Single-Step Protocol for Isolating the Recombinant Extracellular Domain of the Luteinizing Hormone Receptor from the Ovis aries Testis
Abstract
1. Introduction
2. Materials and Methods
2.1. RT-PCR
2.2. Cloning, Expression and Purification of the rLHR-Bed
2.3. Purification: Lysis, Column Refolding and Elution
2.4. Computational Methods
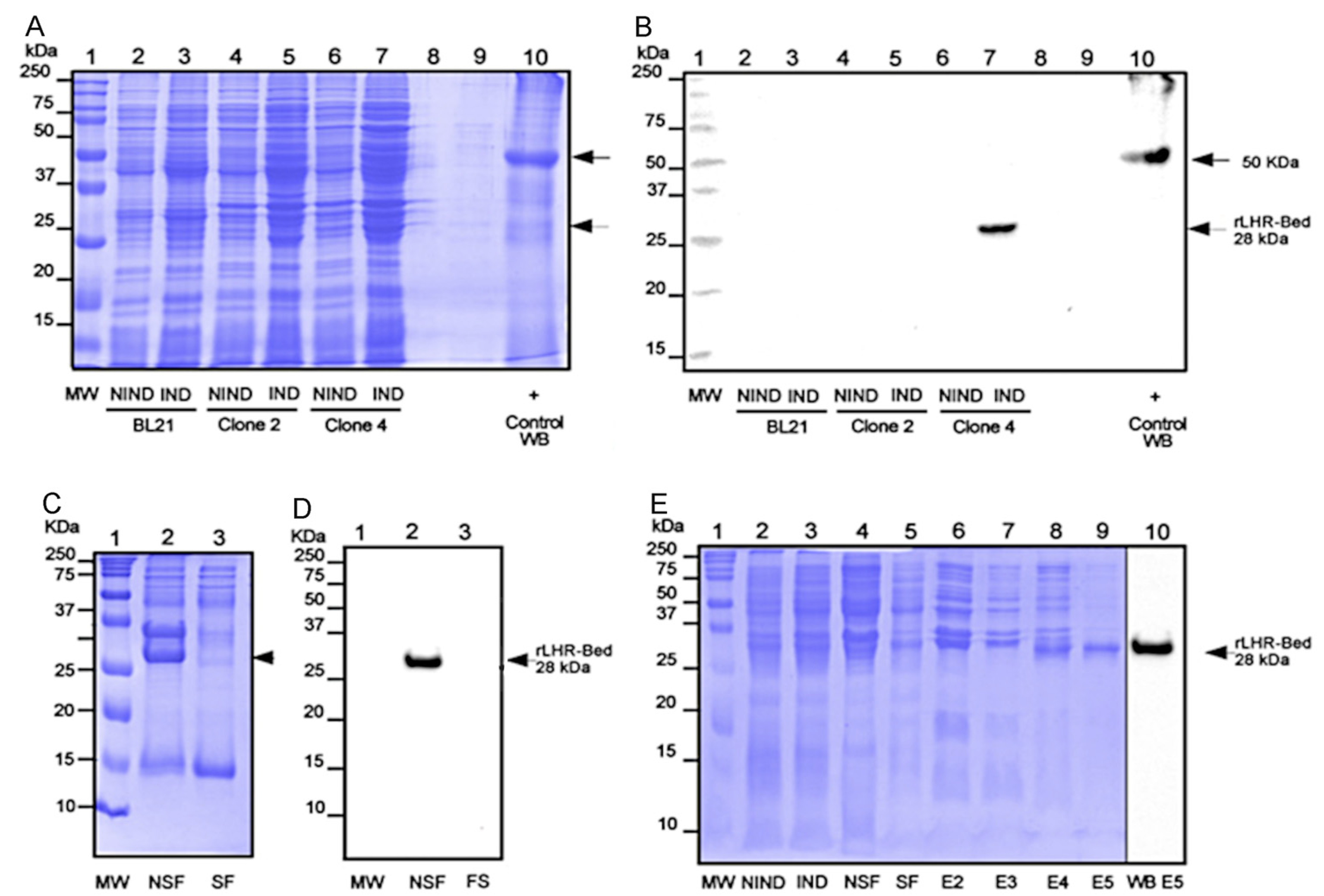
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McFarland, K.C.; Sprengel, R.; Phillips, H.S.; Köhler, M.; Rosemblit, N.; Nikolics, K.; Segaloff, D.L.; Seeburg, P.H. Lutropin-Choriogonadotropin Receptor: An Unusual Member of the G Protein-Coupled Receptor Family. Science 1989, 245, 494–499. [Google Scholar] [CrossRef] [PubMed]
- Bukovsky, A.; Indrapichate, K.; Fujiwara, H.; Cekanova, M.; Ayala, M.E.; Dominguez, R.; Caudle, M.R.; Wimalsena, J.; Elder, R.F.; Copas, P.; et al. Multiple Luteinizing Hormone Receptor (LHR) Protein Variants, Interspecies Reactivity of Anti-LHR MAb Clone 3B5, Subcellular Localization of LHR in Human Placenta, Pelvic Floor and Brain, and Possible Role for LHR in the Development of Abnormal Pregnancy, Pelvic Floor Disorders and Alzheimer’s Disease. Reprod. Biol. Endocrinol. 2003, 1, 1–18. [Google Scholar]
- Puett, D.; Angelova, K.; da Costa, M.R.; Warrenfeltz, S.W.; Fanelli, F. The Luteinizing Hormone Receptor: Insights into Structure-Function Relationships and Hormone-Receptor-Mediated Changes in Gene Expression in Ovarian Cancer Cells. Mol. Cell. Endocrinol. 2010, 329, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Dufau, M.L. The Luteinizing Hormone Receptor. Annu. Rev. Physiol. 1998, 60, 461–496. [Google Scholar] [CrossRef]
- Xie, Y.B.; Wang, H.; Segaloff, D.L. Extracellular Domain of Lutropin/Choriogonadotropin Receptor Expressed in Transfected Cells Binds Choriogonadotropin with High Affinity. J. Biol. Chem. 1990, 265, 21411–21414. [Google Scholar] [CrossRef]
- Pajot-Augy, E.; Couture, L.; Bozon, V.; Remy, J.J.; Biache, G.; Severini, M.; Huet, J.C.; Pernollet, J.C.; Salesse, R. High-Level Expression of Recombinant Porcine LH Receptor in Baculovirus-Infected Insect Cells or Caterpillars. J. Mol. Endocrinol. 1995, 14, 51–66. [Google Scholar] [CrossRef]
- Narayan, P.; Gray, J.; Puett, D. Expression of Functional Lutropin/Choriogonadotropin Receptor in the Baculovirus System. Mol. Cell. Endocrinol. 1996, 117, 95–100. [Google Scholar] [CrossRef]
- Lobel, L.; Pollak, S.; Lustbader, B.; Klein, J.; Lustbader, J.W. Bacterial Expression of a Natively Folded Extracellular Domain Fusion Protein of the HFSH Receptor in the Cytoplasm of Escherichia coli. Protein Expr. Purif. 2002, 25, 124–133. [Google Scholar] [CrossRef]
- Fox, K.M.; Dias, J.A.; Van Roey, P. Three-Dimensional Structure of Human Follicle-Stimulating Hormone. Mol. Endocrinol. 2001, 15, 378–389. [Google Scholar] [CrossRef]
- Ulloa-Aguirre, A.; Zariñán, T.; Jardón-Valadez, E.; Gutiérrez-Sagal, R.; Dias, J.A. Structure-Function Relationships of the Follicle-Stimulating Hormone Receptor. Front. Endocrinol. 2018, 9, 378–389. [Google Scholar] [CrossRef]
- Montero-Pardo, A.; Diaz, D.; Olivares, A.; González-Padilla, E.; Murcia, C.; Gómez-Chavarín, M.; Gutiérrez-Ospina, G.; Perera-Marín, G. Effect of Ovine Luteinizing Hormone (OLH) Charge Isoforms on VEGF and CAMP Production. Anim. Reprod. Sci. 2015, 163, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Spadiut, O.; Posch, G.; Ludwig, R.; Haltrich, D.; Peterbauer, C.K. Evaluation of Different Expression Systems for the Heterologous Expression of Pyranose 2-Oxidase from Trametes Multicolor in E. coli. Microb. Cell Fact. 2010, 9, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A Protein Secondary Structure Prediction Server. Nucleic Acids Res. 2015, 43, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Qing, G.; Ma, L.-C.; Khorchid, A.; Swapna, G.V.T.; Mal, T.K.; Takayama, M.M.; Xia, B.; Phadtare, S.; Ke, H.; Acton, T.; et al. Cold-Shock Induced High-Yield Protein Production in Escherichia Coli. Nat. Biotechnol. 2004, 22, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Seddon, A.M.; Curnow, P.; Booth, P.J. Membrane Proteins, Lipids and Detergents: Not Just a Soap Opera. Biochim. Biophys. Acta (BBA)—Biomembr. 2004, 1666, 105–117. [Google Scholar] [CrossRef]
- Ahram, M.; Litou, Z.I.; Fang, R.; Al-Tawallbeh, G. Estimation of Membrane Proteins in the Human Proteome. Silico Biol. 2006, 6, 379–386. [Google Scholar]
- Palczewski, K.; Kumasaka, T.; Hori, T.; Behnke, C.A.; Motoshima, H.; Fox, B.A.; Trong, I.L.; Teller, D.C.; Okada, T.; Stenkamp, R.E.; et al. Crystal Structure of Rhodopsin: A G Protein-Coupled Receptor. Science 2000, 289, 739–745. [Google Scholar] [CrossRef]
- Babcock, J.J.; Li, M. Deorphanizing the Human Transmembrane Genome: A Landscape of Uncharacterized Membrane Proteins. Acta Pharmacol. Sin. 2014, 35, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M. Strategies for the Purification of Membrane Proteins. Methods Mol. Biol. 2017, 1485, 389–400. [Google Scholar] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- McGee, S.R.; Narayan, P. Precocious Puberty and Leydig Cell Hyperplasia in Male Mice with a Gain of Function Mutation in the LH Receptor Gene. Endocrinology 2013, 154, 3900–3913. [Google Scholar] [CrossRef] [PubMed]
- Roman, E.A.; González Flecha, F.L. Kinetics and Thermodynamics of Membrane Protein Folding. Biomolecules 2014, 4, 354–373. [Google Scholar] [CrossRef]
- Ventura, S.; Villaverde, A. Protein Quality in Bacterial Inclusion Bodies. Trends Biotechnol. 2006, 24, 179–185. [Google Scholar] [CrossRef]
- Osuga, Y.; Kudo, M.; Kaipia, A.; Kobilka, B.; Hsueh, A.J. Derivation of Functional Antagonists Using N-Terminal Extracellular Domain of Gonadotropin and Thyrotropin Receptors. Mol. Endocrinol. 1997, 11, 1659–1668. [Google Scholar] [CrossRef][Green Version]
- Kwon, S.-K.; Kim, S.K.; Lee, D.-H.; Kim, J.F. Comparative Genomics and Experimental Evolution of Escherichia Coli BL21 (DE3) Strains Reveal the Landscape of Toxicity Escape from Membrane Protein Overproduction. Sci. Rep. 2015, 5, 16076. [Google Scholar] [CrossRef]
- San-Miguel, T.; Pérez-Bermúdez, P.; Gavidia, I. Production of Soluble Eukaryotic Recombinant Proteins in E. coli Is Favoured in Early Log-Phase Cultures Induced at Low Temperature. SpringerPlus 2013, 2, 89. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villalpando-Aguilar, J.L.; López-Rosas, I.; Montero-Pardo, A.; Azuara-Liceaga, E.; Valencia-Méndez, J.d.J.; Trejo-Muñoz, C.R.; Kubli-Garfias, C. Single-Step Protocol for Isolating the Recombinant Extracellular Domain of the Luteinizing Hormone Receptor from the Ovis aries Testis. Curr. Issues Mol. Biol. 2022, 44, 5718-5727. https://doi.org/10.3390/cimb44110387
Villalpando-Aguilar JL, López-Rosas I, Montero-Pardo A, Azuara-Liceaga E, Valencia-Méndez JdJ, Trejo-Muñoz CR, Kubli-Garfias C. Single-Step Protocol for Isolating the Recombinant Extracellular Domain of the Luteinizing Hormone Receptor from the Ovis aries Testis. Current Issues in Molecular Biology. 2022; 44(11):5718-5727. https://doi.org/10.3390/cimb44110387
Chicago/Turabian StyleVillalpando-Aguilar, José Luis, Itzel López-Rosas, Arnulfo Montero-Pardo, Elisa Azuara-Liceaga, Javier de Jesús Valencia-Méndez, Cynthia R. Trejo-Muñoz, and Carlos Kubli-Garfias. 2022. "Single-Step Protocol for Isolating the Recombinant Extracellular Domain of the Luteinizing Hormone Receptor from the Ovis aries Testis" Current Issues in Molecular Biology 44, no. 11: 5718-5727. https://doi.org/10.3390/cimb44110387
APA StyleVillalpando-Aguilar, J. L., López-Rosas, I., Montero-Pardo, A., Azuara-Liceaga, E., Valencia-Méndez, J. d. J., Trejo-Muñoz, C. R., & Kubli-Garfias, C. (2022). Single-Step Protocol for Isolating the Recombinant Extracellular Domain of the Luteinizing Hormone Receptor from the Ovis aries Testis. Current Issues in Molecular Biology, 44(11), 5718-5727. https://doi.org/10.3390/cimb44110387







