Abstract
Chronic obstructive pulmonary disease (COPD) is the third leading cause of death worldwide; the main risk factors associated with the suffering are tobacco smoking (TS) and chronic exposure to biomass-burning smoke (BBS). Different biological pathways have been associated with COPD, especially xenobiotic or drug metabolism enzymes. This research aims to identify single nucleotide polymorphisms (SNPs) profiles associated with COPD from two expositional sources: tobacco smoking and BBS. One thousand-five hundred Mexican mestizo subjects were included in the study and divided into those exposed to biomass-burning smoke and smokers. Genome-wide exome genotyping was carried out using Infinium Exome-24 kit arrays v. 1.2. Data quality control was conducted using PLINK 1.07. For clinical and demographic data analysis, Rstudio was used. Eight SNPs were found associated with COPD secondary to TS and seven SNPs were conserved when data were analyzed by genotype. When haplotype analyses were carried out, five blocks were predicted. In COPD secondary to BBS, 24 SNPs in MGST3 and CYP family genes were associated. Seven blocks of haplotypes were associated with COPD-BBS. SNPs in the ARNT2 and CYP46A1 genes are associated with COPD secondary to TS, while in the BBS comparison, SNPs in CYP2C8, CYP2C9, MGST3, and MGST1 genes were associated with increased COPD risk.
1. Introduction
Chronic obstructive pulmonary disease (COPD) is a complex and multifactorial disease; preventable, treatable, and partially reversible, characterized by airflow limitation due to an airway inflammatory process in response to chronic exposure to noxious particles [1,2,3]. Worldwide, COPD is the third leading cause of death, with a prevalence of 251 million people and nearly 3000 million deaths. Of these deaths, 90% were recorded in low- and middle-income countries [3,4].
COPD is associated with environmental exposure risk factors, such as tobacco smoking or biomass-burning smoke exposure (BBS). Other factors are clinical and genetic characteristics that could lead to different phenotypes of COPD [1,2].
The main clinical features of COPD are dyspnea, pulmonary hypertension, hypoxemia, hypercapnia, chest tightness, wheezing, cough, phlegm (sputum), mucus (bronchitis), and low oxygen saturation, among others. [2,5] It has been described that the clinical phenotype can vary depending on the exposure factor. For example, patients with COPD secondary to BBS (COPD-BBS) present higher FEV1 values than smokers with COPD (COPD-S) [6,7]; the predominant phenotype in COPD-BBS is bronchitis, with increased production of mucus and phlegm [8]. In addition, the CAT (COPD assessment test) score indicates that patients with COPD-BBS have a smaller decrease in their quality of life, evidenced by less difficulty when carrying out their daily activities and better physical performance (6-min walk test) [9]. These phenotypic differences in the disease may be due to the heterogeneity of pollutants substances’ content and individuals’ genetic variability.
COPD research has dabbled in the genetics of the disease. So far, it is known that the genetic deficiency of α1-antitrypsin, encoded by the SERPINA1 gene, caused by a single nucleotide polymorphism (SNPs), is the only genetic factor that predisposes to COPD, regardless of environmental risk factors [10]; however, it does not explain the whole variety of the disease in the world population.
Genome-wide association studies (GWAS), in which thousands of genetic variants are included in case-control comparisons and cohort models, have helped identify SNPs’ association with the disease’s clinical characteristics, such as tobacco smoking, cachexia, and decreased lung function [11,12,13]. Some associated SNPs are present in non-coding areas, making it challenging to explain their biological impact on the disease.
Another strategy used in genetic association studies is the analysis of variants in candidate genes by gene functionality or their relationship with other genes within the same interaction network [14,15]. Some reported genes include TNF, CCL2, SERPINA12, SERPINE2, FAM13A, and TFGB1; this has allowed the description of associations between SNPs and the different clinical phenotypes of the disease (emphysema or bronchitis) [16,17]. It is essential to mention that these studies have been carried out in groups of smokers, which has left aside the evaluation of patients with COPD-BBS.
This work aims to describe the association of SNPs in candidate genes related to the processing of xenobiotic, cytotoxic, and drugs with COPD, both secondary to tobacco smoking and BBS, and possible SNPs profiles that can differentiate them.
2. Materials and Methods
2.1. Population Included
For this study, 1500 subjects were included and divided into 2 comparison groups; the first was composed of 900 smokers: 300 patients with COPD secondary to tobacco smoking (COPD-S) and 600 smokers without the disease (SWOC). The second comparison group included 600 subjects exposed to smoke from biomass burning, divided into 220 patients with COPD secondary to BBS (COPD-BBS) and 380 subjects exposed to BBS but without COPD (BBES).
Mexican mestizo subjects over 40 years of age and of indistinct sex were included; for the comparison group of smokers, participants with a tobacco index (TI) > 5 packs/year and no history of exposure to BBS were included. In the exposed to BBS group, subjects with an exposure index to BBS (BEI) >100 h/year were included and were never smokers. COPD patients were defined when the post-bronchodilator FEV1/FVC ratio was <70% (Supplementary Figure S1). Participants with other inflammatory, autoimmune, or respiratory diseases were eliminated. The participants were recruited from the COPD Clinic, smoking cessation support groups of the Tobacco Smoking and COPD Research Department, from COPD early detection campaigns in rural Oaxaca [18], and from suburban areas of Mexico City. The Ethics in Research Committee from Instituto Nacional de Enfermedades Respiratorias Ismael Cosio Villegas approved the protocol under code numbers B14-17, B11-19, and C53-19.
The clinical evaluation of the patients was carried out by specialized chest physicians from the Instituto Nacional de Enfermedades Respiratorias Ismael Cosio Villegas using GOLD guidelines. Ref. [19] Demographic and ancestry data were obtained through a questionnaire. Before taking biological samples, all patients signed an informed consent approved by institutional ethical boards.
2.2. Biological Samples
All participants took a blood sample by forearm puncture, and DNA and plasma were obtained using the previously described methodology [20]. The DNA samples were quantified spectrophotometry through a nanodrop device (Thermo Scientific, Wilmington, DE, USA), and samples with a 260/280 ratio between 1.8 and 2.2 were selected, adjusted to 60 ng/µL, and their integrity was evaluated in 1.5% agarose gels.
2.3. Whole Exome Genotyping
After sample quality control, the groups were composed of 370 smokers (COPD-S = 150, SWOC = 220) and 401 subjects exposed to BBS (COPD-BBS = 101, BBES = 300) (Supplementary Figure S2). The samples were genotyped using the Illumina Infinium Exome-24 Kit arrays v1.2 (5200 Illumina Way, San Diego, CA 92122, USA) with a genotyping capacity of up to 560,000 variants.
We applied functional candidate gene methodology to select only genes related to the metabolism of xenobiotics, cytotoxic products, and drug metabolism. Through bibliography research, we selected 38 genes. After applying the Hardy–Weinberg (p > 1 × 10−9 test and excluding SNPs with MAF < 10%, we chose all SNPs in the proposed genes. We worked with 748 SNPs in both comparison groups (Supplementary Figure S3).
2.4. Data Analysis
PLINK v. 1.07 [21] was used for data quality control (QC). We considered a genotype call rate > 95% and eliminated subjects with >0.05 of missing genotypes; sex discrepancies were considered by X chromosome homozygosity (men > 0.8, women < 0.2), while relatedness was assessed by identity by descent (IBD) considering pi-hat values < 0.25.
Association analysis was carried out using PLINK v. 1.07 applying Fisher’s exact test adjusted by covariates; in the smoker group comparison, we included sex, age, and cigarettes/day (TI) as covariates, while in the biomass group, comparison, age, and biomass burning-smoke exposure index (BEI) was included, utilizing the Bonferroni correction test.
The R language [22] and the Rstudio interface [23] were employed for statistical analysis. Admixture and principal component analysis (PCA) were carried out using packages SNPRelate and gdsfmt from Bioconductor. We included Hapmap population data from Northern Europeans from Utah (CEU), Yoruba in Ibadan from Nigeria (YRI), and native Amerindian populations (AMR) described by Huerta-Chagoya, and we selected 32 ancestry informative markers (AIMs) and used k = 3 [24]. The distribution of demographic variables, exposure data, or lung function was analyzed to determine the type of statistical comparison being made.
2.5. Severity Analysis
Afterward, we stratified COPD patients based on the GOLD states, comparing mild (GOLD 1 + 2) vs. severe forms (GOLD 3 + 4) of the illness to avoid bias by subgrouping. This analysis was carried out for COPD-S and COPD-BBS individually using PLINK v. 1.07 and applying Fisher’s exact test, correcting by covariates, and the Bonferroni multiple testing.
2.6. Multiple Correspondence Analysis
We applied multiple correspondence analysis (MCA) to determine possible grouping between SNPs associated, exposure indexes, and/or FEV1 values. These analyses were carried out using Rstudio [22] and the packages FactoMineR [25] and factoextra [26].
2.7. Calculation of Haplotype Blocks
We included 750 SNPs in the Haplotype blocks analysis. This analysis was carried out using Haploview 4.2 software, [27] applying the analysis algorithm presented by Gabriel et al. [28]. We applied a window of inclusion of 5000 Kb per pair of SNPs. Linkage disequilibrium (LD) was presented using D’ value. Haplotypes association analysis was carried out using R through Fisher’s exact test between cases vs. controls and adjustment by logistic regression, including covariates. Genes’ schemes and SNPs’ positions are included in the Supplementary Material (Supplementary Figures S7 and S8).
3. Results
3.1. Population Studied
After quality control, 745 subjects were included; 354 were in the group of smokers (COPD-S = 141, SWOC = 213) and 391 had been exposed to BBS (COPD-BBS = 98, BBES = 293). The distribution of the variables presented a non-normal distribution, so the demographic, clinical, and exposure variables are presented as a function of the median and quartiles 1 and 3. At the same time, the comparisons were made using the Mann–Whitney U test and χ2 for qualitative variables.
When comparing COPD-S vs. SWOC, significant differences (p < 0.05) were found in the male–female ratio, age, BMI, and TI; because of this, sex, age, and TI were selected for covariate correction. In the BBS comparison group (COPD-BBS vs. BBES), significant differences (p < 0.05) were found in age and BBS exposure index (BEI), so these were included as covariates in the association analysis of this group (Table 1).

Table 1.
Demographic and lung function data of COPD and control subjects.
By ancestry analysis, we found different proportions for both groups of comparison. We found a highly conserved Amerindian composition in the biomass-burning comparison group (COPD_BBS, BBES), while in the smokers’ comparison (COPD_S, SWOC), we found a heterogeneous composition, predominantly Amerindian and Caucasian (Figure 1).
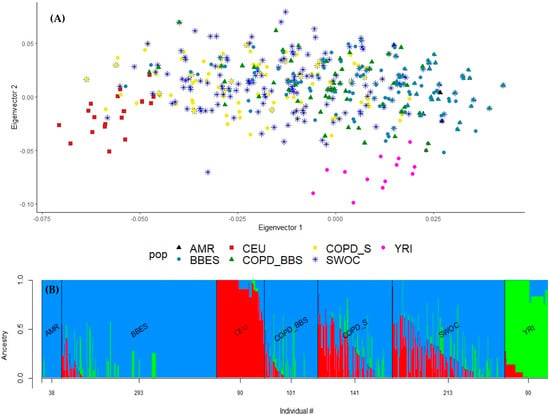
Figure 1.
(A) PCA for the ancestral composition of the included population. We included Hapmap population data: northern Europeans from Utah (CEU), Yoruba in Ibadan from Nigeria (YRI), and native Amerindian (AMR). COPD_BBS: COPD patients exposed to biomass-burning smoke; COPD_S: COPD patients as smokers; SWOC: Smokers without COPD; COPD_BBS: COPD secondary to biomass-burning smoke exposition; BBES: Biomass burning exposed subject. (B) Admixture plot for ancestry composition in subjects included. We included the following Hapmap population data: Northern Europeans from Utah (CEU, in red), Yoruba in Ibadan from Nigeria (YRI, in green), and native Amerindian (AMR, in blue).
3.2. Association Analysis in the Group of Smokers
All the SNPs associated with this stage met the Hardy–Weinberg equilibrium and MAF > 10% (Supplementary Table S1). In the comparison of smokers (COPD-S vs. SWOC), after correction for covariates, 8 SNPs/alleles were found associated (p < 0.05) with COPD secondary to TS, 6 SNPs (rs11572191, rs8133, rs17497857, rs4964059, rs3901896, rs8041826) associated with increased risk (OR > 1.0), and 2 SNPs (rs4147611, rs3742377) with decreased risk (OR < 1.0). Of the SNPs associated with risk, rs11572191 in the CYP2J2 gene presented the highest OR value, with an almost three-fold increased risk of developing COPD secondary to smoking. On the other hand, the ARNT2 and ARNTL2 genes each presented two SNPs associated with increased risk, these being the genes with the highest number of associated SNPs in this comparison group. However, when we applied the Bonferroni correction, no significant associations were retained (Table 2).

Table 2.
Allele association analysis in smokers’ comparison.
Seven SNPs associated with an increased risk of COPD secondary to TS were found in the genotype analysis. Of these SNPs, rs11572191 in CYP2J2, rs17497857 in ARNTL2, rs3901896, and rs8041826 in ARNT2 remained associated. On the other hand, the rs1951576 and rs943881 in CYP46A1 and rs6488842 in MGST1 are new findings by this model analysis. Interestingly, rs11572191 and rs17497857 are associated with heterozygous genotypes (Table 3).

Table 3.
Genotype association analysis in smokers’ comparison.
We extracted the data of COPD-S and COPD-BBS, looking for possible differentiation patterns, including SNPs with MAF > 1% by MCA. Even though we have differential grouping patterns, the variance did not surpass >1% (Supplementary Figure S4A).
We included 336 SNPs for the MCA in smokers’ comparison. By biplots, we did not find any cluster of SNPs that could explain variance >1% (Supplementary Figure S4C). Next, we included all the SNPs associated with the allele analysis but did not get any possible component (Supplementary Figure S5).
The possible participation of other SNPs in the genetic susceptibility was assessed through haplotype blocks, including all associated SNPs, before correction for covariates to maximize the analysis screen. Five blocks of haplotypes were found to form in the ARNTL2 gene, CYP19A1, ARNT2, CYP46A2, and MGST3, all with LD > 85 (Figure 2).
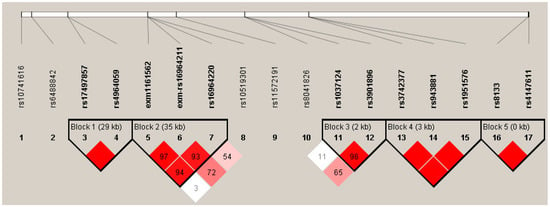
Figure 2.
Haplotype block for SNPs associated in the smokers’ comparison. Five blocks with LD > 80 were predicted, block 1 is conformed of two SNPs in ARNTL2, block 2 by 3 SNPs in CYP19A1, block 3 by SNPs in ARNT2, block 4 with 3 SNPs in CYP46A1, and block 5 by 3 SNPs in MGST3. Color and values in the haplotypes represent D’ LD values. The color intensity corresponds to the higher LD value.
When the association of haplotypes was carried out, we found nine different combinations of SNPs associated with COPD-S in the genes: ARNTL (rs10741616-rs7126796), ARNTL2 (rs11048977-rs1037924-rs17497857-rs7138982), CYP19A1 (rs10046-rs700519-rs6493489-rs2899472-rs2414095-rs700518), ARNT2 (rs1374213-rs3901896-rs7168908-rs2278709), CYP46A1 (rs3742377-rs943881-rs1951576-rs12435918-rs2146238), ARNT (rs10847-rs11552229-rs2228099), and MGST3 (rs8133-rs4147611) (Table 4). Of these combinations, six haplotypes were associated with a lower risk of COPD-S and three to higher risk (OR > 1.5). We found five haplotypes containing SNPs previously associated in the allele or genotype analysis: rs3901896 in ARNT2, rs1951576 in CYP46A1, and rs17497857 in ARNTL2, also rs8133 and rs4147611 in MGST3.

Table 4.
Haplotypes association analysis in smokers’ comparison.
3.3. Severity Analysis
We stratify COPD-S and COPD-BBS subjects according to the GOLD stages (mild stages: GOLD I + II; severe stages: GOLD III + IV). In COPD-S, we found four SNPs: rs12435918 in CYP46A1, rs625456 in GSTM2, and rs1058930 in CYP2C8 associated with severe forms of COPD secondary to tobacco smoking (Supplementary Table S8). For COPD-BBS, we found rs12300289 in ARNTL2, rs10847 in ARNT, and rs2234696 in GSTM3 associated with the severe form of COPD secondary to biomass-burning smoke exposition (Supplementary Table S9).
3.4. Association Analysis in the Group Exposed to BBS
In the BBS exposure comparison group (COPD-BBS vs. BBES), 24 SNPs were found to be significantly associated (p < 0.05), of which twenty were associated with a higher risk of COPD and four with a decreased risk of suffering from the disease. Interestingly, the associated polymorphisms are mainly distributed in the MGST3, MGST1, CYP2C8, and CYP2C9 genes (Table 5). After applying the Bonferroni correction test, only three SNPs remained associated, rs11799886/MGST3 (p = 0.019), rs1856908/CYP2C9 (p = 0.003), and rs1934953/CYP2C8 (p = 0.021).

Table 5.
Allele association analysis in exposed biomass-burning smoke comparison.
When performing the genotype analysis, 23 SNPs associated with the disease were found; three with reduced risk and twenty with a higher COPD risk. In six SNPs, no homozygotes were found for the minor allele and the leading associations were with the heterozygous genotypes. It should be noted that the groups of SNPs in the MGST3, CYP2C8, CYP2C9, and MGST1 genes remained associated. MGST3 presented the highest number of associated SNPs and OR values, presenting a three-fold increased risk of developing the disease. In the case of CYP2C8, although only three SNPs were found to be associated with increased risk, their OR values were also up to four times higher risk of developing COPD secondary to BBS (Table 6).

Table 6.
Genotype association analysis in the exposed to biomass-burning smoke.
For BBS comparison, we included 298 SNPs after filtering by MAF (>1%). We did not find any clusters with more than 2% of the variance (Supplementary Figure S4B). Looking for other clustering patterns, we included only the SNPs associated with COPD-BBS, but no grouping patterns that could explain higher variability were found (<1%) (Supplementary Figure S6).
We found seven blocks of haplotypes in high LD in the genes ARNTL, CYP2R1, MGST1, ARNTL2, GSTP1, CYP1A2, ARNT2, CYP2C18, CYP2C9, CYP2C8, GSTM5, GSTM3, and MGST3 (Figure 3). In block 4, we found the rs1856908 reported in allele and genotype analysis. A haplotype block (block 7) was found in MGST3; this block was found in the smokers’ comparison (rs8133-rs4147611).
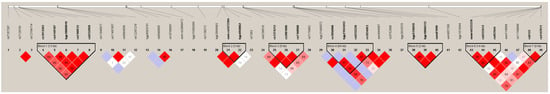
Figure 3.
Haplotypes blocks include all the SNPs associated with BBS comparison. Seven blocks were identified in the biomass comparison. Color and values in the diamonds represent D’ LD values. The intensity of the color is directly proportional to the higher LD value.
Eight combinations of SNPs were associated with a lower risk of suffering COPD (OR < 1) and eighteen were associated with a higher risk (OR > 1.5). The larger SNP combination was composed of 15 variants that range from CYP2C18 to CYP2C9, with the highest OR value at almost eight times higher risk of COPD. MGST3 was the gene with more blocks; we found three haplotypes, and the SNPs included in the haplotypes had been previously reported in alleles and genotypes analyses (Table 7).

Table 7.
Haplotypes association analysis in BBS comparison.
In the analysis by severity, we found four SNPs associated, three with the severe GOLD stages and one with mild COPD stages. When data were corrected by covariates, three out of four SNPs remained associated. However, no SNP retained its association after Bonferroni correction. (Supplementary Table S8). We found five SNPs in the severity analysis of the COPD-BBS group, four associated with a severe form of COPD and one with a mild form of the illness. Although four remained associated after the correction by covariates, no SNP conserves association after Bonferroni adjustment (Supplementary Table S9).
4. Discussion
Although multiple GWAS have described associations with COPD, most studies focus on COPD secondary to tobacco smoking in Caucasian populations from Europe and the USA; we analyzed SNPs in exome regions in the whole human genome by the array genotyping technology looking for variants associated with COPD both secondary to tobacco smoking or biomass-burning smoke in the Mexican mestizo population. The participants were recruited from different campaigns of COPD early detection in Mexico City and the highlands of Oaxaca.
In our group, Perez-Rubio et al. had previously described the genetic component of the population included in this study, demonstrating the contribution of the Amerindian/Caucasian genetic component [29]. All patients had at least three prior generations born in Mexico (parents and grandparents) and were considered Mexican mestizos. We have previously demonstrated that this criterion is a good proxy of Mexican ancestry evaluated by ancestry-informative markers [30].
We found differences in variables, such as sex, age, BMI, and tobacco index, in comparing smokers. Due to these differences, we included these covariates in the association of alleles and haplotypes analyses to avoid false positive findings. For the BBS group, we found differences in age and exposure data. We did not find differences in the men/women ratio, but women are predominantly represented in both groups. Low- to middle-income countries are the principal users of biomass, and each region worldwide reported the use of specific kind of biomass; for example, in China, there is a predominance in the use of charcoal and coal; in Nepal and Kenya, the use of manure from big ruminants is a common practice; in a large variety of Latin America, African and South Asian countries is predominant the use of firewood from a great variety of trees and even agriculture waste [18]. The primary biomass fuel used in Mexico is firewood or mixtures of firewood, manure, and farming waste, especially in rural or suburban areas. The principal population exposed are women and children because women are the principal family members in charge of cooking [18,31].
Rehfuess and collaborators establish that 52% of the world population uses either biomass or solid fuel. Stratifying six geographic areas, they determined that Africa, South Asia, and different areas of Latin America are the principal biomass users [32].
The World Health Organization reported that around 2.5 billion people used any biomass only to cook, and, especially in rural zones, combustion takes place indoors, in closed or poorly ventilated places using improvised stoves or pipes, resulting in an event called “indoor pollution”, affecting mainly women and children, and producing 1.3 million of premature deaths associated with respiratory diseases and infections [31,33].
Candidate genes analysis methodologies are strategies for post-genotyping data in genome-wide studies (GWAS) [34]. In this study, we used genotyping exome array that includes up to 560 thousand specific sequence probes capable of detecting the SNPs in exome regions. We included genes whose biological function was related to xenobiotic and drug metabolism processing.
Xenobiotics are exogenous bodily substances that involve absorption, distribution, and metabolism [35]. Some genes related to the xenobiotic processing are genes of the glutathione transferase family (phase 2 metabolizers) [36], cytochrome P450 (phase 1 metabolizing isoenzymes) [37], aryl hydro-carbon receptors and translocators [38], and ADRB1 genes (β1 adrenergic receptors) [39].
The genes included were CYP4B1, CYP4Z2P, CYP4A11, CYP4 × 1, CYP4Z1, CYP4A22, CYP2J2, CYP26C1, CYP26A1, CYP2C18, CYP2C19, CYP2C9, CYP2C8, CYP17A1, CYP2E1, CYP2R1, CYP27B1, ACYP1, CYP46A1, CYP19A1, CYP11A1, CYP1A1, CYP1A2, GSTM4, GSTM3, GSTM2, GSTM1, MGST3, GSTO1, GSTO2, GSTP1, MGST1, GSTZ1, ADRB1, ARNT2, ARNT, ARNTL2, and ARNTL, and a total of 750 SNPs were selected.
In smokers’ comparison, we found eight SNPs associated with COPD; six SNPs were associated with a higher risk of suffering COPD-S; rs11572191 in CYP2J2; rs8133 in MGST3; rs17497857 and rs4964059 in ARNTL2; and rs3901896 and rs8041826 in ARNT2, all with the minor allele. Only two SNPs were found associated with lower risk; rs4147611 in MGST3 and rs3742377 in CYP46A1.
Our is the first study reporting these sets of SNPs with COPD-S, particularly in a mestizo (admixed) population as the Mexican. Although any polymorphism in our findings was previously described, the genes associated are reported in different studies as associated factors to lung diseases. Four SNPs in CYP2J2 were found to be associated with the Chinese Han population with COPD-S [40], and even in the Russian population, SNPs in CYP2J2 are associated with bronchitis secondary to smoking [41]. Other investigations have demonstrated that SNPs in CYP2J2 could be involved in lung ischemia and reperfusion injury, especially in smokers [42,43]. Although our investigation focuses on COPD, lung injury and hypertension are common in subjects with COPD. Additionally, CYP2J2 is related to asthma models and cancer. Refs. [44,45,46] CYP2C9 has been included in studies related to adenocarcinoma and other forms of lung cancer [45,47,48].
Even though there are few reports of MGST3 and COPD, some SNPs have been associated with attenuating smokers’ accelerated decline in FEV1/FVC [49]. No other reports of lung diseases have been reported.
ARNT genes encode proteins capable of binding to aryl hydro-carbon receptors to translocate them to the cell nucleus as transcription factors related to gene promoters such as HIF1α. The principal studies between ARNT genes suggest a possible relation with small-cell cancer [50,51,52,53].
The protein encoded by MGST1 (microsomal glutathione S-transferase 1) is a membrane-associated protein with peroxidase activity which avoids lipid damage against reactive oxygen species (ROS), cytotoxic, and drugs. The principal association between MGST1 and lung diseases includes different types of cancer, such as adenocarcinoma or non-small cell lung cancer [54,55]. Woldhuis et al. proposed that microsomal glutathione S-transferase 1 could be related to cell senescence and extracellular matrix reorganization [56]. Recently, ferroptosis has been described as a programmed dead type with a higher lipid peroxide concentration in other illnesses. MGST1 is differentially expressed in alveolar type 2 cells [57]. In the case of MGST3, sets of polymorphism attenuated lung function decline in European-American smokers [49].
In genotypes, only higher risk associated SNPs were found associated with the illness; among these, four were previously described in allele analysis; rs11572191, rs17497857, rs3901896, and rs8041826. Three more SNPs were found in genotype analysis, the GG of rs1951576 and CC genotype of rs943881 in gene CYP46A1 and TT of rs6488842 in gene MGST1. The alleles associated with a low risk of COPD were possibly not found in the genotype phase due to the low frequency of minor alleles; not enough homozygous minor allele genotypes were found.
There is limited information regarding the severity data related to the SNPs and genes associated with xenobiotic metabolism. Studies in emphysema have demonstrated that the expression of GSTM3 was upregulated in mild illness [58], while other studies describe SNPs associated with a lower FEV1/FVC ratio [59]. GSTM3 is a gene in which protein product is related to eliminating electrophilic compounds and carcinogens. We found rs2234696 in GSTM3 to be associated with the severe form of COPD in smokers, and while there are no reports regarding the SNP, we can state that the SNP could affect the structure of the protein codified by the gene, thus preventing its biological function.
Haplotypes analysis is used to elucidate possible associations in groups of SNPs in different regions of genes [60]. For the comparison with smokers, we found five blocks with high LD (>85) in ARNTL1, CYP19A1, ARNT2, CYP46A1, and MGST3. Multiple genes have been associated with complex diseases like COPD but with moderate OR [61]. Including multiple analyses as polygenic risk scores has demonstrated that a combination of genetic variants could explain the multiples association and even reach the haplotype analysis [62]. In the haplotype blocks, we found five combinations of SNPs associated with allele analysis, suggesting a probably critical role in COPD pathophysiology.
We found more SNPs associated with COPD-BBS than COPD-S; at the allele level, the principal findings include SNPs in MGST3, CYP2C8, and MGST1. Few studies have been made in the genetic field about COPD-BBS. Our current study is the first in exome-wide genotyping.
Principal studies with COPD-BBS in Latin America emphasize clinical description; other studies include Chinese and Chilean populations but focus on genes such as PRDM15 and CXCL10, respectively [63,64]. Additionally, we found a greater number of SNPs in COPD-BBS than in COPD-S, which could suggest a possible major complex in developing COPD-BBS.
In haplotype analysis, we found seven blocks in high LD; among these findings, the larger haplotype block was found in MGST1, and the leading role of the protein encoded is related to extracellular matrix reorganization. A clinical characteristic of COPD-BBS is anthracofibrosis, bronchial caliber diminution, and increased mucus production [65]. This bronchial remodeling could be related to genes such as MGST1, but we cannot demonstrate it due to the limited investigation of the cellular effects of the BBS.
Additionally, we found two different haplotypes in ARNT2 associated higher risk of COPD-BBS. Previous reports about COPD focus on tobacco smoking, and some of the most significant results involve AHR and ARNT family genes. The evidence demonstrates an important role of AHR in attenuating inflammation related to neutrophils [66] and in lung remodeling by different genes such as MMP9 [67]. Studies in animal models have demonstrated a possible relationship between the aryl hydrocarbon receptors and CYP genes, especially in asthma, which control inflammatory processes [68]. We found many haplotypes in CYP and ARNT genes, which could support the biological relation.
Even though we found SNPs associated specifically with COPD-S or COPD-BBS, we also found similar SNPs and haplotypes, such as ARNT2 and MGST3. This result could suggest the participation of a molecular shared component. Using in silico databases, such as GTEx, we found four SNPs (rs6681 and rs9333378 in MGST3, rs10789501 in CYP4A22, rs117987520 in CYP11A1) that affect the expression levels in the genes where they are located.
With the MCA, we included the SNPs associated with each subtype of COPD, but we did not find clear subgroups. Some studies have demonstrated that multivariate analysis as MCA and polygenic risk score calculation could give more information regarding the effect of exposure/clinical variables and genetic variants as SNPs [69].
Other phenomena reported in our investigation are the SNPs associated with a lower risk of COPD. In previous investigations, we have described similar associations with other SNPs in different genes [70,71]. This effect is described in different illnesses, called the “Hispanic Paradox”, a theory that describes the role of the genetic background of Amerindians which could lead to lower severity or better prognosis in illnesses, including COPD [72,73].
Our is the first exome-wide association study in Mexican mestizos with COPD, classified by tobacco smoking and biomass burning-smoke exposition. We demonstrated the highly conserved composition of the Mexican Amerindian population. Although we found differences in demographics and exposure, we corrected data by logistic regression. Nevertheless, our study is not exempt from limitations; first of all, we need more clinical data, such as the number of exacerbations or predominant phenotypes (bronchitis or emphysema). We also require other auxiliary tools, for instance, expression-related or immunohistochemical. Additionally, we need to include more COPD patients to strengthen the severity analysis.
5. Conclusions
Single-nucleotide variants in CYP2C8, CYP2C9, and MGST3 genes are associated with the risk of COPD secondary to biomass-burning smoke exposure. In addition, shared haplotype blocks in MGST3 and ARNT2 were found in both tobacco smokers and biomass-burning smoke-exposed subjects.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/article/10.3390/cimb45020053/s1. Supplementary Figure S1. Sample selection criteria for both comparison groups, Supplementary Figure S2. Sample selection flowchart for both comparison groups, Supplementary Figure S3. SNPs filtering flowchart for SNPs association analysis, Supplementary Figure S4. Multiple Correspondence Analysis (MCA), Supplementary Figure S5. Multiple correspondence analysis (MCA) for smokers’ comparison, Supplementary Figure S6. Multiple correspondence analysis (MCA) for biomass’ comparison, Supplementary Figure S7. Gene schemes with the SNPs associated in haplotype analysis in smokers’ comparison, Supplementary Figure S8. Gene schemes with the SNPs associated with haplotype analysis in biomass comparison, Supplementary Table S1. List of SNPs included in the analysis, Supplementary Table S2. Molecular data for SNPs associated in the smokers’ comparison, Supplementary Table S3. Molecular data for SNPs associated in exposed biomass-burning smoke comparison, Supplementary Table S4. Alleles data for smokers’ comparison with no correction, Supplementary Table S5. Genotype data for smokers’ comparison with no correction, Supplementary Table S6. Alleles data in exposed biomass-burning smoke comparison with no correction, Supplementary Table S7. Genotype data in exposed biomass-burning smoke comparison with no correction, Supplementary Table S8. Analysis by severity in patients with COPD-S, Supplementary Table S9. Analysis by severity in patients with COPD-BBES.
Author Contributions
Conceptualization, G.P.-R., A.R.-V., R.S., J.P.-R. and R.F.-V.; Data curation, G.P.-R., J.C.F.-L. and R.S.; Formal analysis, E.A.-O., G.P.-R., A.R.-V. and J.C.F.-L.; Funding acquisition, R.S. and R.F.-V.; Investigation, E.A.-O., A.R.-V., F.C.-V. and J.P.-R.; Methodology, E.A.-O., M.E.R.-D., M.d.L.M.-G. and R.F.-V.; Project administration, R.d.J.H.-Z., R.S., J.P.-R. and R.F.-V.; Resources, A.R.-V., R.d.J.H.-Z., M.E.R.-D., F.C.-V., M.d.L.M.-G. and R.F.-V.; Software, E.A.-O., G.P.-R., J.C.F.-L. and R.S.; Supervision, A.R.-V., M.d.L.M.-G. and R.F.-V.; Validation, E.A.-O., G.P.-R., R.d.J.H.-Z., R.S., J.P.-R. and R.F.-V.; Visualization, E.A.-O. and J.C.F.-L.; Writing—original draft, E.A.-O., G.P.-R., J.P.-R. and R.F.-V.; Writing—review and editing, E.A.-O. and R.F.-V. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the budget allocated to research (RFV-HLA Laboratory) from the Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas (INER).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Ethics Committee of Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas (protocol numbers C35-19).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data analyzed in this study are available in ClinVar under the submission code SUB11745124 (SCV002540796–SCV002540803) and SUB11745177 (SCV002540844–SCV002540866).
Acknowledgments
The authors acknowledge the support received from physicians and technicians from the COPD clinic at INER to confirm the diagnosis, the acquisition of data on lung function, and the clinical care of the study participants.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Global Inititiative for Chronic Obstructive Lung Disease (GOLD). Global Inititiative for Chronic Obstructive Lung Disease; GOLD: Deer Park, IL, USA, 2017; pp. 1–139. [Google Scholar]
- de Oca, M.M.; López Varela, M.V.; Acuña, A.; Schiavi, E.; Rey, M.A.; Jardim, J.; Casas, A.; Tokumoto, A.; Torres Duque, C.A.; Ramírez-Venegas, A.; et al. ALAT-2014 Chronic Obstructive Pulmonary Disease (COPD) Clinical Practice Guidelines: Questions and Answers. Arch. Bronconeumol. 2015, 51, 403–416. [Google Scholar] [CrossRef]
- OMS | Enfermedad Pulmonar Obstructiva Crónica (EPOC). World Health Organization. 2015. Available online: http://www.who.int/mediacentre/factsheets/fs315/es/ (accessed on 18 August 2016).
- Mathers, C.D.; Loncar, D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006, 3, 2011–2030. [Google Scholar] [CrossRef] [PubMed]
- Alvar Agusti, R.B.; Chen, R.; Gerard, C.; Frith, P.; Halpin, D. Global Initiative for Chronic Obstructive Lung Disease Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Pulmonary Disease (2018 Report). 2018, p. 142. Available online: https://goldcopd.org/wp-content/uploads/2018/11/GOLD-2019-v1.7-FINAL-14Nov2018-WMS.pdf (accessed on 14 July 2019).
- Mukherjee, S.; Roychoudhury, S.; Siddique, S.; Banerjee, M.; Bhattacharya, P.; Lahiri, T.; Ray, M.R. Respiratory symptoms, lung function decrement and chronic obstructive pulmonary disease in pre-menopausal Indian women exposed to biomass smoke. Inhal. Toxicol. 2014, 26, 866–872. [Google Scholar] [CrossRef] [PubMed]
- Golpe, R.; Mengual-Macenlle, N.; Sanjuán-López, P.; Cano-Jiménez, E.; Castro-Añón, O.; Pérez-De-Llano, L.A. Prognostic Indices and Mortality Prediction in COPD Caused by Biomass Smoke Exposure. Lung 2015, 193, 497–503. [Google Scholar] [CrossRef]
- Shaikh, M.; Sood, R.G.; Sarkar, M.; Thakur, V. Quantitative Computed Tomography (CT) Assessment of Emphysema in Patients with Severe Chronic Obstructive Pulmonary Disease (COPD) and its Correlation with Age, Sex, Pulmonary Function Tests, BMI, Smoking, and Biomass Exposure. Pol. J. Radiol. 2017, 82, 760–766. [Google Scholar] [CrossRef]
- Ghosh, B.; Gaike, A.H.; Pyasi, K.; Brashier, B.; Das, V.V.; Londhe, J.D.; Juvekar, S.; Shouche, Y.S.; Donnelly, L.E.; Salvi, S.S.; et al. Bacterial load and defective monocyte-derived macrophage bacterial phagocytosis in biomass smoke-related COPD. Eur. Respir. J. 2018, 53, 1702273. [Google Scholar] [CrossRef]
- Stoller, J.K.; Aboussouan, L.S. Alpha1-antitrypsin deficiency. Lancet 2005, 365, 2225–2236. [Google Scholar] [CrossRef]
- Wan, E.S.; Cho, M.H.; Boutaoui, N.; Klanderman, B.J.; Sylvia, J.S.; Ziniti, J.P.; Won, S.; Lange, C.; Pillai, S.G.; Anderson, W.H.; et al. Genome-wide association analysis of body mass in chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2011, 45, 304–310. [Google Scholar] [CrossRef]
- Kim, D.K.; Hersh, C.P.; Washko, G.R.; Hokanson, J.E.; Lynch, D.A.; Newell, J.D.; Murphy, J.R.; Crapo, J.D.; Silverman, E.K. Epidemiology, radiology, and genetics of nicotine dependence in COPD. Respir. Res. 2011, 12, 9. [Google Scholar] [CrossRef]
- Thorgeirsson, T.E.; Gudbjartsson, D.F.; Surakka, I.; Vink, J.M.; Amin, N.; Geller, F.; Sulem, P.; Rafnar, T.; Esko, T.; Walter, S.; et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 2010, 42, 448–453. [Google Scholar] [CrossRef]
- Teramoto, S. Recent progress in genetic background of chronic obstructive pulmonary disease (COPD). Nihon Rinsho 2016, 74, 733–742. [Google Scholar]
- Jackson, V.E.; Ntalla, I.; Sayers, I.; Morris, R.; Whincup, P.; Casas, J.P.; Amuzu, A.; Choi, M.; Dale, C.; Kumari, M.; et al. Exome-wide analysis of rare coding variation identifies novel associations with COPD and airflow limitation in MOCS3, IFIT3 and SERPINA12. Thorax 2016, 71, 501–509. [Google Scholar] [CrossRef]
- Cha, S.I.; Kang, H.-G.; Choi, J.E.; Kim, M.J.; Park, J.; Lee, W.K.; Kim, C.H.; Jung, T.H.; Park, J.Y. SERPINE2 polymorphisms and chronic obstructive pulmonary disease. J. Korean Med Sci. 2009, 24, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Yanbaeva, D.G.; Dentener, M.A.; Spruit, M.A.; Houwing-Duistermaat, J.J.; Kotz, D.; Passos, V.L.; Wouters, E.F. IL6 and CRP haplotypes are associated with COPD risk and systemic inflammation: A case-control study. BMC Med. Genet. 2009, 10, 23. [Google Scholar] [CrossRef]
- Ramírez-Venegas, A.; Velázquez-Uncal, M.; Pérez-Hernández, R.; Guzmán-Bouilloud, N.E.; Falfán-Valencia, R.; Mayar-Maya, M.E.; Aranda-Chávez, A.; Sansores, R.H. Prevalence of COPD and respiratory symptoms associated with biomass smoke exposure in a suburban area. Int. J. Chronic Obstr. Pulm. Dis. 2018, 13, 1727–1734. [Google Scholar] [CrossRef] [PubMed]
- GOLD 2021. Global Initiative for Chronic Obstructive Lung Disease: Pocket Guide To COPD Diagnosis, Management, and Prevention, A Guide for Health Care Professionals. Gold 2021. pp. 1–33. Available online: https://goldcopd.org/wp-content/uploads/2020/11/GOLD-2021-POCKET-GUIDE-v1.0-16Nov20_WMV.pdf (accessed on 27 April 2021).
- Ambrocio-Ortiz, E.; Pérez-Rubio, G.; Abarca-Rojano, E.; Montaño, M.; Ramos, C.; Hernández-Zenteno, R.D.; Del Angel-Pablo, A.D.; Reséndiz-Hernández, J.M.; Ramírez-Venegas, A.; Falfán-Valencia, R. Influence of proinflammatory cytokine gene polymorphisms on the risk of COPD and the levels of plasma protein. Cytokine 2018, 111, 364–370. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. European Environment Agency. 2020. Available online: https://www.eea.europa.eu/data-and-maps/indicators/oxygen-consuming-substances-in-rivers/r-development-core-team-2006 (accessed on 5 February 2022).
- R Studio Team. Available online: http://www.rstudio.com/ (accessed on 4 February 2022).
- Huerta-Chagoya, A.; Moreno-Macías, H.; Fernández-López, J.C.; Ordóñez-Sánchez, M.L.; Rodríguez-Guillén, R.; Contreras, A.V.; Hidalgo-Miranda, A.; Alfaro-Ruíz, L.A.; Salazar-Fernandez, E.P.; Moreno-Estrada, A.; et al. A Panel of 32 AIMs Suitable for Population Stratification Correction and Global Ancestry Estimation in Mexican Mestizos 06 Biological Sciences 0604 Genetics. BMC Genet. 2019, 20, 5. Available online: https://bmcgenomdata.biomedcentral.com/articles/10.1186/s12863-018-0707-7 (accessed on 8 January 2023). [CrossRef] [PubMed]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. Available online: https://cran.r-project.org/package=FactoMineR (accessed on 19 September 2022). [CrossRef]
- Kassambara, A.; Mundt, F. Factoextra: Extract and visualize the results of multivariate data analyses. 2017, pp. 1–76. Available online: http//www.sthda.com/english/rpkgs/factoextraBugReports; https://cran.r-project.org/package=factoextra (accessed on 19 September 2022).
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The structure of haplotype blocks in the human genome. Science 2002, 296, 2225–2229. Available online: http://www.sciencemag.org/cgi/doi/10.1126/science.1069424 (accessed on 4 September 2019). [CrossRef]
- Pérez-Rubio, G.; Falfán-Valencia, R.; Fernández-López, J.; Ramírez-Venegas, A.; Hernández-Zenteno, R.; Flores-Trujillo, F.; Silva-Zolezzi, I. Genetic Factors Associated with COPD Depend on the Ancestral Caucasian/Amerindian Component in the Mexican Population. Diagnostics 2021, 11, 599. [Google Scholar] [CrossRef]
- Pérez-Rubio, G.; Silva-Zolezzi, I.; Fernández-López, J.C.; Camarena, Á.; Velázquez-Uncal, M.; Morales-Mandujano, F.; Hernández-Zenteno, R.D.J.; Flores-Trujillo, F.; Sánchez-Romero, C.; Velázquez-Montero, A.; et al. Genetic Variants in IL6R and ADAM19 are Associated with COPD Severity in a Mexican Mestizo Population. COPD 2016, 13, 610–615. [Google Scholar] [CrossRef] [PubMed]
- OMS. Contaminación del aire de Interiores y Salud; OMS: Geneva, Switzerland, 2018. [Google Scholar] [CrossRef]
- Rehfuess, E.A.; Puzzolo, E.; Stanistreet, D.; Pope, D.; Bruce, N.G. Enablers and Barriers to Large-Scale Uptake of Improved Solid Fuel Stoves: A Systematic Review. Environ. Health Perspect. 2014, 122, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Padilla, J.R.; Regalado-Pineda, J.; Morán-Mendoza, A.O. The domestic inhalation of the smoke from firewood and of other biological materials. A risk for the development of respiratory diseases. Gac. Med. de Mex. 1999, 135, 19–29. [Google Scholar]
- Espin-Garcia, O.; Craiu, R.V.; Bull, S.B. Two-phase sample selection strategies for design and analysis in post-genome-wide association fine-mapping studies. Stat. Med. 2021, 40, 6792–6817. [Google Scholar] [CrossRef]
- Los Xenobióticos. Available online: https://centrosconacyt.mx/noticia/los-xenobioticos/ (accessed on 9 October 2021).
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef]
- van Schaik, R.H.N. CYP450 pharmacogenetics for personalizing cancer therapy. Drug Resist. Updates 2008, 11, 77–98. [Google Scholar] [CrossRef]
- Shinde, R.; McGaha, T.L. The Aryl Hydrocarbon Receptor: Connecting Immunity to the Microenvironment. Trends Immunol. 2018, 39, 1005–1020. [Google Scholar] [CrossRef]
- Strosberg, A.D. Structure, function, and regulation of adrenergic receptors. Protein Sci. 1993, 2, 1198–1209. [Google Scholar] [CrossRef]
- Lu, H.; Yang, Y.; Chen, X.; Wu, C.; Zhao, J.; Feng, Q.; Zhou, X.; Xu, D.; Li, Q.; Niu, H.; et al. Influence of the CYP2J2 Gene Polymorphisms on Chronic Obstructive Pulmonary Disease Risk in the Chinese Han Population. Arch. Bronconeumol. 2020, 56, 697–703. [Google Scholar] [CrossRef]
- Akhmadishina, L.A.; Korytina, G.F.; Victorova, T.V. Polymorphic markers of the CYP1B1 (4326C > G), CYP2F1 (c.14_15insC), CYP2J2 (-76G > T), and CYP2S1 (13106C > T and 13255A > G) genes and genetic predisposition to chronic respiratory diseases induced by smoking and occupational factors. Genetika 2011, 47, 1402–1410. [Google Scholar] [PubMed]
- Kamata, S.; Fujino, N.; Yamada, M.; Grime, K.; Suzuki, S.; Ota, C.; Tando, Y.; Okada, Y.; Sakurada, A.; Noda, M.; et al. Expression of cytochrome P450 mRNAs in Type II alveolar cells from subjects with chronic obstructive pulmonary disease. Pharmacol. Res. Perspect. 2018, 6, e00405. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, J.; Peng, W.; Shu, Y.; Mo, X. Cytochrome P450 Epoxygenase 2J2 Protects Against Lung Ischemia/Reperfusion Injury by Activating the P13K/Akt/GSK-3-β/NF-kB Signaling Pathway During Deep Hypothermic Low Flow in Mice. J. Surg. Res. 2020, 253, 8–17. [Google Scholar] [CrossRef]
- Hukkanen, J.; Pelkonen, O.; Hakkola, J.; Raunio, H. Expression and regulation of xenobiotic-metabolizing cytochrome P450 (CYP) enzymes in human lung. Crit. Rev. Toxicol. 2002, 32, 391–411. [Google Scholar] [CrossRef] [PubMed]
- Rubin, K.; Ewing, P.; Bäckström, E.; Abrahamsson, A.; Bonn, B.; Kamata, S.; Grime, K. Pulmonary metabolism of substrates for key drug-metabolizing enzymes by human alveolar type ii cells, human and rat lung microsomes, and the isolated perfused rat lung model. Pharmaceutics 2020, 12, 117. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zheng, G.; Yang, S.; Ping, W.; Fu, X.; Zhang, N.; Wang, D.W.; Wang, J. CYP2J2 and EETs protect against oxidative stress and apoptosis in vivo and in vitro following lung ischemia/reperfusion. Cell. Physiol. Biochem. 2014, 35, 1663–1680. [Google Scholar] [CrossRef] [PubMed]
- London, S.J.; Daly, A.K.; Leathart, J.B.; Navidi, W.C.; Idle, J.R. Lung cancer risk in relation to the CYP2C9*1/CYP2C9*2 genetic polymorphism among African-Americans and Caucasians in Los Angeles County, California. Pharmacogenetics 1996, 6, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Sausville, L.N.; Gangadhariah, M.H.; Chiusa, M.; Mei, S.; Wei, S.; Zent, R.; Luther, J.M.; Shuey, M.M.; Capdevila, J.H.; Falck, J.R.; et al. The cytochrome P450 slow metabolizers CYP2C92 and CYP2C93 directly regulate tumorigenesis via reduced epoxyeicosatrienoic acid production. Cancer Res. 2018, 78, 4865–4877. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Bentley, A.R.; Kritchevsky, S.B.; Harris, T.B.; Newman, A.B.; Bauer, D.C.; Meibohm, B.; Cassano, P.A. Genetic variation in antioxidant enzymes, cigarette smoking, and longitudinal change in lung function. Free Radic. Biol. Med. 2013, 63, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Awji, E.G.; Chand, H.; Bruse, S.; Smith, K.R.; Colby, J.K.; Mebratu, Y.; Levy, B.D.; Tesfaigzi, Y. Wood smoke enhances cigarette smoke-induced inflammation by inducing the aryl hydrocarbon receptor repressor in airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2015, 52, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ji, N.; Wang, Z.; Wu, C.; Sun, Z.; Li, Y.; Hu, F.; Wang, Z.; Huang, M.; Zhang, M. Fine particulate matter (PM25) promoted the invasion of lung cancer cells via an ARNT2/PP2A/STAT3/MMP2 Pathway. J. Biomed. Nanotechnol. 2018, 14, 2172–2184. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Yang, E.; Liao, H.; Wang, Z.; Den, Z.; Ren, H. ARNT2 is downregulated and serves as a potential tumor suppressor gene in non-small cell lung cancer. Tumor Biol. 2015, 36, 2111–2119. [Google Scholar] [CrossRef]
- Drutel, G.; Kathmann, M.; Héron, A.; Gros, C.; Macé, S.; Schwartz, J.; Arrang, J. Two splice variants of the hypoxia-inducible factor HIF-1α as potential dimerization partners of ARNT2 in neurons. Eur. J. Neurosci. 2000, 12, 3701–3708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, C.; Gao, Y.; Wang, Y.; Ding, Z.; Zhang, Y.; Shen, W.; Zheng, Y.; Wan, Y. A Novel Long Non-coding RNA, MSTRG.51053.2 Regulates Cisplatin Resistance by Sponging the miR-432-5p in Non-small Cell Lung Cancer Cells. Front. Oncol. 2020, 10, 215. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Ge, C.; Li, R.; Zhang, Z.; Fu, Q.; Li, Z.; Lin, Z.; Liu, L.; Xue, Y.; Xu, Y.; et al. Knockdown of microsomal glutathione S-transferase 1 inhibits lung adenocarcinoma cell proliferation and induces apoptosis. Biomed. Pharmacother. 2019, 121, 109562. [Google Scholar] [CrossRef]
- Woldhuis, R.R.; De Vries, M.; Timens, W.; van den Berge, M.; DeMaria, M.; Oliver, B.G.G.; Heijink, I.H.; Brandsma, C.-A. Link between increased cellular senescence and extracellular matrix changes in COPD. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2020, 319, L48–L60. [Google Scholar] [CrossRef]
- Sauler, M.; McDonough, J.E.; Adams, T.S.; Kothapalli, N.; Barnthaler, T.; Werder, R.B.; Schupp, J.C.; Nouws, J.; Robertson, M.J.; Coarfa, C.; et al. Characterization of the COPD Alveolar Niche Using Single-Cell RNA Sequencing. Nat. Commun. 2022, 13, 394. Available online: https://www.nature.com/articles/s41467-022-28062-9 (accessed on 20 March 2020). [CrossRef]
- Francis, S.M.S.; Larsen, J.E.; Pavey, S.J.; Bowman, R.V.; Hayward, N.K.; Fong, K.M.; Yang, I.A. Expression Profiling Identifies Genes Involved in Emphysema Severity. Respir. Res. 2009, 10, 81. Available online: http://respiratory-research.biomedcentral.com/articles/10.1186/1465-9921-10-81 (accessed on 9 January 2023). [CrossRef]
- Kukkonen, M.K.; Hämäläinen, S.; Kaleva, S.; Vehmas, T.; Huuskonen, M.S.; Oksa, P.; Vainio, H.; Piirilä, P.; Hirvonen, A. Genetic Polymorphisms of Xenobiotic-Metabolizing Enzymes Influence the Risk of Pulmonary Emphysema. Pharm. Genom. 2011, 21, 876–883. Available online: https://journals.lww.com/01213011-201112000-00012 (accessed on 9 January 2023). [CrossRef] [PubMed]
- D’Amelio, A.M.; Monroy, C.; El-Zein, R.; Etzel, C.J. Using haplotype analysis to elucidate significant associations between genes and Hodgkin lymphoma. Leuk. Res. 2012, 36, 1359–1364. [Google Scholar] [CrossRef]
- Hou, Z.F.; Yuan, Z.H.; Chang, K.; Cao, Y.H.; Guan, F.X.; Gao, Y. NLRP3 rs1539019 is significantly associated with chronic obstructive pulmonary disease in a Chinese Han population: A case-control study. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 5821–5828. [Google Scholar] [PubMed]
- Mahesworo, B.; Budiarto, A.; Hidayat, A.A.; Pardamean, B. Cancer Risk Score Prediction Based on a Single-Nucleotide Polymorphism Network. Healthc. Inform. Res. 2022, 28, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, Q.; Dong, L.; Xiong, M.; Jiang, H.; Guo, M.; Zhao, L.; Yuan, L.; Li, Z.; Liu, H.; et al. The effects of CXCL10 polymorphisms on COPD susceptibility. Mol. Genet. Genom. 2017, 293, 649–655. [Google Scholar] [CrossRef]
- Hosgood, H.D., III; Díaz-Peña, R.; Blansky, D.; Jaime, S.; Parra, V.; Boekstegers, F.; Bermejo, J.L.; García-Valero, J.; Montes, J.F.; Valdivia, G.; et al. PRDM15 Is Associated with Risk of Chronic Obstructive Pulmonary Disease in a Rural Population in Chile. Respiration 2020, 99, 307–315. [Google Scholar] [CrossRef]
- Ramírez-Venegas, A.; Torres-Duque, C.A.; Guzmán-Bouilloud, N.E.; González-García, M.; Sansores, R.H. Small Airway Disease in COPD Associated to Biomass Exposure. Rev. Investig. Clin. 2019, 71, 70–78. [Google Scholar] [CrossRef]
- Rico de Souza, A.; Traboulsi, H.; Wang, X.; Fritz, J.H.; Eidelman, D.H.; Baglole, C.J. The Aryl Hydrocarbon Receptor Attenuates Acute Cigarette Smoke-Induced Airway Neutrophilia Independent of the Dioxin Response Element. Front. Immunol. 2021, 12, 630427. [Google Scholar] [CrossRef]
- Guerrina, N.; Traboulsi, H.; de Souza, A.R.; Bossé, Y.; Thatcher, T.H.; Robichaud, A.; Ding, J.; Li, P.Z.; Simon, L.; Pareek, S.; et al. Aryl hydrocarbon receptor deficiency causes the development of chronic obstructive pulmonary disease through the integration of multiple pathogenic mechanisms. FASEB J. 2021, 35, e21376. [Google Scholar] [CrossRef]
- Alessandrini, F.; de Jong, R.; Wimmer, M.; Maier, A.-M.; Fernandez, I.; Hils, M.; Buters, J.T.; Biedermann, T.; Zissler, U.M.; Hoffmann, C.; et al. Lung Epithelial CYP1 Activity Regulates Aryl Hydrocarbon Receptor Dependent Allergic Airway Inflammation. Front. Immunol. 2022, 13, 901194. [Google Scholar] [CrossRef]
- Park, H.A.; Edelmann, D.; Canzian, F.; Harrison, T.A.; Hua, X.; Shi, Q.; Silverman, A.; Schneider, M.; Goldberg, R.M.; Alberts, S.R.; et al. Predictive Polygenic Score for Outcome after First-Line Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients Using Supervised Principal Component Analysis. Cancer Epidemiol. Biomark. Prev. 2022, 31, 2087–2091. [Google Scholar] [CrossRef]
- Ortega-Martínez, A.; Pérez-Rubio, G.; Ambrocio-Ortiz, E.; Nava-Quiroz, K.J.; Hernández-Zenteno, R.D.J.; Abarca-Rojano, E.; Rodríguez-Llamazares, S.; Hernández-Pérez, A.; García-Gómez, L.; Ramírez-Venegas, A.; et al. The SNP rs13147758 in the HHIP Gene Is Associated With COPD Susceptibility, Serum, and Sputum Protein Levels in Smokers. Front. Genet. 2020, 11, 882. [Google Scholar] [CrossRef]
- Ambrocio-Ortiz, E.; Pérez-Rubio, G.; Ramírez-Venegas, A.; Hernández-Zenteno, R.; Paredes-López, A.; Sansores, R.; Ramírez-Díaz, M.; Cruz-Vicente, F.; Martínez-Gómez, M.; Falfán-Valencia, R. Protective Role of Genetic Variants in HSP90 Genes-Complex in COPD Secondary to Biomass-Burning Smoke Exposure and Non-Severe COPD Forms in Tobacco Smoking Subjects. Curr. Issues Mol. Biol. 2021, 43, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Powell, R.; Barr, R.G. Response letter to: The Hispanic paradox further unraveled? Thorax 2014, 69, 185–186. [Google Scholar] [CrossRef] [PubMed]
- Mroziewicz, M.; Tyndale, R.F. Pharmacogenetics: A tool for identifying genetic factors in drug dependence and response to treatment. Addict. Sci. Clin. Pract. 2010, 5, 17–29. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).