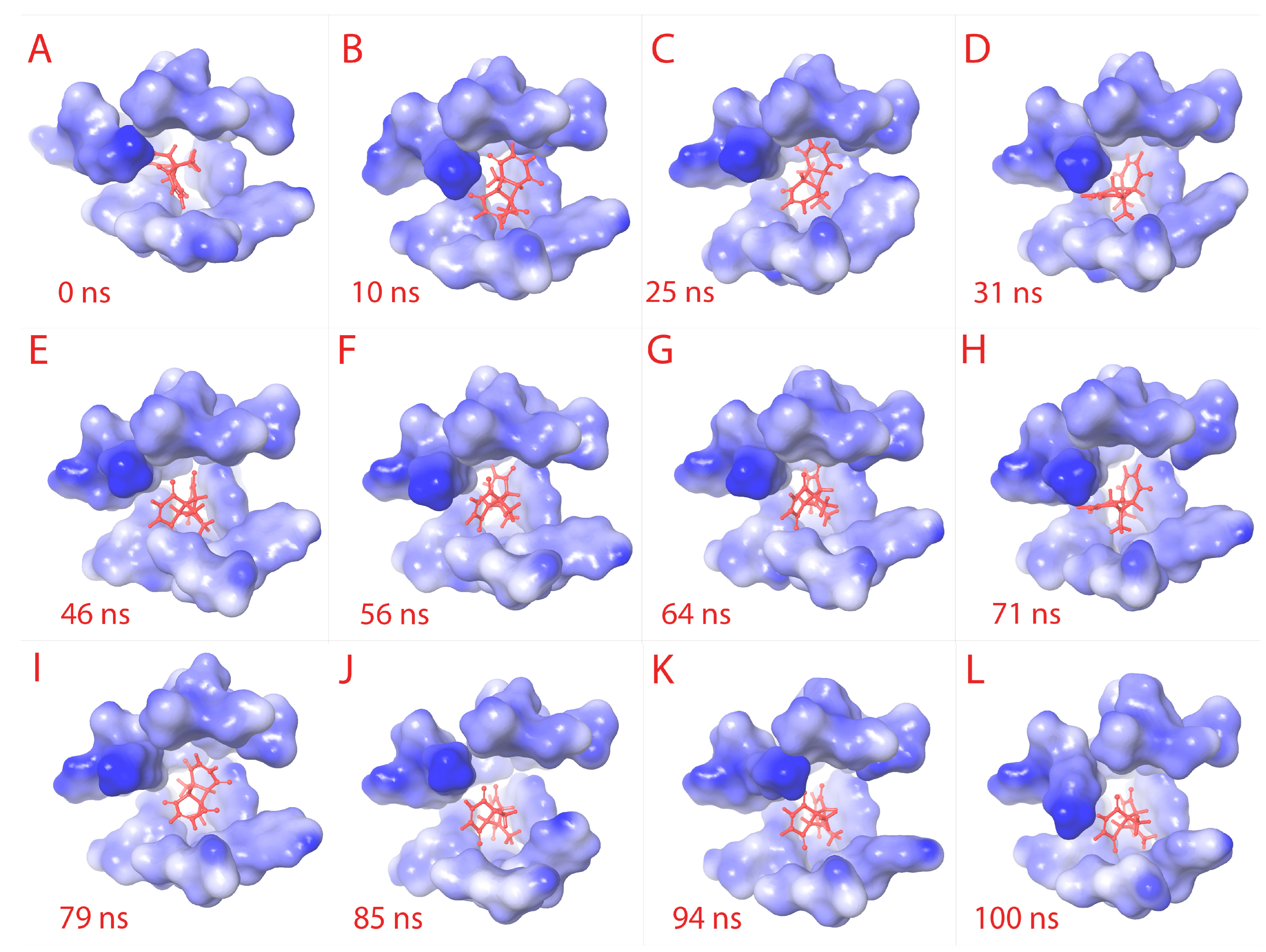
3.5.3. Meta Dynamics Studies to Gain Insights on the Unbinding Potential of CoECG-M1
3.6. Quantum Insight into the Stability, Reactivity, and Structural Characteristics of the Principal Compound
4. Discussion
- Bamlanivimab: a monoclonal antibody that targets the HR1 domain of the spike protein and blocks its interaction with the human ACE2 receptor [76].
- Casirivimab and Imdevimab: a combination of two monoclonal antibodies that target the HR1 and HR2 domains of the spike protein [77].
- VIR-7831: a monoclonal antibody that targets the HR1 domain of the spike protein [78].
- REGN-COV2: a combination of two monoclonal antibodies that target different domains of the spike protein [79].
- Fc-engineered antibodies: a class of antibodies that have been modified to enhance their ability to neutralize the virus by targeting the HR1 and HR2 domains of the spike protein [80].
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ibrahim, S.; Lowe, J.R.; Bramante, C.T.; Shah, S.; Klatt, N.R.; Sherwood, N.; Aronne, L.; Puskarich, M.; Tamariz, L.; Palacio, A.; et al. Metformin and Covid-19: Focused Review of Mechanisms and Current Literature Suggesting Benefit. Front. Endocrinol. 2021, 12, 587801. [Google Scholar] [CrossRef]
- Low, Z.Y.; Yip, A.J.W.; Lal, S.K. Repositioning Ivermectin for Covid-19 treatment: Molecular mechanisms of action against SARS-CoV-2 replication. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166294. [Google Scholar] [CrossRef]
- Lenze, E.J.; Mattar, C.; Zorumski, C.F.; Stevens, A.; Schweiger, J.; Nicol, G.E.; Miller, J.P.; Yang, L.; Yingling, M.; Avidan, M.S.; et al. Fluvoxamine vs Placebo and Clinical Deterioration in Outpatients with Symptomatic COVID-19: A Randomized Clinical Trial. JAMA 2020, 324, 2292–2300. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Herbrich, R.; Escalera, S. A Guide to the NeurIPS 2018 Competitions. In The NeurIPS ‘18 Competition; Springer International Publishing: New York, NY, USA, 2019; pp. 1–9. [Google Scholar]
- Welling, M. Herding dynamical weights to learn. In Proceedings of the 26th Annual International Conference on Machine Learning, Montreal, QC, Canada, 14–18 June 2009; ACM: New York, NY, USA, 2009. [Google Scholar]
- Jing, B.; Corso, G.; Chang, J.; Barzilay, R.; Jaakkola, T. Torsional Diffusion for Molecular Conformer Generation. arXiv 2022, arXiv:2206.01729. [Google Scholar]
- Drotár, P.; Jamasb, A.R.; Day, B.; Cangea, C.; Liò, P. Structure-Aware Generation of Drug-like Molecules. 2021. Available online: https://www.mlsb.io/papers_2021/MLSB2021_Structure-aware_generation_of_drug-like.pdf (accessed on 13 December 2022).
- Du, Y.; Fu, T.; Sun, J.; Liu, S. MolGenSurvey: A Systematic Survey in Machine Learning Models for Molecule Design. arXiv 2022, arXiv:2203.14500. [Google Scholar]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Luo, S.; Hu, W. Diffusion Probabilistic Models for 3D Point Cloud Generation. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021. [Google Scholar]
- Abdool Karim, S.S.; Devnarain, N. Time to Stop Using Ineffective Covid-19 Drugs. N. Engl. J. Med. 2022, 387, 654–655. [Google Scholar] [CrossRef]
- Xing, L.; Xu, X.; Xu, W.; Liu, Z.; Shen, X.; Zhou, J.; Xu, L.; Pu, J.; Yang, C.; Huang, Y.; et al. A Five-Helix-Based SARS-CoV-2 Fusion Inhibitor Targeting Heptad Repeat 2 Domain against SARS-CoV-2 and Its Variants of Concern. Viruses 2022, 14, 597. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Amp. Mol. Biol. 2003, 10, 980. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H.; Markley, J.L. The worldwide Protein Data Bank (wwPDB): Ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007, 35, D301–D303. [Google Scholar] [CrossRef]
- wwPDB consortium. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.; Abiona, O.; Graham, B.S.; McLellan, J.S. Prefusion 2019-nCoV Spike Glycoprotein with a Single Receptor-Binding Domain Up; Worldwide Protein Data Bank, 2020. [Google Scholar]
- Sigrist, C.J.A.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef]
- de Castro, E.; Sigrist, C.J.A.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Anderson, J.B.; Derbyshire, M.K.; DeWeese-Scott, C.; Gonzales, N.R.; Gwadz, M.; Hao, L.; He, S.; Hurwitz, D.I.; Jackson, J.D.; et al. CDD: A conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007, 35, D237–D240. [Google Scholar] [CrossRef]
- Igashov, I.; Jamasb, A.R.; Sadek, A.; Sverrisson, F.; Schneuing, A.; Liò, P.; Blundell, T.L.; Bronstein, M.; Correia, B. Decoding Surface Fingerprints for Protein-Ligand Interactions; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2022. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef]
- Schrödinger, LLC. Schrödinger; Release 2022-2: Maestro, [(Licensed on 22 January 2022)]; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pK a prediction and protonation state generation for drug-like molecules. J. Comput.-Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2015, 12, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.J.; Sacerdoti, F.D.; Salmon, J.K.; Shan, Y.; Shaw, D.E.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; et al. Molecular dynamics—Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing—SC ‘06, Tampa, FL, USA, 11–17 November 2006; ACM Press: New York, NY, USA, 2006. [Google Scholar]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Uttarkar, A.; Kishore, A.P.; Srinivas, S.M.; Rangappa, S.; Kusanur, R.; Niranjan, V. Coumarin derivative as a potent drug candidate against triple negative breast cancer targeting the frizzled receptor of wingless-related integration site signaling pathway. J. Biomol. Struct. Dyn. 2022, 41, 1561–1573. [Google Scholar] [CrossRef] [PubMed]
- Skariyachan, S.; Ravishankar, R.; Gopal, D.; Muddebihalkar, A.G.; Uttarkar, A.; Praveen, P.K.U.; Niranjan, V. Response regulator GacA and transcriptional activator RhlR proteins involved in biofilm formation of Pseudomonas aeruginosa are prospective targets for natural lead molecules: Computational modelling, molecular docking and dynamic simulation studies. Infect. Genet. Evol. 2020, 85, 104448. [Google Scholar] [CrossRef] [PubMed]
- Gopal, D.; Muddebihalkar, A.G.; Skariyachan, S.; Uttarkar, A.C.; Kaveramma, P.; Praveen, U.; Shankar, R.R.; Venkatesan, T.; Niranjan, V. Mitogen activated protein kinase-1 and cell division control protein-42 are putative targets for the binding of novel natural lead molecules: A therapeutic intervention against Candida albicans. J. Biomol. Struct. Dyn. 2019, 38, 4584–4599. [Google Scholar] [CrossRef]
- Uttarkar, A.; Niranjan, V. Brefeldin A variant via combinatorial screening acts as an effective antagonist inducing structural modification in EPAC2. Mol. Simul. 2022, 48, 1592–1603. [Google Scholar] [CrossRef]
- Ylilauri, M.; Pentikäinen, O.T. MMGBSA As a Tool to Understand the Binding Affinities of Filamin–Peptide Interactions. J. Chem. Inf. Model. 2013, 53, 2626–2633. [Google Scholar] [CrossRef]
- Li, J.; Abel, R.; Zhu, K.; Cao, Y.; Zhao, S.; Friesner, R.A. The VSGB 2.0 model: A next generation energy model for high resolution protein structure modeling. Proteins 2011, 79, 2794–2812. [Google Scholar] [CrossRef] [PubMed]
- Setlur, A.S.; Karunakaran, C.; Pandey, S.; Sarkar, M. Molecular interaction studies of thymol via molecular dynamic simulations and free energy calculations using multi-target approach against Aedes aegypti proteome to decipher its role as mosquito repellent. Mol. Simul. 2023, 49, 325–340. [Google Scholar] [CrossRef]
- Niranjan, V. Well-Tempered Metadynamics Protocol v2; ZappyLab, Inc.: Berkeley, CA, USA, 2022. [Google Scholar] [CrossRef]
- Wang, J.; Ishchenko, A.; Zhang, W.; Razavi, A.; Langley, D. A highly accurate metadynamics-based Dissociation Free Energy method to calculate protein-protein and protein-ligand binding potencies. Sci. Rep. 2022, 12, 2024. [Google Scholar] [CrossRef] [PubMed]
- Bochevarov, A.D.; Harder, E.; Hughes, T.F.; Greenwood, J.R.; Braden, D.A.; Philipp, D.M.; Rinaldo, D.; Halls, M.D.; Zhang, J.; Friesner, R.A. Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013, 113, 2110–2142. [Google Scholar] [CrossRef]
- Hehre, W.J.; Ditchfield, R.; Pople, J.A. Self-Consistent Molecular Orbital Methods. XII. Furth. Ext. Gaussian—Type Basis Sets Use Mol. Orbital Stud. Org. Molecules. J. Chem. Phys. 1972, 56, 2257–2261. [Google Scholar] [CrossRef]
- Niranjan, V.; Uttarkar, A.; Murali, K.; Niranjan, S.; Gopal, J.; Kumar, J. Mycobacterium Time-Series Genome Analysis Identifies AAC2’ as a Potential Drug Target with Naloxone Showing Potential Bait Drug Synergism. Molecules 2022, 27, 6150. [Google Scholar] [CrossRef]
- The UniProt Consortium, UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Cao, W.; Dong, C.; Kim, S.; Hou, D.; Tai, W.; Du, L.; Im, W.; Zhang, X.F. Biomechanical characterization of SARS-CoV-2 spike RBD and human ACE2 protein-protein interaction. Biophys. J. 2021, 120, 1011–1019. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Chan, W.-E.; Chuang, C.-K.; Yeh, S.-H.; Chang, M.-S.; Chen, S.S.L. Functional characterization of heptad repeat 1 and 2 mutants of the spike protein of severe acute respiratory syndrome coronavirus. J. Virol. 2006, 80, 3225–3237. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [PubMed]
- Bento, A.P.; Gaulton, A.; Hersey, A.; Bellis, L.J.; Chambers, J.; Davies, M.; Krüger, F.A.; Light, Y.; Mak, L.; McGlinchey, S.; et al. The ChEMBL bioactivity database: An update. Nucleic Acids Res. 2014, 42, D1083–D1090. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. PubChem Substance Record for SID 475724928, SID 475724928, Source: Vidya Lab, Department of Biotechnology, RV College of Engineering, Bengaluru. PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/substance/475724928 (accessed on 7 February 2023).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 2717, Chlormezanone. PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Chlormezanone (accessed on 7 February 2023).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 4165, Meticrane. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Meticrane (accessed on 7 February 2023).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 57697, Pemirolast. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Pemirolast (accessed on 7 February 2023).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 636372, Aceglatone. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Aceglatone (accessed on 7 February 2023).
- Kufareva, I.; Abagyan, R. Methods of protein structure comparison. In Methods in Molecular Biology; Humana Press: Clifton, NJ, USA, 2012; Volume 857, pp. 231–257. [Google Scholar] [CrossRef]
- Ahmad, S.; Bhanu, P.; Kumar, J.; Pathak, R.K.; Mallick, D.; Uttarkar, A.; Niranjan, V.; Mishra, V. Molecular dynamics simulation and docking analysis of NF-κB protein binding with sulindac acid. Bioinformation 2022, 18, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Niranjan, V.; Jayaprasad, S.; Uttarkar, A.; Kusanur, R.; Kumar, J. Design of Novel Coumarin Derivatives as NUDT5 Antagonists That Act by Restricting ATP Synthesis in Breast Cancer Cells. Molecules 2022, 28, 89. [Google Scholar] [CrossRef]
- WHO Solidarity Trial Consortium. Remdesivir and three other drugs for hospitalised patients with COVID-19: Final results of the WHO Solidarity randomised trial and updated meta-analyses. Lancet 2022, 399, 1941–1953. [Google Scholar] [CrossRef]
- Maziarz, M.; Stencel, A. The failure of drug repurposing for COVID-19 as an effect of excessive hypothesis testing and weak mechanistic evidence. Hist. Philos. Life Sci. 2022, 44, 47. [Google Scholar] [CrossRef]
- Martinez, M.A. Lack of Effectiveness of Repurposed Drugs for COVID-19 Treatment. Front. Immunol. 2021, 12, 635371. [Google Scholar] [CrossRef]
- Vanden Eynde, J.J. COVID-19: Failure of the DisCoVeRy Clinical Trial, and Now-New Hopes? Pharmaceuticals 2021, 14, 664. [Google Scholar] [CrossRef]
- Akshay, U.; Vidya, N.; Shivam, P.; Srividya, S. Study of SARS-nCoV2 Indian Isolates Gaining Insights into Mutation Frequencies, Protein Stability and Prospective Effect on Pathogenicity. Coronaviruses 2021, 2, e170821190411. [Google Scholar] [CrossRef]
- Xia, X. Domains and Functions of Spike Protein in Sars-Cov-2 in the Context of Vaccine Design. Viruses 2021, 13, 109. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.-F.; Xu, W.; Liu, S.-W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Du, L. SARS-CoV-2 spike protein: A key target for eliciting persistent neutralizing antibodies. Signal Transduct. Target. Ther. 2021, 6, 95. [Google Scholar] [CrossRef]
- Taylor, P.C.; Adams, A.C.; Hufford, M.M.; de la Torre, I.; Winthrop, K.; Gottlieb, R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021, 21, 382–393. [Google Scholar] [CrossRef]
- Su, H.; Zhou, F.; Huang, Z.; Ma, X.; Natarajan, K.; Zhang, M.; Huang, Y.; Su, H. Molecular Insights into Small-Molecule Drug Discovery for SARS-CoV-2. Angew. Chem. Int. Ed. 2021, 60, 9789–9802. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Z.; Cao, X.; Liu, X.; He, H.; Yang, Y. Design and synthesis of pyridine-substituted itraconazole analogues with improved antifungal activities, water solubility and bioavailability. Bioorganic Med. Chem. Lett. 2011, 21, 4779–4783. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 55283, Itraconazole. PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Itraconazole (accessed on 7 February 2023).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 222757, Estradiol benzoate. PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Estradiol-benzoate (accessed on 7 February 2023).
- Yang, C.; Pan, X.; Huang, Y.; Cheng, C.; Xu, X.; Wu, Y.; Xu, Y.; Shang, W.; Niu, X.; Wan, Y.; et al. Drug Repurposing of Itraconazole and Estradiol Benzoate against COVID-19 by Blocking SARS-CoV-2 Spike Protein-Mediated Membrane Fusion. Adv. Ther. 2021, 4, 2000224. [Google Scholar] [CrossRef]
- Destras, G.; Assaad, S.; Bal, A.; Bouscambert-Duchamp, M.; Avrillon, V.; Simon, B.; Valette, M.; Blay, J.-Y.; Lina, B.; Frobert, E.; et al. Bamlanivimab as monotherapy in two immunocompromised patients with COVID-19. Lancet Microbe 2021, 2, e424. [Google Scholar] [CrossRef]
- Deeks, E.D. Casirivimab/Imdevimab: First Approval. Drugs 2021, 81, 2047–2055. [Google Scholar] [CrossRef]
- Cathcart, A.L.; Havenar-Daughton, C.; Lempp, F.A.; Ma, D.; Schmid, M.A.; Agostini, M.L.; Guarino, B.; Di iulio, J.; Rosen, L.E.; Tucker, H.; et al. The Dual Function Monoclonal Antibodies VIR-7831 and VIR-7832 Demonstrate Potent In Vitro and In Vivo Activity against SARS-CoV-2; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2021. [Google Scholar]
- Baum, A.; Ajithdoss, D.; Copin, R.; Zhou, A.; Lanza, K.; Negron, N.; Ni, M.; Wei, Y.; Mohammadi, K.; Musser, B.; et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science 2020, 370, 1110–1115. [Google Scholar] [CrossRef] [PubMed]
- Yamin, R.; Jones, A.T.; Hoffmann, H.-H.; Schäfer, A.; Kao, K.S.; Francis, R.L.; Sheahan, T.P.; Baric, R.S.; Rice, C.M.; Ravetch, J.V.; et al. Fc-engineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature 2021, 599, 465–470. [Google Scholar] [CrossRef] [PubMed]
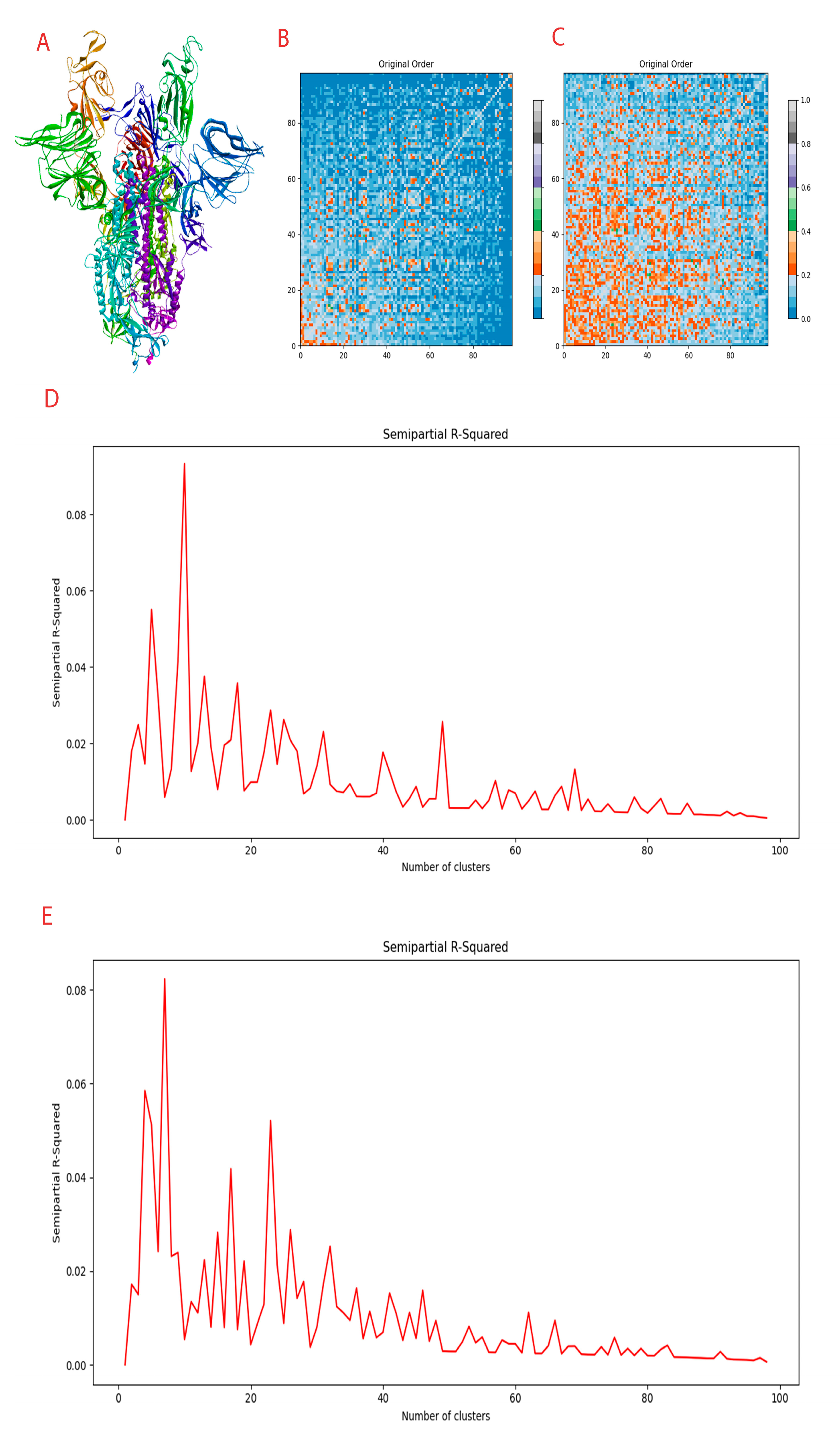
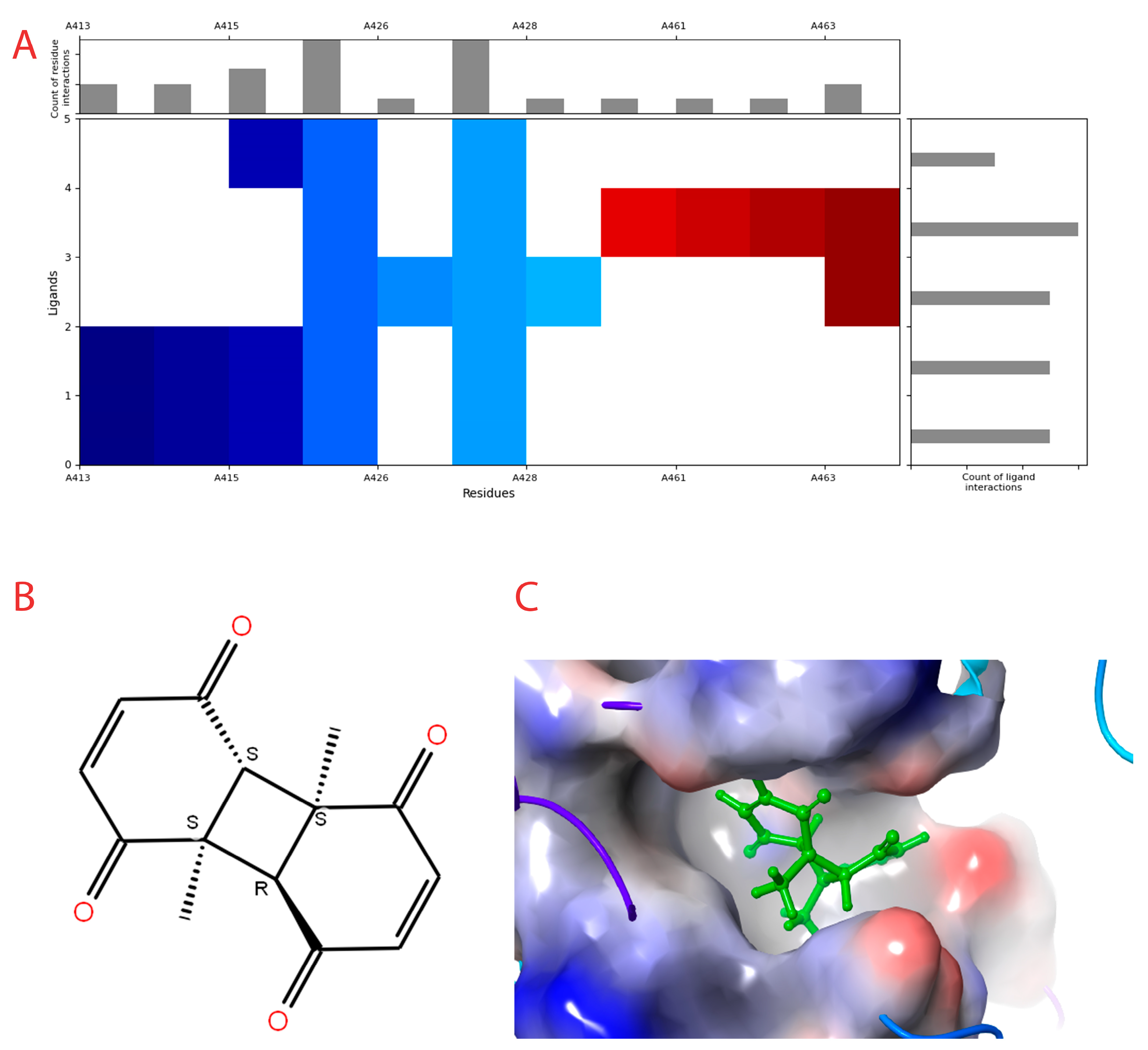
| Sl No | Amino Acids | Function |
|---|---|---|
| 1 | 280–301 | Putative superantigen; may bind T-cell receptor alpha/TRAC |
| 2 | 319–541 | Receptor-binding domain (RBD) |
| 3 | 403–405 | Integrin-binding motif; |
| 4 | 437–508 | Receptor-binding motif; binding to human ACE2 |
| 5 | 448–456 | Immunodominant HLA epitope recognized by the CD8+; called NF9 peptide |
| 6 | 681–684 | Putative superantigen; may bind T-cell receptor beta/TRBC1 |
| 7 | 816–837 | Fusion peptide 1 |
| 8 | 835–855 | Fusion peptide 2 |
| 9 | 920–970 | Heptad repeat 1 |
| 10 | 1163–1202 | Heptad repeat 2 |
| Sl No. | Structure | Smiles | Docking Score in kcal/mol |
|---|---|---|---|
| 1 |  | O=C(O)C1CC(O)C(=O)C1(O)O | −4.346 |
| 2 | 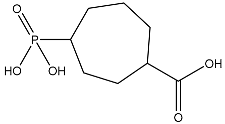 | O=C(O)C1CCC(P(=O)(O)O)CCC1 | −4.192 |
| 3 | 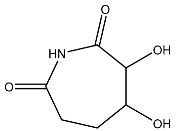 | O=C1C(O)C(O)CC(C(=O)N1) | −4.112 |
| 4 | 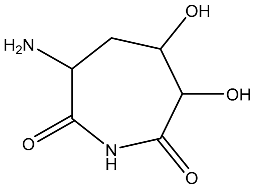 | O=C1C(C(CC(N)C(N1)=O)O)O | −3.900 |
| 5 | 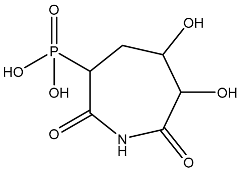 | O=C1C(O)C(O)CC(C(=O)N1)P(=O)(O)O | −3.842 |
| 6 | 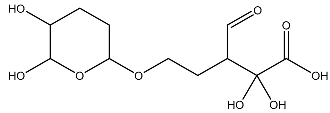 | O=C(O)C(O)(O)C(C=O)CCOC(CC1)OC(O)C1O | −3.722 |
| 7 | 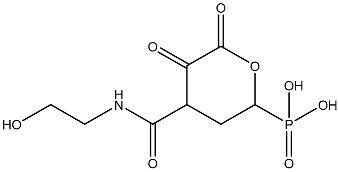 | OCCNC(=O)C1CC(P(=O)(O)O)OC(=O)C1=O | −3.616 |
| 8 | 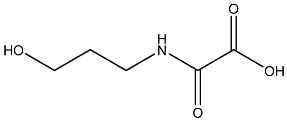 | O=C(O)C(=O)NCCCO | −3.508 |
| 9 |  | O=C(O)N1CCN(C1O)C(=O)C(O2)CC(C23)CCC3=O | −3.503 |
| 10 | 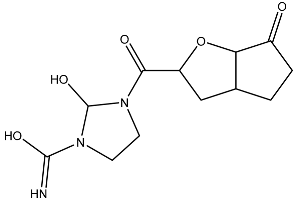 | N=C(N1CCN(C(C2OC3C(CCC3=O)C2)=O)C1O)O | −3.438 |
| 11 | 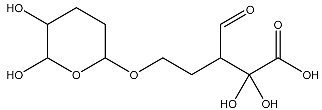 | O=C(O)C(O)(O)C(C=O)CCOC(CC1)OC(O)C1O | −3.234 |
| 12 | 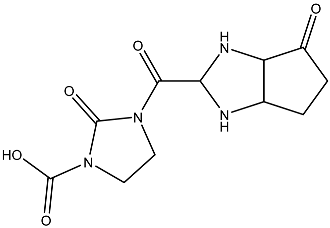 | O=C(N1CCN(C(C2NC3C(CCC3=O)N2)=O)C1=O)O | −2.676 |
| 13 | 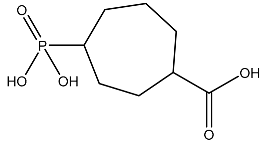 | O=C(O)C1CCC(P(=O)(O)O)CCC1 | −2.225 |
| 14 | 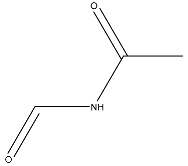 | O=C(O)N1CCN)C(O2)CC(C23)CCC3=O | −2.211 |
| 15 | 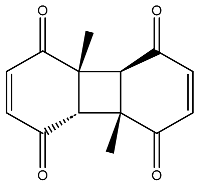 | O=C1C=CC(=O)[C@@H]([C@]12C)[C@]3(C)[C@H]2C(=O)C=CC3=O | −6.930 |
| Sl No. | Property | Value | Range (95% of Drugs) |
|---|---|---|---|
| 1 | Molecular Weight | 244.246 | 130.0/725.0 |
| 2 | Dipole Moment | 1.678 | 1.0/12.5 |
| 3 | Total SASA | 424.755 | 300.0/1000.0 |
| 4 | Hydrophobic SASA | 125.120 | 0.0/750.0 |
| 5 | Hydrophilic SASA | 154.362 | 7.0/330.0 |
| 6 | Carbon Pi SASA | 145.273 | 0.0/450.0 |
| 7 | Weakly Polar SASA | 0.000 | 0.0/175.0 |
| 8 | Molecular Volume | 747.138 | 500.0/2000.0 |
| 9 | vdW Polar SA | 101.833 | 7.0/200.0 |
| 10 | Rotatable Bonds | 0.000 | 0.0/15.0 |
| 11 | Hydrogen Bonds | 0.000 (Donor)/8.000 (Acceptor) | 0.0/6.0 (Donor)/2.0/20.0 (Acceptor) |
| 12 | Globularity | 0.937 | 0.75/0.95 |
| 13 | Ionization Potential | 10.847 (eV) | 7.9/10.5 |
| 14 | Electron Affinity | 1.316 (eV) | −0.9/1.7 |
| 15 | QP Polarizability | 25.134M (Å3) | 13.0/70.0 |
| 16 | QP log P (hexadecane/gas) | 7.710M | 4.0/18.0 |
| 17 | QP log P (octanol/gas) | 12.663 | 8.0/35.0 |
| 18 | QP log P (water/gas) | 10.659 | 4.0/45.0 |
| 19 | QP log P (octanol/water) | −0.625 | −2.0/6.5 |
| 20 | QP log S (aqueous solubility) | −0.078 | −6.5/0.5 |
| 21 | QP log S (conformation independent) | −0.770 | −6.5/0.5 |
| 22 | QP log K (serum protein binding) | −1.353 | −1.5/1.5 |
| 23 | Caco-2 Permeability (nm/s) | 340 | <25 poor, >500 great |
| Sl No. | Classification | Target | Prediction | Probability |
|---|---|---|---|---|
| 1 | Organ toxicity | Hepatotoxicity | Inactive | 0.75 |
| 2 | Toxicity endpoints | Carcinogenicity | Inactive | 0.51 |
| 3 | Toxicity endpoints | Immunotoxicity | Inactive | 0.57 |
| 4 | Toxicity endpoints | Mutagenicity | Inactive | 0.62 |
| 5 | Toxicity endpoints | Cytotoxicity | Inactive | 0.79 |
| 6 | Tox21-Nuclear receptor signalling pathways | Aryl hydrocarbon Receptor (AhR) | Inactive | 0.84 |
| 7 | Tox21-Nuclear receptor signalling pathways | Androgen Receptor (AR) | Inactive | 0.93 |
| 8 | Tox21-Nuclear receptor signalling pathways | Androgen Receptor Ligand Binding Domain (AR-LBD) | Inactive | 0.98 |
| 9 | Tox21-Nuclear receptor signalling pathways | Aromatase | Inactive | 0.87 |
| 10 | Tox21-Nuclear receptor signalling pathways | Estrogen Receptor Alpha (ER) | Inactive | 0.68 |
| 11 | Tox21-Nuclear receptor signalling pathways | Estrogen Receptor Ligand Binding Domain (ER-LBD) | Inactive | 0.85 |
| 12 | Tox21-Nuclear receptor signalling pathways | Peroxisome Proliferator-Activated Receptor Gamma (PPAR-Gamma) | Inactive | 0.97 |
| 13 | Tox21-Stress response pathways | Nuclear factor (erythroid-derived 2)-like 2/antioxidant responsive element (nrf2/ARE) | Inactive | 0.90 |
| 14 | Tox21-Stress response pathways | Heat shock factor response element (HSE) | Inactive | 0.90 |
| 15 | Tox21-Stress response pathways | Mitochondrial Membrane Potential (MMP) | Inactive | 0.57 |
| 16 | Tox21-Stress response pathways | Phosphoprotein (Tumour Suppressor) p53 | Inactive | 0.77 |
| 17 | Tox21-Stress response pathways | ATPase family AAA domain-containing protein 5 (ATAD5) | Inactive | 0.90 |
| Sl No. | Measure | Run 1 | Run 2 |
|---|---|---|---|
| 1 | Protein RMSD | 5.8 angstroms | 7.2 angstroms |
| 2 | Ligand RMSD | 0.35 angstrom | 0.28 angstrom |
| 3 | Secondary structure elements | 37.8% | 38.7% |
| 4 | H-Bond interactions | Lys 1038, Arg 1107 and Asn 1108 | Arg 1107 and Asn 1108 |
| 5 | Radius of gyration | 2.64 angstroms | 2.64 angstroms |
| Time (ns) | MD Run 1 | MD Run 2 | ||
|---|---|---|---|---|
| DG Bind (kcal/mol) | DG Bind NS (kcal/mol) | DG Bind (kcal/mol) | DG Bind NS (kcal/mol) | |
| 10 | −38.37404477 | −40.6192763 | −39.51735788 | −39.82294397 |
| 20 | −38.42947824 | −39.1927792 | −41.90389158 | −43.09712232 |
| 30 | −42.21866335 | −42.59491568 | −37.4298756 | −37.95905421 |
| 40 | −41.55274598 | −42.46953731 | −47.63490585 | −48.53847099 |
| 50 | −41.29063169 | −41.50944017 | −36.60442487 | −37.80706116 |
| 60 | −39.52055104 | −40.4680222 | −36.70081353 | −37.67308507 |
| 70 | −35.63802159 | −36.09386049 | −36.82613064 | −37.68762383 |
| 80 | −41.09375804 | −42.18120988 | −37.641591 | −37.99902137 |
| 90 | −48.9177109 | −50.1835614 | −37.55085127 | −37.15958535 |
| 100 | −44.66043935 | −46.04530244 | −36.10956839 | −36.99196607 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niranjan, V.; Uttarkar, A.; Ramakrishnan, A.; Muralidharan, A.; Shashidhara, A.; Acharya, A.; Tarani, A.; Kumar, J. De Novo Design of Anti-COVID Drugs Using Machine Learning-Based Equivariant Diffusion Model Targeting the Spike Protein. Curr. Issues Mol. Biol. 2023, 45, 4261-4284. https://doi.org/10.3390/cimb45050271
Niranjan V, Uttarkar A, Ramakrishnan A, Muralidharan A, Shashidhara A, Acharya A, Tarani A, Kumar J. De Novo Design of Anti-COVID Drugs Using Machine Learning-Based Equivariant Diffusion Model Targeting the Spike Protein. Current Issues in Molecular Biology. 2023; 45(5):4261-4284. https://doi.org/10.3390/cimb45050271
Chicago/Turabian StyleNiranjan, Vidya, Akshay Uttarkar, Ananya Ramakrishnan, Anagha Muralidharan, Abhay Shashidhara, Anushri Acharya, Avila Tarani, and Jitendra Kumar. 2023. "De Novo Design of Anti-COVID Drugs Using Machine Learning-Based Equivariant Diffusion Model Targeting the Spike Protein" Current Issues in Molecular Biology 45, no. 5: 4261-4284. https://doi.org/10.3390/cimb45050271
APA StyleNiranjan, V., Uttarkar, A., Ramakrishnan, A., Muralidharan, A., Shashidhara, A., Acharya, A., Tarani, A., & Kumar, J. (2023). De Novo Design of Anti-COVID Drugs Using Machine Learning-Based Equivariant Diffusion Model Targeting the Spike Protein. Current Issues in Molecular Biology, 45(5), 4261-4284. https://doi.org/10.3390/cimb45050271


_Kim.png)



