Developing Biliary Atresia-like Model by Treating Human Liver Organoids with Polyinosinic:Polycytidylic Acid (Poly (I:C))
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Growing of Liver Organoids
2.3. Treatment of Non-BA Control Liver Organoids with Poly I:C
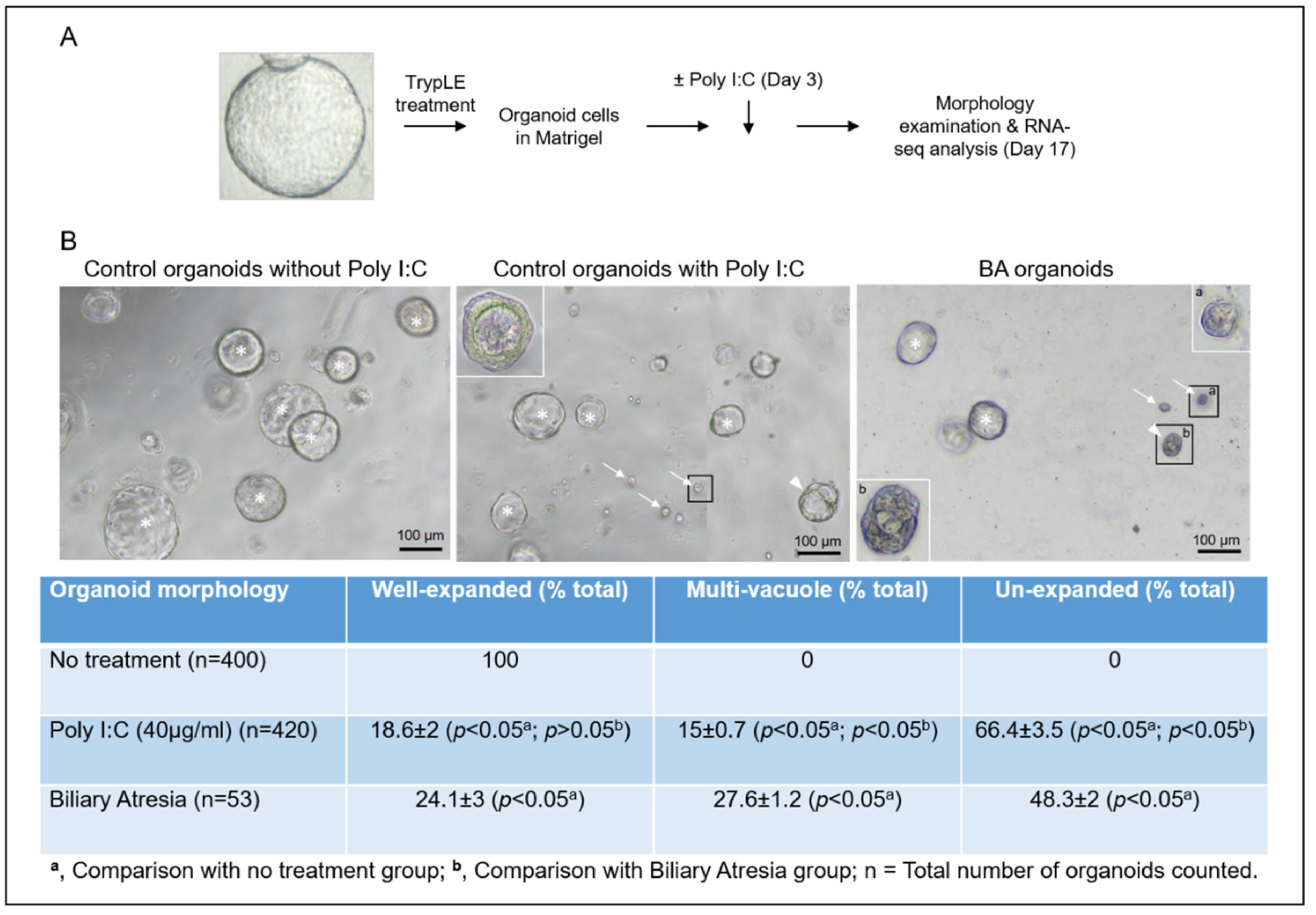
2.4. Counting of Organoids
2.5. RNA-Sequencing Analysis
2.6. Transcriptome Analysis
3. Results
3.1. Poly I:C Treated Liver Organoids Were Morphologically Similar to BA Liver Organoids
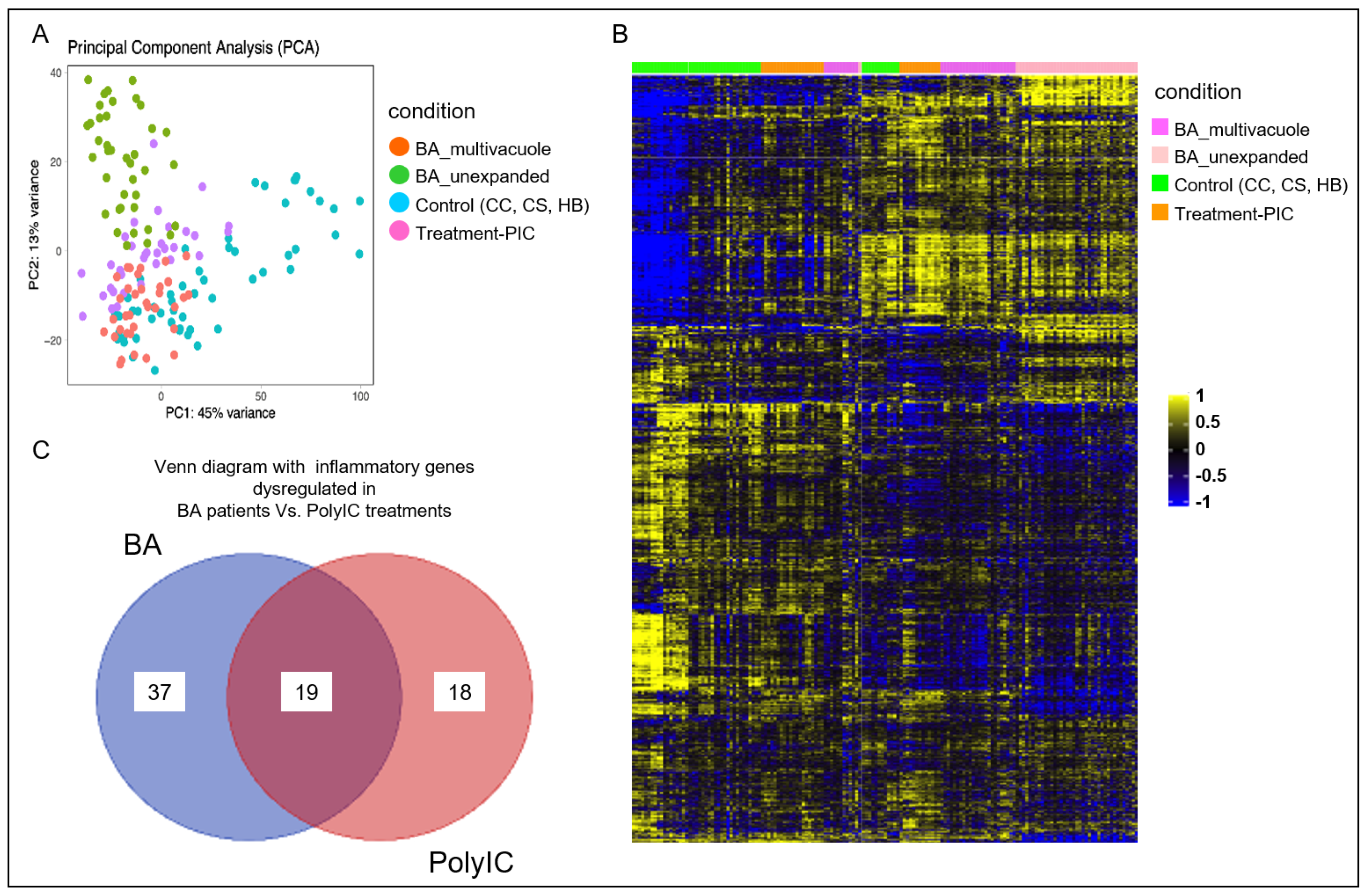
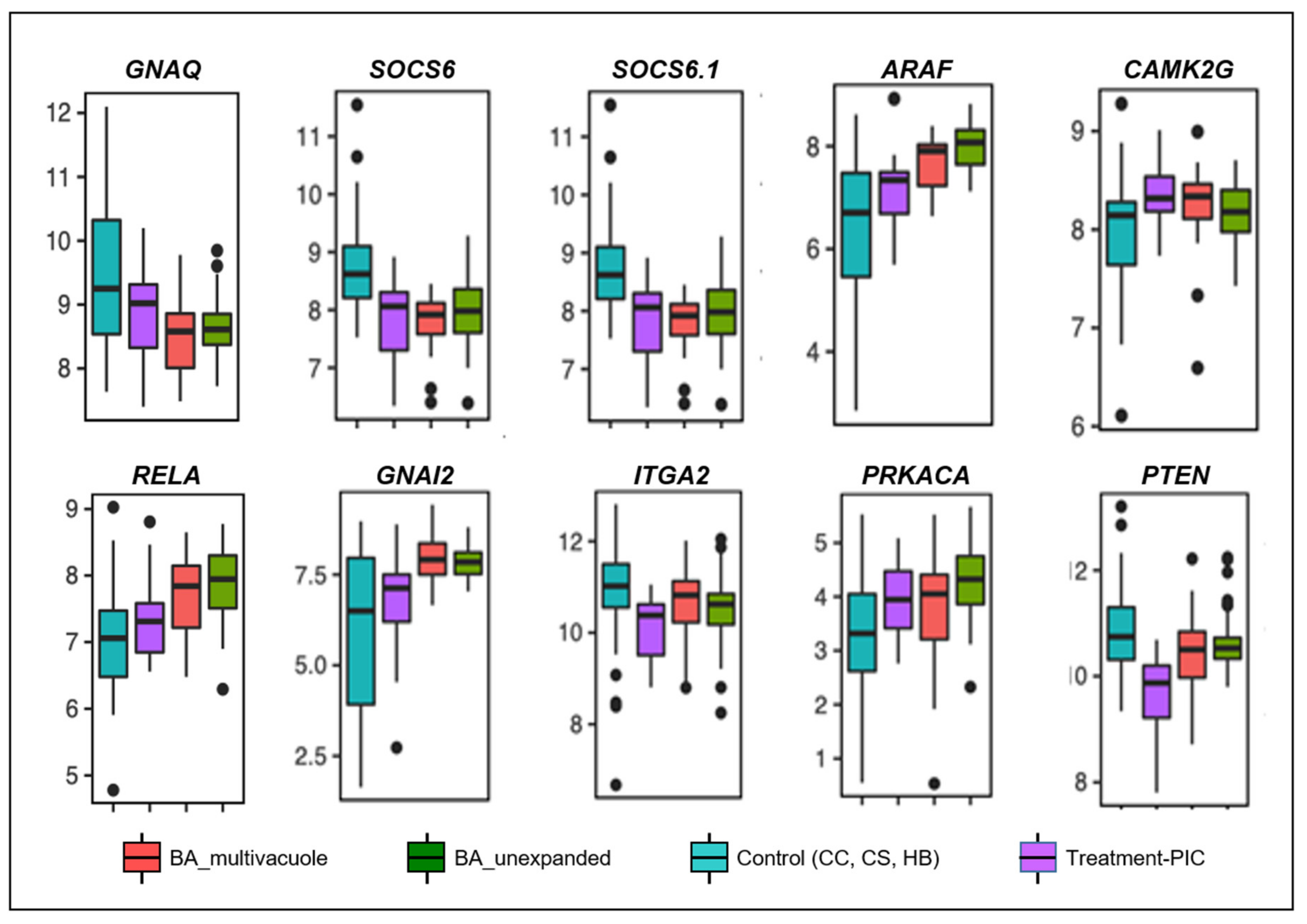
3.2. Poly I:C Treated Organoids and BA Organoids Displayed Similar Transcriptomic Signatures
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Scottoni, F.; Davenport, M. Biliary atresia: Potential for a new decade. Semin. Pediatr. Surg. 2020, 29, 150940. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.H.Y.; Zheng, S.; Tam, P.K.H. Biliary atresia: East versus west. Semin. Pediatr. Surg. 2020, 29, 150950. [Google Scholar] [CrossRef] [PubMed]
- Tam, P.K.H.; Chung, P.H.Y.; St Peter, S.D.; Gayer, C.P.; Ford, H.R.; Tam, G.C.H.; Wong, K.K.Y.; Pakarinen, M.P.; Davenport, M. Advances in paediatric gastroenterology. Lancet 2017, 390, 1072–1082. [Google Scholar] [CrossRef]
- Chung, P.H.Y.; Chan, E.K.W.; Yeung, F.; Chan, A.C.Y.; Mou, J.W.C.; Lee, K.H.; Hung, J.W.S.; Leung, M.W.Y.; Tam, P.K.H.; Wong, K.K.Y. Life long follow up and management strategies of patients living with native livers after Kasai portoenterostomy. Sci. Rep. 2021, 11, 11207. [Google Scholar] [CrossRef]
- Chen, S.; Li, P.; Wang, Y.; Yin, Y.; de Ruiter, P.E.; Verstegen, M.M.A.; Peppelenbosch, M.P.; van der Laan, L.J.W.; Pan, Q. Rotavirus Infection and Cytopathogenesis in Human Biliary Organoids Potentially Recapitulate Biliary Atresia Development. mBio 2020, 11, e01968-20. [Google Scholar] [CrossRef] [PubMed]
- Garrido, M.; Escobar, C.; Zamora, C.; Rejas, C.; Varas, J.; Párraga, M.; Martin, S.S.; Montedónico, S. Bile duct ligature in young rats: A revisited animal model for biliary atresia. Eur. J. Histochem. 2017, 61, 2803. [Google Scholar] [CrossRef]
- Hertel, P.M.; Estes, M.K. Rotavirus and biliary atresia: Can causation be proven? Curr. Opin. Gastroenterol. 2012, 28, 10–17. [Google Scholar] [CrossRef]
- Picelli, S.; Björklund, Å.; Faridani, O.; Sagasser, S.; Winberg, G.; Sandberget, R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 2013, 10, 1096–1098. [Google Scholar] [CrossRef]
- Babu, R.O.; Lui, V.C.H.; Chen, Y.; Yiu, R.S.W.; Ye, Y.; Niu, B.; Wu, Z.; Zhang, R.; Na Yu, M.O.; Chung, P.H.Y.; et al. Beta-amyloid deposition around hepatic bile ducts is a novel pathobiological and diagnostic feature of biliary atresia. J. Hepatol. 2020, 73, 1391–1403. [Google Scholar] [CrossRef]
- Babraham Bioinformatics. FastQC A Quality Control tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 November 2021).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, W169–W175. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Badran, Y.R.; Dedeoglu, F.; Leyva Castillo, J.M.; Bainter, W.; Ohsumi, T.K.; Bousvaros, A.; Goldsmith, J.D.; Geha, R.S.; Chou, J. Human RELA haploinsufficiency results in autosomal-dominant chronic mucocutaneous ulceration. J. Exp. Med. 2017, 214, 1937–1947. [Google Scholar] [CrossRef]
- Buckler, J.L.; Liu, X.; Turka, L.A. Regulation of T-cell responses by PTEN. Immunol. Rev. 2008, 224, 239–248. [Google Scholar] [CrossRef]
- Nelson, D.S.; Quispel, W.; Badalian-Very, G.; van Halteren, A.G.S.; van den Bos, C.; Bovée, J.V.M.G.; Tian, S.Y.; Van Hummelen, P.; Ducar, M.; MacConaill, L.E.; et al. Somatic activating ARAF mutations in Langerhans cell histiocytosis. Blood 2014, 123, 3152–3155. [Google Scholar] [CrossRef]
- Berthon, A.S.; Szarek, E.; Stratakis, C.A. PRKACA: The catalytic subunit of protein kinase A and adrenocortical tumors. Front. Cell Dev. Biol. 2015, 3, 26. [Google Scholar] [CrossRef] [PubMed]
- Palencia-Campos, A.; Aoto, P.C.; Machal, E.M.F.; Rivera-Barahona, A.; Soto-Bielicka, P.; Bertinetti, D.; Baker, B.; Vu, L.; Piceci-Sparascio, F.; Torrente, I.; et al. Germline and Mosaic Variants in PRKACA and PRKACB Cause a Multiple Congenital Malformation Syndrome. Am. J. Hum. Genet. 2020, 107, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.A.; Boucher, G.; Lees, C.W.; Franke, A.; D’amato, M.; Taylor, K.D.; Lee, J.C.; Goyette, P.; Imielinski, M.; Latiano, A.; et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011, 43, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Iida, A.; Saito, S.; Sekine, A.; Tabei, W.; Kataoka, Y.; Nakamura, Y. Identification of 20 novel SNPs in the guanine nucleotide binding protein alpha 12 gene locus. J. Hum. Genet. 2004, 49, 445–448. [Google Scholar] [CrossRef][Green Version]
- Yang, L.; Wu, B.; Wu, Z.; Xu, Y.; Wang, P.; Li, M.; Xu, R.; Liang, Y. CAMKIIγ is a targetable driver of multiple myeloma through CaMKIIγ/Stat3 axis. Aging (Albany N. Y.) 2020, 12, 13668–13683. [Google Scholar] [CrossRef]
- Saito, T.; Hishiki, T.; Terui, K.; Mitsunaga, T.; Terui, E.; Nakata, M.; Yoshida, H. Toll-like receptor mRNA expression in liver tissue from patients with biliary atresia. J. Pediatr. Gastroenterol. Nutr. 2011, 53, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Riepenhoff-Talty, M.; Schaekel, K.; Clark, H.F.; Mueller, W.; Uhnoo, I.; Rossi, T.; Fisher, J.; Ogra, P.L. Group A rotaviruses produce extrahepatic biliary obstruction in orally inoculated newborn mice. Pediatr. Res. 1993, 33, 394–399. [Google Scholar]
- Li, Y.; Xu, X.L.; Zhao, D.; Pan, L.N.; Huang, C.W.; Guo, L.J.; Lu, Q.; Wang, J. TLR3 ligand Poly IC Attenuates Reactive Astrogliosis and Improves Recovery of Rats after Focal Cerebral Ischemia. CNS Neurosci. Ther. 2015, 21, 905–913. [Google Scholar] [CrossRef]
| Diagnosis | Gender | Age at Sample Collection | Total Bilirubin (umol/L) | AST (U/L) | ALT (U/L) | GGT(U/L) |
|---|---|---|---|---|---|---|
| Choledochal cyst | F | 8 months | 18 | 42 | 29 | 41 |
| Choledochal cyst | F | 6 months | 10 | 38 | 20 | 30 |
| Hepatoblastoma | F | 20 months | 9 | 52 | 32 | 18 |
| Hepatoblastoma | M | 8 months | 12 | 47 | 30 | 22 |
| Biliary atresia | F | 63 days | 121 | 68 | 100 | 398 |
| Biliary atresia | F | 61 days | 98 | 121 | 112 | 285 |
| Gene | Action/Related Pathway | Associated Disorders |
|---|---|---|
| RELA (RELA Proto-Oncogene, NF-KB Subunit) | Adaptive immunity and responses to pathogens via NF-κB activation. | Inflammatory bowel diseases |
| SOCS6, SOCS6.1 (Suppressor Of Cytokine Signaling 6) | Inducible by cytokine stimulation, negative regulators of cytokine signaling in a classical negative feedback loop. | Chronic myelogenous leukemia (CML) and erythroleukemia |
| ARAF (A-Raf Proto-Oncogene, Serine/Threonine Kinase) | Dendritic cells activation and function. | Langerhans cell histiocytosis |
| CAMK2G (calcium/calmodulin dependent protein kinase II gamma) | Immature T cell lifespan and T cell memory formation. | T cell lymphoma |
| GNA12 (G Protein Subunit Alpha 12) | Activation of cAMP-Dependent PKA | Familial hyperaldosteronism, Inflammatory bowel diseases |
| ITGA2 (Integrin Subunit Alpha 2) | Encodes human platelet antigens (HPA) on the surface of platelets and are immunogenic structures. | Neonatal alloimmune thrombocytopenia, Bleeding Disorder, Platelet-Type, 9. |
| PRKACA (Protein Kinase CAMP-Activated Catalytic Subunit Alpha) | Encodes catalytic protein kinase isoform | Adrenocortical tumor |
| PTEN (Phosphatase and tensin homolog) | T cells development | Autoimmune disease and lymphoma |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chung, P.H.-Y.; Babu, R.O.; Wu, Z.; Wong, K.K.-Y.; Tam, P.K.-H.; Lui, V.C.-H. Developing Biliary Atresia-like Model by Treating Human Liver Organoids with Polyinosinic:Polycytidylic Acid (Poly (I:C)). Curr. Issues Mol. Biol. 2022, 44, 644-653. https://doi.org/10.3390/cimb44020045
Chung PH-Y, Babu RO, Wu Z, Wong KK-Y, Tam PK-H, Lui VC-H. Developing Biliary Atresia-like Model by Treating Human Liver Organoids with Polyinosinic:Polycytidylic Acid (Poly (I:C)). Current Issues in Molecular Biology. 2022; 44(2):644-653. https://doi.org/10.3390/cimb44020045
Chicago/Turabian StyleChung, Patrick Ho-Yu, Rosana Ottakandathil Babu, Zhongluan Wu, Kenneth Kak-Yuen Wong, Paul Kwong-Hang Tam, and Vincent Chi-Hang Lui. 2022. "Developing Biliary Atresia-like Model by Treating Human Liver Organoids with Polyinosinic:Polycytidylic Acid (Poly (I:C))" Current Issues in Molecular Biology 44, no. 2: 644-653. https://doi.org/10.3390/cimb44020045
APA StyleChung, P. H.-Y., Babu, R. O., Wu, Z., Wong, K. K.-Y., Tam, P. K.-H., & Lui, V. C.-H. (2022). Developing Biliary Atresia-like Model by Treating Human Liver Organoids with Polyinosinic:Polycytidylic Acid (Poly (I:C)). Current Issues in Molecular Biology, 44(2), 644-653. https://doi.org/10.3390/cimb44020045







