Cloning, Expression, and Functional Characterization of FUT1, a Key Gene for Histo-Blood Group Antigens Synthesis in Crassostrea gigas
Abstract
1. Introduction
2. Materials and Methods
2.1. Oyster Samples
2.2. Total RNA Extraction and Reverse Transcription
2.3. Cloning of Full-Length Gene
2.4. Sequence and Phylogenetic Analyses
2.5. RT-qPCR
2.6. Expression and Western Blot Analysis of CgFUT1
2.7. Purification of Recombinant Proteins Produced in E. coli
2.8. Validation of the Enzyme Function of CgFUT1
2.9. Expression of CgFUT1 in CHO Cells and Immunofluorescence Assays
3. Results
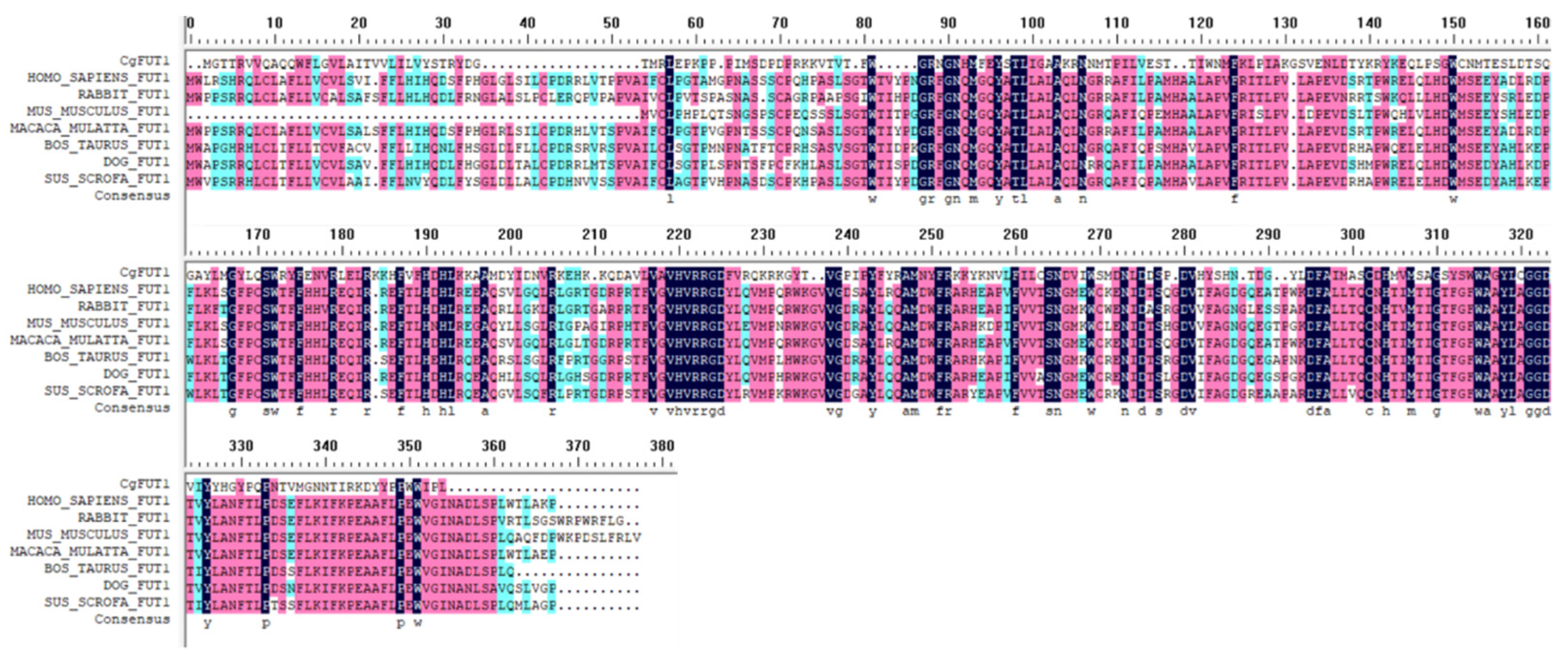
3.1. Molecular Characterization of CgFUT1
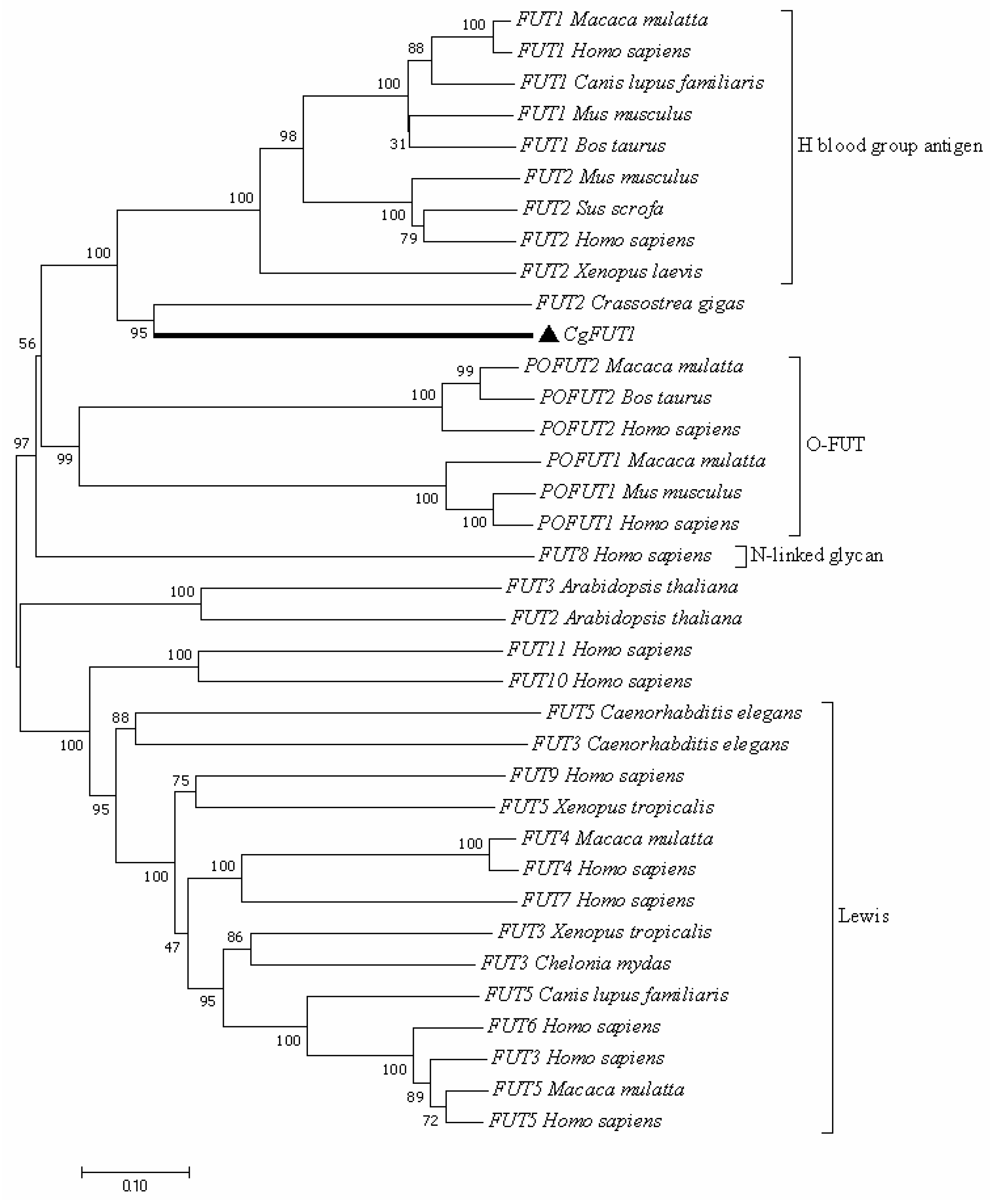
3.2. Homologous and Phylogenetic Analysis of CgFUT1
3.3. Tissue Distribution of CgFUT1
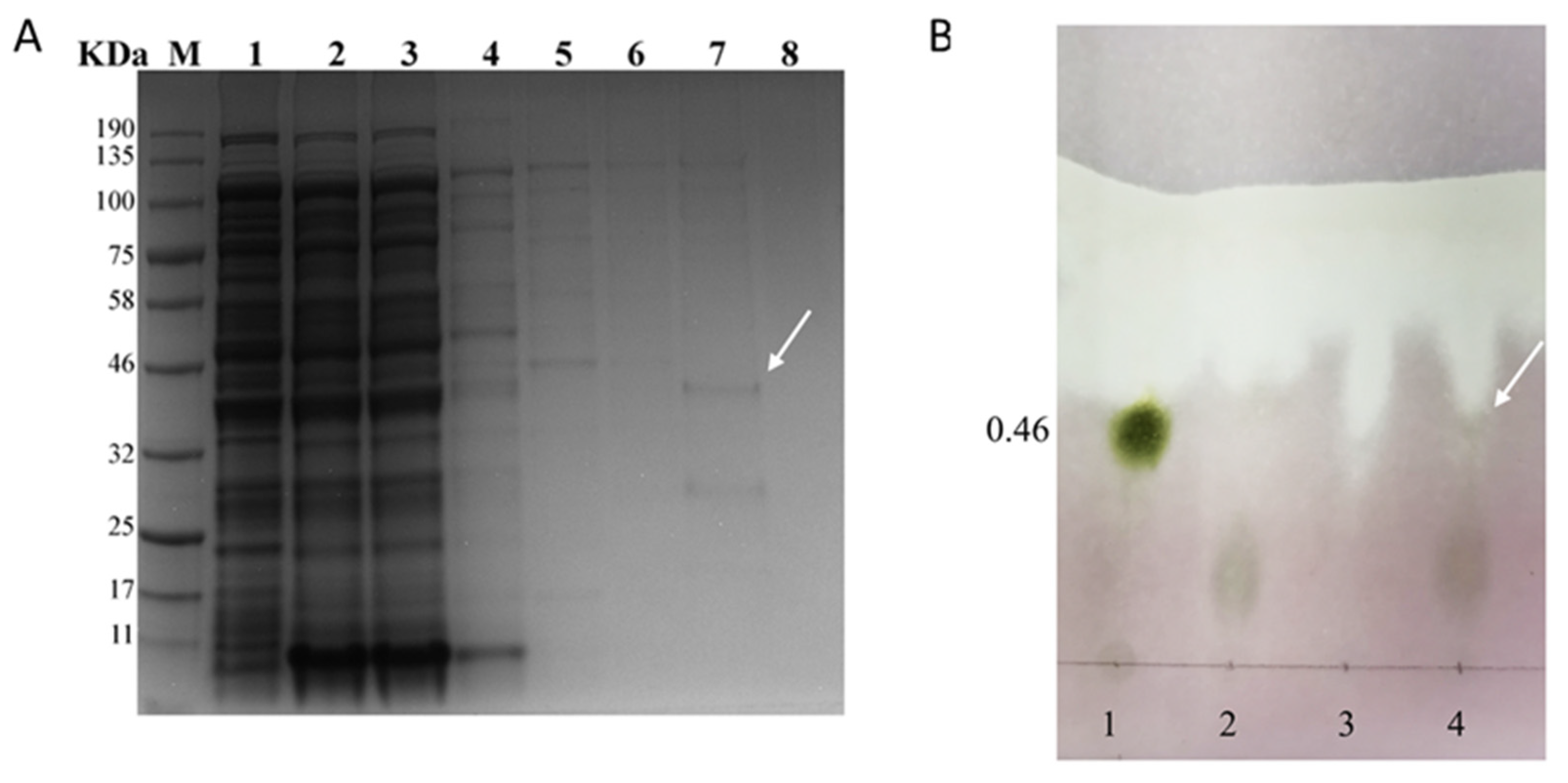
3.4. Expression and Western Blot Analysis of CgFUT1
3.5. CgFUT1 Recombinant Protein Purification and Enzyme Function Validation
3.6. Expression and Functional Characterization of CgFUT1 in CHO Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cannon, J.L.; Bonifacio, J.; Bucardo, F.; Buesa, J.; Bruggink, L.; Chan, M.C.-W.; Fumian, T.M.; Giri, S.; Gonzalez, M.D.; Hewitt, J.; et al. Global Trends in Norovirus Genotype Distribution among Children with Acute Gastroenteritis. Emerg. Infect. Infect. Dis. 2021, 27, 1438–1445. [Google Scholar] [CrossRef]
- Hassan, E.; Baldridge, M.T. Norovirus encounters in the gut: Multifaceted interactions and disease outcomes. Mucosal Immunol. 2019, 12, 1259–1267. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.L.F.; Perin, J.; Aryee, M.J.; Boschi-Pinto, C.; Black, R.E. Diarrhea incidence in low- and middle-income countries in 1990 and 2010: A systematic review. BMC Public Health 2012, 12, 220. [Google Scholar]
- Teunis, P.F.M.; Moe, C.L.; Liu, P.; Miller, S.E.; Lindesmith, L.; Pendu, B.J.L.; Calderon, R.L. Norwalk virus: How infectious is it? J. Med. Virol. 2008, 80, 1468–1476. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Jiang, X. Norovirus and its histo-blood group antigen receptors: An answer to a historical puzzle. Trends Microbiol. 2005, 13, 285–293. [Google Scholar] [CrossRef]
- Atmar, R.L.; Ramani, S.; Estes, M.K. Human noroviruses: Recent advances in a 50-year history. Curr. Opin. Infect. Dis. 2018, 31, 422–432. [Google Scholar] [CrossRef]
- Razafimahefa, R.M.; Ludwig-Begall, L.F.; Thiry, E.E. Cockles and mussels, alive, alive, oh—The role of bivalve molluscs as transmission vehicles for human norovirus infections. Transbound Emerg. Dis. 2020, 67, 9–25. [Google Scholar] [CrossRef] [PubMed]
- McLeod, C.; Polo, D.; Le Saux, J.-C.; Le Guyader, F.S. Depuration and Relaying: A Review on Potential Removal of Norovirus from Oysters. Compr. Rev. Food Sci. Food Saf. 2017, 16, 692–706. [Google Scholar] [CrossRef]
- Esseili, M.A.; Gao, X.; Boley, P.; Hou, Y.; Saif, L.J.; Brewer-Jensen, P.; Lindesmith, L.C.; Baric, R.S.; Atmar, R.L.; Wang, Q. Human Norovirus Histo-Blood Group Antigen (HBGA) Binding Sites Mediate the Virus Specific Interactions with Lettuce Carbohydrates. Viruses 2019, 11, 833. [Google Scholar] [CrossRef]
- Huang, P.; Farkas, T.; Marionneau, S.; Zhong, W.; Ruvoën-Clouet, N.; Morrow, A.L.; Altaye, M.; Pickering, L.K.; Newburg, D.S.; LePendu, J.; et al. Noroviruses bind to human ABO, lewis, and secretor Histo-Blood group antigens: Identification of 4 distinct Strain-Specific patterns. J. Infect. Dis. 2003, 188, 19–31. [Google Scholar] [CrossRef]
- Le Guyader, F.S.; Loisy, F.; Atmar, R.L.; Hutson, A.M.; Estes, M.K.; Ruvoën-Clouet, N.; Pommepuy, M.; Le Pendu, J. Norwalk Virus–specific Binding to Oyster Digestive Tissues. Emerg. Infect. Dis. J. 2006, 12, 931. [Google Scholar] [CrossRef]
- Tian, P.; Bates, A.; Jensen, H.; Mandrell, R. Norovirus binds to blood group A-like antigens in oyster gastrointestinal cells. Lett. Appl. Microbiol. 2006, 43, 645–651. [Google Scholar] [CrossRef]
- Ruiz-Palacios, G.M.; Cervantes, L.E.; Ramos, P.; Chavez-Munguia, B.; Newburg, D.S. Campylobacter jejuni binds intestinal H(O) antigen (Fucα1, 2Galβ1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 2003, 278, 14112–14120. [Google Scholar] [CrossRef]
- Rossez, Y.; Maes, E.; Darroman, T.L.; Gosset, P.; Ecobichon, C.; Curt, M.J.C.; Boneca, I.G.; Michalski, J.-C.; Robbe-Masselot, C. Almost all human gastric mucin O-glycans harbor blood group a, B or H antigens and are potential binding sites for Helicobacter pylori. Glycobiology 2012, 22, 1193–1206. [Google Scholar] [CrossRef]
- Lomas-Francis, C.; Westhoff, C.M. Red cell antigens and antibodies. Hematol./Oncol. Clin. N. Am. 2022, 36, 283–291. [Google Scholar] [CrossRef]
- Cooling, L. Blood Groups in Infection and Host Susceptibility. Clin. Microbiol. Rev. 2015, 28, 801–870. [Google Scholar] [CrossRef]
- Nordgren, J.; Svensson, L. Genetic susceptibility to human norovirus infection: An update. Viruses 2019, 11, 226. [Google Scholar] [CrossRef]
- Huang, P.; Farkas, T.; Zhong, W.; Tan, M.; Thornton, S.; Morrow, A.L.; Jiang, X. Norovirus and Histo-Blood group antigens: Demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J. Virol. 2005, 79, 6714–6722. [Google Scholar] [CrossRef]
- Feng, C.; Ghosh, A.; Amin, M.N.; Giomarelli, B.; Shridhar, S.; Banerjee, A.; Fernández-Robledo, J.A.; Bianchet, M.; Wang, L.-X.; Wilson, I.; et al. The Galectin CvGal1 from the Eastern Oyster (Crassostrea virginica) Binds to Blood Group a Oligosaccharides on the Hemocyte Surface. J. Biol. Chem. 2013, 288, 24394–24409. [Google Scholar] [CrossRef]
- Pu, F.; Yang, B.; Ke, C. Characterization of reference genes for qPCR analysis in various tissues of the Fujian oyster Crassostrea angulate. Chin. J. Oceanol. Limnol. 2015, 33, 838–845. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Liu, X.-W.; Peng, P.; Yi, W.; Chen, X.; Wang, F.; Cao, H. Diversity-Oriented Enzymatic Modular Assembly of ABO Histo-blood Group Antigens. ACS Catal. 2016, 6, 8140–8144. [Google Scholar] [CrossRef]
- Paulson, J.C.; Colley, K.J. Glycosyltransferases: Structure, localization, and control of cell type-specific glycosylation. J. Biol. Chem. 1989, 264, 17615–17618. [Google Scholar] [CrossRef] [PubMed]
- de Vries, T.; Storm, J.; Rotteveel, F.; Verdonk, G.; van Duin, M.; van den Eijnden, D.H.; Joziasse, D.H.; Bunschoten, H. Production of soluble human α3-fucosyltransferase (FucT VII) by membrane targeting and in vivo proteolysis. Glycobiology 2001, 11, 711–717. [Google Scholar] [CrossRef][Green Version]
- El-Battari, A.; Prorok, M.; Angata, K.; Mathieu, S.; Zerfaoui, M.; Ong, E.; Suzuki, M.; Lombardo, D.; Fukuda, M. Different glycosyltransferases are differentially processed for secretion, dimerization, and autoglycosylation. Glycobiology 2003, 13, 941–953. [Google Scholar] [CrossRef]
- Cicéron, F.; Rocha, J.; Kousar, S.; Hansen, S.F.; Chazalet, V.; Gillon, E.; Breton, C.; Lerouxel, O. Expression, purification and biochemical characterization of AtFUT1, a xyloglucan-specific fucosyltransferase from Arabidopsis thaliana. Biochimie 2016, 128–129, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Becker, D.J.; Lowe, J.B. Fucose: Biosynthesis and biological function in mammals. Glycobiology 2003, 13, 41R–53R. [Google Scholar] [CrossRef]
- Castillo, B.; Dasgupta, A.; Klein, K.; Tint, H.; Wahed, A. Chapter 5—Red cell antigens and antibody. In Transfusion Medicine for Pathologists; Elsevier: Amsterdam, The Netherlands, 2018; pp. 69–112. [Google Scholar]
- Wang, D.; Wu, Q.; Kou, X.; Yao, L.; Zhang, J. Distribution of norovirus in oyster tissues. J. Appl. Microbiol. 2008, 105, 1966–1972. [Google Scholar] [CrossRef]
- Münster, J.; Ziegelmüller, P.; Spillner, E.; Bredehorst, R. High level expression of monomeric and dimeric human α1,3-fucosyltransferase V. J. Biotechnol. 2006, 121, 448–457. [Google Scholar] [CrossRef]
- Christensen, L.L.; Jensen, U.B.; Bross, P.; Ørntoft, T.F. The C-terminal N-glycosylation sites of the human α1,3/4-fucosyltransferase III, -V, and -VI (hFucTIII, -V and -VI) are necessary for the expression of full enzyme activity. Glycobiology 2000, 10, 931–939. [Google Scholar] [CrossRef]
- Li, E.; Gibson, R.; Kornfeld, S. Structure of an unusual complex-type oligosaccharide isolated from Chinese hamster ovary cells. Arch. Biochem. Biophys. 1980, 199, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Prieto, P.A.; Larsen, R.D.; Cho, M.; Rivera, H.N.; Shilatifard, A.; Lowe, J.B.; Cummings, R.D.; Smith, D.F. Expression of human h-type α1,2-Fucosyltransferase encoding for blood group H(O) antigen in chinese hamster ovary cells: Evidence for preferential fucosylation and truncation of polylactosamine sequences. J. Biol. Chem. 1997, 272, 2089–2097. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Engelbrektson, A.L.; Jiang, X.; Zhong, W.; Mandrell, R.E. Norovirus recognizes Histo-Blood group antigens on gastrointestinal cells of clams, mussels, and oysters: A possible mechanism of bioaccumulation. J. Food Prot. 2007, 70, 2140–2147. [Google Scholar] [CrossRef] [PubMed]
- Almand, E.A.; Moore, M.D.; Outlaw, J.; Jaykus, L.-A. Human norovirus binding to select bacteria representative of the human gut microbiota. PLoS ONE 2017, 12, e0173124. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.; Hanisch, F.-G.; Wegner, K.M.; Schroten, H. Pandemic GII.4 sydney and epidemic GII.17 kawasaki308 noroviruses display distinct specificities for Histo-Blood group antigens leading to different transmission vector dynamics in pacific oysters. Front. Microbiol. 2018, 9, 2826. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Sano, D.; Suenaga, A.; Yoshimura, T.; Fuzawa, M.; Nakagomi, T.; Nakagomi, O.; Okabe, S. Histo-Blood group Antigen-Like substances of human enteric bacteria as specific adsorbents for human noroviruses. J. Virol. 2013, 87, 9441–9451. [Google Scholar] [CrossRef]
- Li, D.; Breiman, A.; Le Pendu, J.; Uyttendaele, M. Binding to histo-blood group antigen-expressing bacteria protects human norovirus from acute heat stress. Front. Microbiol. 2015, 6, 659. [Google Scholar] [CrossRef]




| Primer | Nucleotide Sequence (5′-3′) | Purpose | Efficiency | Length (bp) |
|---|---|---|---|---|
| FT1A | AAGCACAGCAATGGTTCC | Cloning partial sequence | – | 899 |
| RT1A | CCGTATTGTGTTGTTTCCC | |||
| RRT1-A | GATTACGCCAAGCTTTGGGGGTTTGGGTTCCAGTCTCAT | RACE PCR | – | – |
| FRT1-A | GATTACGCCAAGCTTTCCTACTCTTGGTGGGCGGGGTA | |||
| Q-FT1-A | CGATGGAACCATGAGACTGGAACC | RT-qPCR | 99.0% | 141 |
| Q-RT1-A | CTCCTGGCTGCTCCTATCAGAGTAG | |||
| F-TUB | GAGGGTGCTGAACTGGTGG | 99.3% | 281 | |
| R-TUB | CAGAAGGTTTCGTCGGTGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gui, B.; Yao, L.; Qu, M.; Zhang, W.; Li, M.; Jiang, Y.; Wang, L. Cloning, Expression, and Functional Characterization of FUT1, a Key Gene for Histo-Blood Group Antigens Synthesis in Crassostrea gigas. Curr. Issues Mol. Biol. 2023, 45, 4200-4213. https://doi.org/10.3390/cimb45050267
Gui B, Yao L, Qu M, Zhang W, Li M, Jiang Y, Wang L. Cloning, Expression, and Functional Characterization of FUT1, a Key Gene for Histo-Blood Group Antigens Synthesis in Crassostrea gigas. Current Issues in Molecular Biology. 2023; 45(5):4200-4213. https://doi.org/10.3390/cimb45050267
Chicago/Turabian StyleGui, Binbin, Lin Yao, Meng Qu, Weiran Zhang, Mingyu Li, Yanhua Jiang, and Lianzhu Wang. 2023. "Cloning, Expression, and Functional Characterization of FUT1, a Key Gene for Histo-Blood Group Antigens Synthesis in Crassostrea gigas" Current Issues in Molecular Biology 45, no. 5: 4200-4213. https://doi.org/10.3390/cimb45050267
APA StyleGui, B., Yao, L., Qu, M., Zhang, W., Li, M., Jiang, Y., & Wang, L. (2023). Cloning, Expression, and Functional Characterization of FUT1, a Key Gene for Histo-Blood Group Antigens Synthesis in Crassostrea gigas. Current Issues in Molecular Biology, 45(5), 4200-4213. https://doi.org/10.3390/cimb45050267




