A Critical Review: Recent Advancements in the Use of CRISPR/Cas9 Technology to Enhance Crops and Alleviate Global Food Crises
Abstract
:1. Introduction
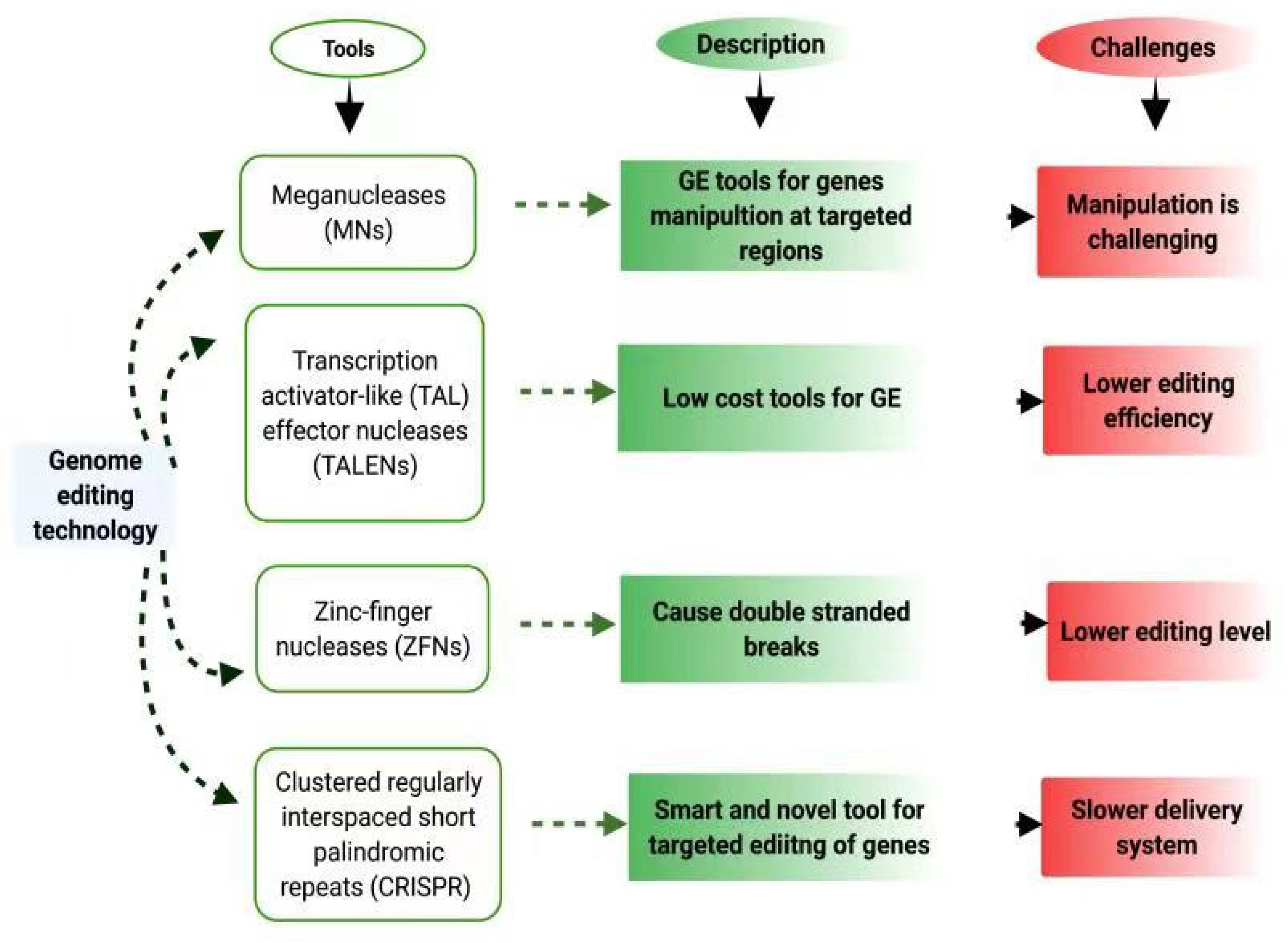
2. Genome Editing Tools
2.1. CRISPR/Cas9 and Its Brief Overview
| Role | ZFNs | TALENs | MNs | CRISPR/Cas9 | References |
|---|---|---|---|---|---|
| Efficacy of target recognition | Higher | Higher | Higher | Higher | [66] |
| Kind of Action | Double-stranded break in target DNA | Double-strandedbreak in target DNA | Direct conversions in targeted regions | Double-stranded break in target DNA | [67] |
| Mutagenesis | Higher | Middle | Middle | Lower | [66] |
| Multiplexing | Difficult | Difficult | Difficult | Possible | [68,69] |
| Target range | Unlimited | Unlimited | Unlimited | Limited by PAM | [70] |
| Effects | Lower | Lower | Lower | Lower | [71] |
| Cost | Higher | Higher | Higher | Low | [72] |
| Crop Improvement | Low | Low | Low | Higher | [72] |
| Range | Narrow | Narrow | Narrow | Broad | [72] |
| Dimerization | Required | Not required | Not required | Not required | [71] |
| Types | One | One | One | Many | [71] |
| Future use | Medium | Medium | Medium | High | [72] |
2.2. CRISPR/Cas9 Mechanism and Function
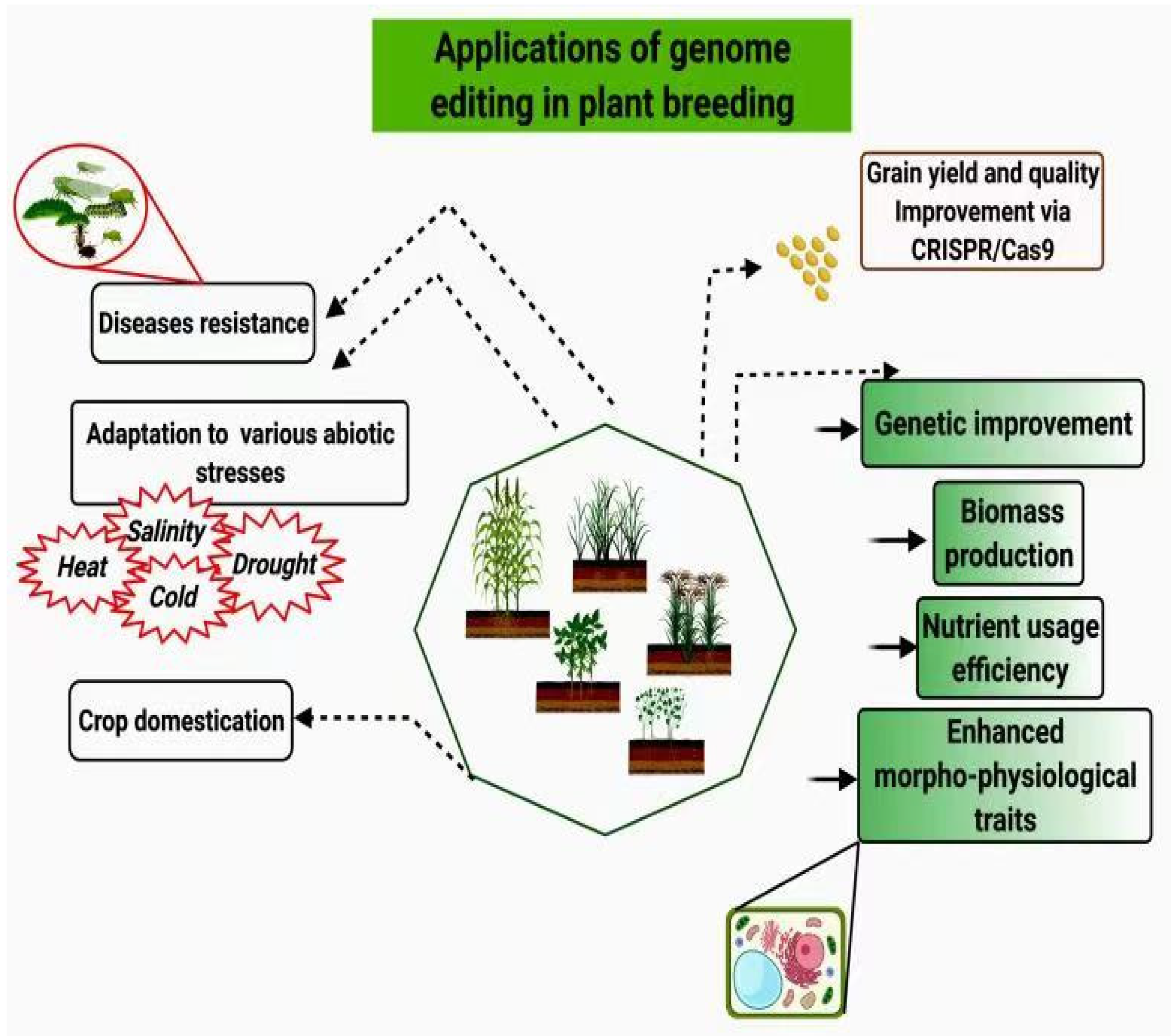
3. Application of CRISPR/Cas9 for Crop Improvement
3.1. Use of CRISPR/Cas9 for Improvement of Yield and Quality
3.2. Development of Disease Resistant Varieties Using CRISPR/Cas9
3.3. Development of Climate-Smart Crops Using CRISPR/Cas9
4. Novel Breakthroughs
4.1. Production of Mutant Libraries
4.2. Base Editing
4.3. Prime Editing
4.4. Transgene Free Editing of the Genome
4.5. Multiplex Genetic Engineering
5. Utilization of CRISPR/Cas9 for Crop Domestication
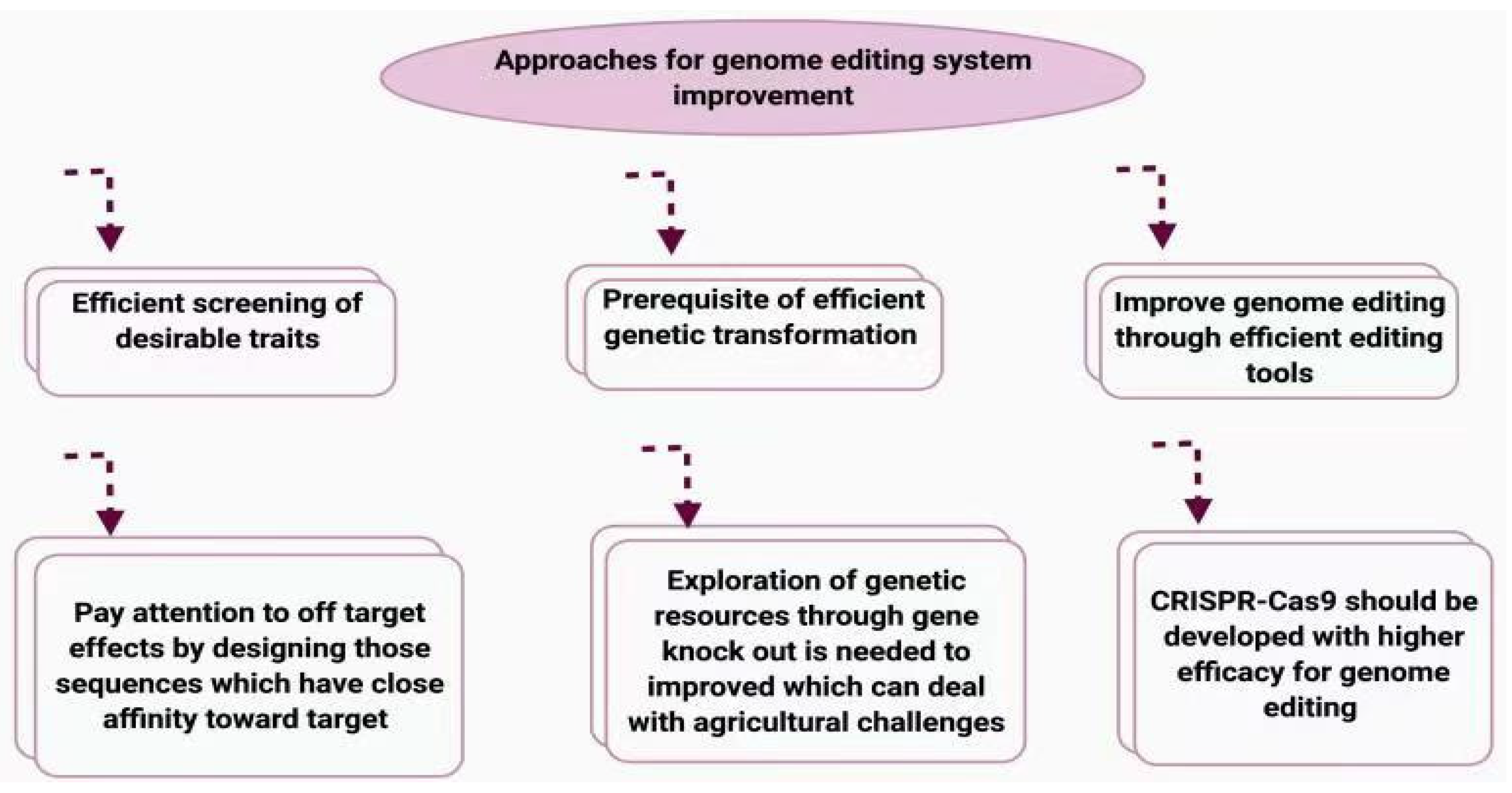
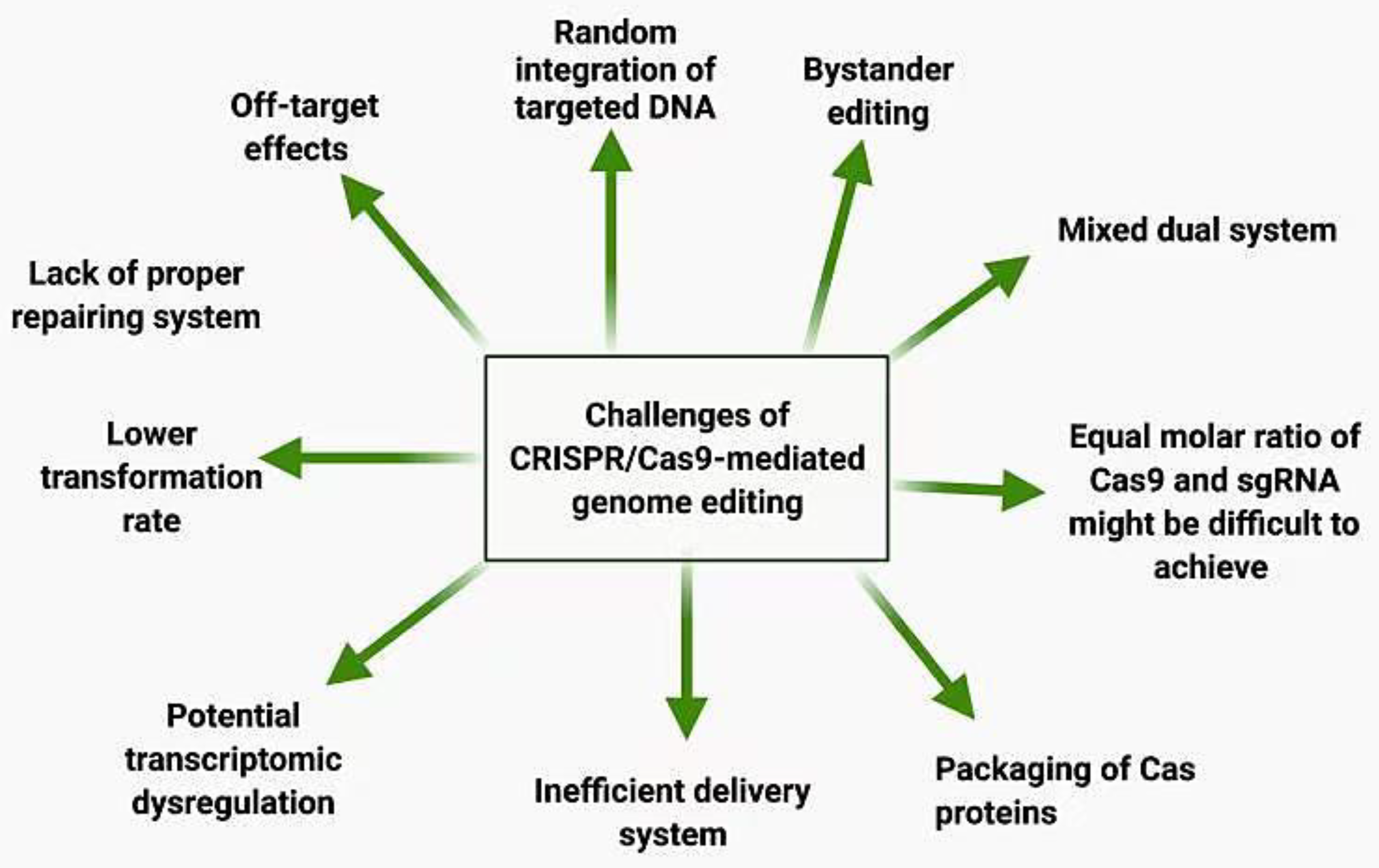
6. Challenges and Limitations of CRISPR/Cas9 Application
7. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barrett, C.B. Overcoming global food security challenges through science and solidarity. Ame. J. Agric. Econ. 2021, 103, 422–447. [Google Scholar] [CrossRef]
- Adhikari, P.; Poudel, M. CRISPR-Cas9 in agriculture: Approaches, applications, future perspectives, and associated challenges. Malay. J. Halal Res. 2020, 3, 6–16. [Google Scholar] [CrossRef]
- Clarke, J.L.; Zhang, P. Plant biotechnology for food security and bioeconomy. Plant Mol. Biol. 2013, 83, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FAOSTAT. Food and Agriculture Data. Available online: http://www.fao.org/faostat/en/#home (accessed on 25 May 2021).
- Stamm, P.; Ramamoorthy, R.; Kumar, P.P. Feeding the extra billions: Strategies to improve crops and enhance future food security. Plant Biotech. Rep. 2011, 5, 107–120. [Google Scholar] [CrossRef]
- Jones, J.D.; Witek, K.; Verweij, W.; Jupe, F.; Cooke, D.; Dorling, S.; Tomlinson, L.; Smoker, M.; Perkins, S.; Foster, S. Elevating crop disease resistance with cloned genes. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, Z.; Khan, S.H.; Ahmad, A. Challenges and future perspective of CRISPR/Cas technology for crop improvement. In CRISPR Crops; Springer: Singapore, 2021; pp. 289–306. [Google Scholar]
- Razzaq, A.; Saleem, F.; Kanwal, M.; Mustafa, G.; Yousaf, S.; Imran Arshad, H.M.; Hameed, M.K.; Khan, M.S.; Joyia, F.A. Modern trends in plant genome editing: An inclusive review of the CRISPR/Cas9 toolbox. Int. J. Mol. Sci. 2019, 20, 4045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasheed, A.; Hassan, M.; Aamer, M.; Bian, J.; Xu, Z.; He, X.; Wu, Z. Iron toxicity, tolerance and quantitative trait loci mapping in rice; a review. Appl. Ecol. Environ. Res. 2020, 18, 7483–7498. [Google Scholar] [CrossRef]
- Rasheed, A.; Hassan, M.U.; Aamer, M.; Batool, M.; Sheng, F.; Ziming, W.; Huijie, L. A critical review on the improvement of drought stress tolerance in rice (Oryza sativa L.). Not. Bot. Horti Agrobot. Cluj-Napoca 2020, 48, 1756–1788. [Google Scholar] [CrossRef]
- Rasheed, A.; Fahad, S.; Hassan, M.U.; Tahir, M.M.; Aamer, M.; Wu, Z. A review on aluminum toxicity and quantitative trait loci maping in rice (Oryza sative L). Appl. Ecol. Environ. Res. 2020, 18, 3951–3961. [Google Scholar] [CrossRef]
- Rasheed, A.; Fahad, S.; Aamer, M.; Hassan, M.U.; Tahir, M.M.; Wu, Z. Role of genetic factors in regulating cadmium uptake, transport and accumulation mechanisms and quantitative trait loci mapping in rice. a review. Appl. Ecol. Environ. Res. 2020, 18, 4005–4023. [Google Scholar] [CrossRef]
- Rasheed, A.; Hassan, M.U.; Fahad, S.; Aamer, M.; Batool, M.; Ilyas, M.; Shang, F.; Wu, Z.; Li, H. Heavy metals stress and plants defense responses. In Sustainable Soil and Land Management and Climate Change; CRC Press: Boca Raton, FL, USA; Taylor and Francis Group: Abingdon, UK, 2021; pp. 57–82. [Google Scholar]
- Rasheed, A.; Wassan, G.M.; Khanzada, H.; Solangi, A.M.; Aamer, M.; Ruicai, H.; Jianmin, B.; Ziming, W. QTL underlying iron toxicity tolerance at seedling stage in backcross recombinant inbred lines (BRILs) population of rice using high density genetic map. Not. Bot. Horti Agrobot. Cluj-Napoca 2021, 49, 12158. [Google Scholar] [CrossRef]
- Rasheed, A.; Wassan, G.M.; Khanzada, H.; Solangi, A.M.; Han, R.; Li, H.; Bian, J.; Wu, Z. Identification of genomic regions at seedling related traits in response to aluminium toxicity using a new high-density genetic map in rice (Oryza sativa L.). Genet. Res. Crop Evol. 2021, 68, 1889–1903. [Google Scholar] [CrossRef]
- Samanta, M.K.; Dey, A.; Gayen, S. CRISPR/Cas9: An advanced tool for editing plant genomes. Trans. Res. 2016, 25, 561–573. [Google Scholar] [CrossRef]
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for crop improvement: An update review. Front. Plant Sci. 2018, 9, 985. [Google Scholar] [CrossRef] [PubMed]
- Gillani, S.F.; Rasheed, A.; Majeed, Y.; Tariq, H.; Yunling, P. Recent advancements on use of CRISPR/Cas9 in maize yield and quality improvement. Not. Bot. Horti Agrobot. Cluj-Napoca 2021, 49, 12459. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas III, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotech. 2013, 31, 397–405. [Google Scholar] [CrossRef] [Green Version]
- Puchta, H. Applying CRISPR/Cas for genome engineering in plants: The best is yet to come. Curr. Opin. Plant Biol. 2017, 36, 1–8. [Google Scholar] [CrossRef]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 1–5. [Google Scholar]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Ann. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Zhou, J.; Li, D.; Wang, G.; Wang, F.; Kunjal, M.; Joldersma, D.; Liu, Z. Application and future perspective of CRISPR/Cas9 genome editing in fruit crops. J. Integr. Plant Biol. 2020, 62, 269–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pourcel, C.; Salvignol, G.; Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663. [Google Scholar] [CrossRef] [Green Version]
- Leibowitz, M.L.; Papathanasiou, S.; Doerfler, P.A.; Blaine, L.J.; Sun, L.; Yao, Y.; Zhang, C.-Z.; Weiss, M.J.; Pellman, D. Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat. Genet. 2021, 53, 889–905. [Google Scholar] [CrossRef]
- Zhan, X.; Lu, Y.; Zhu, J.K.; Botella, J.R. Genome editing for plant research and crop improvement. J. Integr. Plant Biol. 2021, 63, 3–33. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Sharma, S.; Vakhlu, J. Evolution and Biology of CRISPR System: A New Era Tool for Genome Editing in Plants. Bot. Rev. 2021, 1–22. [Google Scholar] [CrossRef]
- Sharma, A.; Badola, P.K.; Trivedi, P.K. CRISPR-Cas9 System for agriculture crop improvement. In Genome Engineering for Crop Improvement; Wiley: Hoboken, NJ, USA, 2021; pp. 97–111. [Google Scholar]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef]
- Namo, F.M.; Belachew, G.T. Genome editing technologies for crop improvement: Current status and future prospective. Plant Cell Biotech. Mol. Biol. 2021, 22, 1–19. [Google Scholar]
- Mishra, R.; Zhao, K. Genome editing technologies and their applications in crop improvement. Plant Biotech. Rep. 2018, 12, 57–68. [Google Scholar] [CrossRef]
- Townsend, J.A.; Wright, D.A.; Winfrey, R.J.; Fu, F.; Maeder, M.L.; Joung, J.K.; Voytas, D.F. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 2009, 459, 442–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Maeder, M.L.; Unger-Wallace, E.; Hoshaw, J.P.; Reyon, D.; Christian, M.; Li, X.; Pierick, C.J.; Dobbs, D.; Peterson, T. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. USA 2010, 107, 12028–12033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puchta, H. The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J. Exp. Bot. 2005, 56, 1–14. [Google Scholar] [CrossRef] [Green Version]
- D’Halluin, K.; Vanderstraeten, C.; Van Hulle, J.; Rosolowska, J.; Van Den Brande, I.; Pennewaert, A.; D’Hont, K.; Bossut, M.; Jantz, D.; Ruiter, R. Targeted molecular trait stacking in cotton through targeted double-strand break induction. Plant Biotech. J. 2013, 11, 933–941. [Google Scholar] [CrossRef] [Green Version]
- Djukanovic, V.; Smith, J.; Lowe, K.; Yang, M.; Gao, H.; Jones, S.; Nicholson, M.G.; West, A.; Lape, J.; Bidney, D. Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P 450-like gene (MS 26) using a re-designed I–C reI homing endonuclease. Plant J. 2013, 76, 888–899. [Google Scholar] [CrossRef]
- Durai, S.; Mani, M.; Kandavelou, K.; Wu, J.; Porteus, M.H.; Chandrasegaran, S. Zinc finger nucleases: Custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005, 33, 5978–5990. [Google Scholar] [CrossRef]
- Carlson, D.F.; Fahrenkrug, S.C.; Hackett, P.B. Targeting DNA with fingers and TALENs. Mol. Ther. Nucleic Acids 2012, 1, e3. [Google Scholar] [CrossRef]
- Carroll, D.; Morton, J.J.; Beumer, K.J.; Segal, D.J. Design, construction and in vitro testing of zinc finger nucleases. Nat. Protoc. 2006, 1, 1329–1341. [Google Scholar] [CrossRef]
- Minczuk, M.; Papworth, M.A.; Miller, J.C.; Murphy, M.P.; Klug, A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 2008, 36, 3926–3938. [Google Scholar] [CrossRef] [Green Version]
- Gaj, T.; Guo, J.; Kato, Y.; Sirk, S.J.; Barbas, C.F. Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat. Methods 2012, 9, 805–807. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.-X.; Zhang, Y.; Yin, H. Genome editing with mRNA encoding ZFN, TALEN, and Cas9. Mol. Ther. 2019, 27, 735–746. [Google Scholar] [CrossRef] [Green Version]
- Schornack, S.; Meyer, A.; Römer, P.; Jordan, T.; Lahaye, T. Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J. Plant Physiol. 2006, 163, 256–272. [Google Scholar] [CrossRef] [PubMed]
- Römer, P.; Hahn, S.; Jordan, T.; Strauss, T.; Bonas, U.; Lahaye, T. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 2007, 318, 645–648. [Google Scholar] [CrossRef] [Green Version]
- Shan, Q.; Zhang, Y.; Chen, K.; Zhang, K.; Gao, C. Creation of fragrant rice by targeted knockout of the Os BADH 2 gene using TALEN technology. Plant Biotech. J. 2015, 13, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Králová, M.; Bergougnoux, V.; Frébort, I. CRISPR/Cas9 genome editing in ergot fungus Clavicepspurpurea. J. Biotechnol. 2021, 325, 341–354. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.-L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotech. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Gao, H.; Smith, J.; Yang, M.; Jones, S.; Djukanovic, V.; Nicholson, M.G.; West, A.; Bidney, D.; Falco, S.C.; Jantz, D. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2010, 61, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, F.; Li, X.; Baller, J.A.; Qi, Y.; Starker, C.G.; Bogdanove, A.J.; Voytas, D.F. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 2013, 161, 20–27. [Google Scholar] [CrossRef] [Green Version]
- Gurushidze, M.; Hensel, G.; Hiekel, S.; Schedel, S.; Valkov, V.; Kumlehn, J. True-breeding targeted gene knock-out in barley using designer TALE-nuclease in haploid cells. PLoS ONE 2014, 9, e92046. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genom. 2014, 41, 63–68. [Google Scholar] [CrossRef]
- Kusano, H.; Onodera, H.; Kihira, M.; Aoki, H.; Matsuzaki, H.; Shimada, H. A simple Gateway-assisted construction system of TALEN genes for plant genome editing. Sci. Rep. 2016, 6, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Yang, F.; Zhang, J.; Liu, H.; Rahman, S.; Islam, S.; Ma, W.; She, M. Application of CRISPR/Cas9 in Crop quality improvement. Int. J. Mol. Sci. 2021, 22, 4206. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbull, C.; Lillemo, M.; Hvoslef-Eide, T.A. Global Regulation of Genetically Modified Crops Amid the Gene Edited Crop Boom–A Review. Front. Plant Sci. 2021, 12, 258. [Google Scholar] [CrossRef] [PubMed]
- Symington, L.S.; Gautier, J. Double-strand break end resection and repair pathway choice. Ann. Rev. Genet. 2011, 45, 247–271. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [Green Version]
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009, 155, 733–740. [Google Scholar] [CrossRef] [Green Version]
- Brouns, S.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.; Snijders, A.P.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; Van Der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef] [Green Version]
- Garneau, J.E.; Dupuis, M.-È.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadán, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Rahim, J.; Gulzar, S.; Zahid, R.; Rahim, K. A Systematic review on the comparison of molecular gene editing tools. Int. J. Innov. Sci. Res. Tech. 2021, 6, 1–8. [Google Scholar]
- Eş, I.; Gavahian, M.; Marti-Quijal, F.J.; Lorenzo, J.M.; Khaneghah, A.M.; Tsatsanis, C.; Kampranis, S.C.; Barba, F.J. The application of the CRISPR-Cas9 genome editing machinery in food and agricultural science: Current status, future perspectives, and associated challenges. Biotech. Adv. 2019, 37, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Sauer, N.J.; Mozoruk, J.; Miller, R.B.; Warburg, Z.J.; Walker, K.A.; Beetham, P.R.; Schöpke, C.R.; Gocal, G.F. Oligonucleotide-directed mutagenesis for precision gene editing. Plant Biotech. J. 2016, 14, 496–502. [Google Scholar] [CrossRef] [Green Version]
- Upadhyay, S.K.; Kumar, J.; Alok, A.; Tuli, R. RNA-guided genome editing for target gene mutations in wheat. G3 Genes Genomes Genet. 2013, 3, 2233–2238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhardwaj, A.; Nain, V. TALENs—An indispensable tool in the era of CRISPR: A mini review. J. Genet. Eng. Biotechnol. 2021, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Khandagale, K.; Nadaf, A. Genome editing for targeted improvement of plants. Plant Biotechnol. Rep. 2016, 10, 327–343. [Google Scholar] [CrossRef]
- Prajapat, R.K.; Mathur, M.; Upadhyay, T.K.; Lal, D.; Maloo, S.; Sharma, D. Genome Editing for Crop Improvement. In Crop Improvement; CRC Press: Boca Raton, FL, USA, 2021; pp. 111–123. [Google Scholar]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [Green Version]
- Hoikkala, V.; Ravantti, J.; Díez-Villaseñor, C.; Tiirola, M.; Conrad, R.A.; McBride, M.J.; Moineau, S.; Sundberg, L.-R. Cooperation between different CRISPR-Cas types enables adaptation in an RNA-targeting system. Mbio 2021, 12, e03338-20. [Google Scholar] [CrossRef]
- Liao, C.; Beisel, C.L. The tracrRNA in CRISPR biology and technologies. Annu. Rev. Genet. 2021, 55. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotech. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Carabias, A.; Fuglsang, A.; Temperini, P.; Pape, T.; Sofos, N.; Stella, S.; Erlendsson, S.; Montoya, G. Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3. Nat. Commun. 2021, 12, 4476. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Shen, B.; Zhang, C.; Huang, X.; Zhang, Y. sgRNAcas9: A software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS ONE 2014, 9, e100448. [Google Scholar] [CrossRef]
- Stemmer, M.; Thumberger, T.; del Sol Keyer, M.; Wittbrodt, J.; Mateo, J.L. CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 2015, 10, e0124633. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, A.; Murik, O.; Bowler, C.; Tirichine, L. PhytoCRISP-Ex: A web-based and stand-alone application to find specific target sequences for CRISPR/CAS editing. BMC Bioinf. 2016, 17, 261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Zhang, Y.; Yu, H.; Pan, D.; Wang, Y.; Wang, Y.; Li, F.; Liu, C.; Nan, H.; Chen, W. Programmed genome editing by a miniature CRISPR-Cas12f nuclease. J. Nat. Chem. Biol. 2021, 17, 1132–1138. [Google Scholar] [CrossRef]
- Chari, R.; Yeo, N.C.; Chavez, A.; Church, G.M. sgRNA Scorer 2.0: A species-independent model to predict CRISPR/Cas9 activity. ACS Synth. Biol. 2017, 6, 902–904. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Liu, H.; Liu, J.; Cheng, S.; Peng, Y.; Zhang, Q.; Yan, J.; Liu, H.-J.; Chen, L.-L. CRISPR-Local: A local single-guide RNA (sgRNA) design tool for non-reference plant genomes. Bioinformatics 2018, 35, 2501–2503. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, G.; Li, J.; Zhang, X.; Huang, S.; Xiang, S.; Hu, X.; Liu, C. CRISPRlnc: A manually curated database of validated sgRNAs for lncRNAs. Nucleic Acids Res. 2019, 47, D63–D68. [Google Scholar] [CrossRef] [Green Version]
- Braatz, J.; Harloff, H.-J.; Mascher, M.; Stein, N.; Himmelbach, A.; Jung, C. CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 2017, 174, 935–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [Green Version]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.D.; Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotech. 2013, 31, 691–693. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotech. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Gao, C. The future of CRISPR technologies in agriculture. Nat. Rev. Mol. Cell Biol. 2018, 19, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, Y.G. CRISPR/Cas9-based multiplex genome editing in monocot and dicot plants. Curr. Protoc. Mol. Biol. 2016, 115. [Google Scholar] [CrossRef] [PubMed]
- Ricroch, A.; Clairand, P.; Harwood, W. Use of CRISPR systems in plant genome editing: Toward new opportunities in agriculture. Emerg. Top. Life Sci. 2017, 1, 169–182. [Google Scholar]
- Zhang, D.; Li, Z.; Li, J.-F. Targeted gene manipulation in plants using the CRISPR/Cas technology. J. Genet. Genom. 2016, 43, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Zhang, J.; Botella, J.R.; Ma, C.; Kong, F.; Liu, B.; Zhu, J.-K. Perspectives on the application of genome-editing technologies in crop breeding. Mol. Plant 2019, 12, 1047–1059. [Google Scholar] [CrossRef] [Green Version]
- Schindele, A.; Dorn, A.; Puchta, H. CRISPR/Cas brings plant biology and breeding into the fast lane. Curr. Opin. Biotechnol. 2020, 61, 7–14. [Google Scholar] [CrossRef]
- Tan, Y.-y.; Du, H.; Wu, X.; Liu, Y.-h.; Jiang, M.; Song, S.-y.; Wu, L.; Shu, Q.-y. Gene editing: An instrument for practical application of gene biology to plant breeding. J. Zhejiang Univ.-Sci. B 2020, 21, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Malzahn, A.A.; Sretenovic, S.; Qi, Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 2019, 5, 778–794. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, C.; Zhang, H.; Zhu, H. CRISPR/Cas9-mediated gene editing revolutionizes the improvement of horticulture food crops. J. Agric. Food Chem. 2021. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.; Kim, J.; Cho, S.W.; Choi, Y.; Kim, J.-S.; Coupland, G. Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 2015, 241, 271–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Mao, Y.; Ha, S.; Liu, W.; Botella, J.R.; Zhu, J.-K. A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 2016, 35, 1519–1533. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Wang, G.; Ma, S.; Xie, X.; Wu, X.; Zhang, X.; Wu, Y.; Zhao, P.; Xia, Q. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 2015, 87, 99–110. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N. The CRISPR/C as9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotech. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]
- Xu, R.-F.; Li, H.; Qin, R.-Y.; Li, J.; Qiu, C.-H.; Yang, Y.-C.; Ma, H.; Li, L.; Wei, P.-C.; Yang, J.-B. Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 2015, 5, 11491. [Google Scholar] [CrossRef] [Green Version]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.; Østergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, L.; Wang, C.; Fu, Y.; Wang, J.; Liu, Q.; Zhang, X.; Yan, C.; Qian, Q.; Wang, K. QTL editing confers opposing yield performance in different rice varieties. J. Integr. Plant Biol. 2018, 60, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Usman, B.; Nawaz, G.; Zhao, N.; Liao, S.; Qin, B.; Liu, F.; Liu, Y.; Li, R. Programmed Editing of Rice (Oryza sativa L.) OsSPL16 gene using CRISPR/Cas9 improves grain yield by modulating the expression of pyruvate enzymes and cell cycle proteins. Int. J. Mol. Sci. 2021, 22, 249. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Guo, S.; Tian, S.; Zhang, J.; Ren, Y.; Li, M.; Gong, G.; Zhang, H.; Xu, Y. CRISPR/Cas9-mediated mutagenesis of ClBG1 decreased seed size and promoted seed germination in watermelon. Hort. Res. 2021, 8, 1–12. [Google Scholar] [CrossRef]
- Monsur, M.B.; Ni, C.; Xiangjin, W.; Lihong, X.; Guiai, J.; Shaoqing, T.; Sreenivasulu, N.; Gaoneng, S.; Peisong, H. Improved eating and cooking quality of indica rice cultivar YK17 via adenine base editing of waxya allele of granule-bound starch synthase I (GBSS I). Rice Sci. 2021, 1, 427–430. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Z.; Bai, J.; Tao, X.; Wang, L.; Zhang, H.; Zhu, J.-K. Disruption of MIR396e and MIR396f improves rice yield under nitrogen-deficient conditions. Nat. Sci. Rev. 2020, 7, 102–112. [Google Scholar] [CrossRef] [Green Version]
- Miao, C.; Wang, Z.; Zhang, L.; Yao, J.; Hua, K.; Liu, X.; Shi, H.; Zhu, J.-K. The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat. Commun. 2019, 10, 3822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.; Li, R. Generation of high yielding and fragrant rice (Oryza sativa L.) Lines by CRISPR/Cas9 targeted mutagenesis of three homoeologs of cytochrome P450 gene family and OsBADH2 and transcriptome and proteome profiling of revealed changes triggered by mutations. Plants 2020, 9, 788. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yang, Y.; Guo, M.; Zhong, C.; Yan, C.; Sun, S. Targeted mutagenesis of amino acid transporter genes for rice quality improvement using the CRISPR/Cas9 system. Crop J. 2020, 8, 457–464. [Google Scholar] [CrossRef]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 With the CRISPR–Cas9 System. Front. Plant Sci. 2020, 10, 1663. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Jiao, G.; Sun, Y.; Chen, J.; Zhong, Y.; Yan, L.; Jiang, D.; Ma, Y.; Xia, L. Modification of starch composition, structure and properties through editing of TaSBEIIa in both winter and spring wheat varieties by CRISPR/Cas9. Plant Biotech. J. 2020, 19, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Okuzaki, A.; Ogawa, T.; Koizuka, C.; Kaneko, K.; Inaba, M.; Imamura, J.; Koizuka, N. CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 2018, 131, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Ellison, E.E.; Nagalakshmi, U.; Gamo, M.E.; Huang, P.-J.; Dinesh-Kumar, S.; Voytas, D.F. Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat. Plants 2020, 6, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Morineau, C.; Bellec, Y.; Tellier, F.; Gissot, L.; Kelemen, Z.; Nogué, F.; Faure, J.D. Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Botech. J. 2017, 15, 729–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, Y.; Cai, S.; Hu, L.; Yang, Y.; Amoo, O.; Fan, C.; Zhou, Y. CRISPR/Cas9-mediated genome editing reveals differences in the contribution of INDEHISCENT homologues to pod shatter resistance in Brassica napus L. Theor. Appl. Genet. 2019, 132, 2111–2123. [Google Scholar] [CrossRef]
- Zaman, Q.U.; Chu, W.; Hao, M.; Shi, Y.; Sun, M.; Sang, S.-F.; Mei, D.; Cheng, H.; Liu, J.; Li, C. CRISPR/Cas9-Mediated Multiplex Genome Editing of JAGGED Gene in Brassica napus L. Biomolecules 2019, 9, 725. [Google Scholar] [CrossRef] [Green Version]
- Soyk, S.; Müller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jiménez-Gómez, J.M.; Lippman, Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017, 49, 162–168. [Google Scholar] [CrossRef]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef] [Green Version]
- Malnoy, M.; Viola, R.; Jung, M.-H.; Koo, O.-J.; Kim, S.; Kim, J.-S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Toda, E.; Koiso, N.; Takebayashi, A.; Ichikawa, M.; Kiba, T.; Osakabe, K.; Osakabe, Y.; Sakakibara, H.; Kato, N.; Okamoto, T. An efficient DNA-and selectable-marker-free genome-editing system using zygotes in rice. Nat. Plants 2019, 5, 363. [Google Scholar] [CrossRef]
- Espley, R.V.; Leif, D.; Plunkett, B.; McGhie, T.; Henry-Kirk, R.; Hall, M.; Johnston, J.W.; Punter, M.P.; Boldingh, H.; Nardozza, S. Red to brown: An elevated anthocyanic response in apple drives ethylene to advance maturity and fruit flesh browning. Front. Plant Sci. 2019, 10, 1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klap, C.; Yeshayahou, E.; Bolger, A.M.; Arazi, T.; Gupta, S.K.; Shabtai, S.; Usadel, B.; Salts, Y.; Barg, R. Tomato facultative parthenocarpy results from Sl AGAMOUS-LIKE 6 loss of function. Plant Biotech. J. 2017, 15, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Wang, N. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 2014, 9, e93806. [Google Scholar]
- Lemmon, Z.H.; Reem, N.T.; Dalrymple, J.; Soyk, S.; Swartwood, K.E.; Rodriguez-Leal, D.; Van Eck, J.; Lippman, Z.B. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 2018, 4, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Naim, F.; Dugdale, B.; Kleidon, J.; Brinin, A.; Shand, K.; Waterhouse, P.; Dale, J. Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transg. Res. 2018, 27, 451–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Gallagher, J.; Arevalo, E.D.; Chen, R.; Skopelitis, T.; Wu, Q.; Bartlett, M.; Jackson, D. Enhancing grain-yield-related traits by CRISPR–Cas9 promoter editing of maize CLE genes. Nat. Plants 2021, 7, 287–294. [Google Scholar] [CrossRef]
- Ueta, R.; Abe, C.; Watanabe, T.; Sugano, S.S.; Ishihara, R.; Ezura, H.; Osakabe, Y.; Osakabe, K. Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 2017, 7, 507. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Chen, L.; Liu, X.; Guo, C.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotech. J. 2018, 16, 176–185. [Google Scholar] [CrossRef] [Green Version]
- Sashidhar, N.; Harloff, H.J.; Potgieter, L.; Jung, C. Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotech. J. 2020, 18, 2241–2250. [Google Scholar] [CrossRef] [Green Version]
- Badhan, S.; Ball, A.S.; Mantri, N. First report of CRISPR/Cas9 mediated DNA-free editing of 4CL and RVE7 genes in chickpea protoplasts. Int. J. Mol. Sci. 2021, 22, 396. [Google Scholar] [CrossRef]
- Zheng, M.; Zhang, L.; Tang, M.; Liu, J.; Liu, H.; Yang, H.; Fan, S.; Terzaghi, W.; Wang, H.; Hua, W. Knockout of two Bna MAX 1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotech. J. 2020, 18, 644–654. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Jia, S.; Yobi, A.; Ge, Z.; Sato, S.J.; Zhang, C.; Angelovici, R.; Clemente, T.E.; Holding, D.R. Editing of an alpha-kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol. 2018, 177, 1425–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, M.; Turesson, H.; Nicolia, A.; Fält, A.-S.; Samuelsson, M.; Hofvander, P. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 2017, 36, 117–128. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 2016, 7, 377. [Google Scholar] [CrossRef] [Green Version]
- Tian, S.; Jiang, L.; Gao, Q.; Zhang, J.; Zong, M.; Zhang, H.; Ren, Y.; Guo, S.; Gong, G.; Liu, F. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2017, 36, 399–406. [Google Scholar] [CrossRef]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genom. Biol. 2015, 16, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 2018, 94, 513–524. [Google Scholar] [CrossRef] [Green Version]
- Qi, W.; Zhu, T.; Tian, Z.; Li, C.; Zhang, W.; Song, R. High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotech. 2016, 16, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.-L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, N.; Alok, A.; Kaur, N.; Pandey, P.; Awasthi, P.; Tiwari, S. CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genom. 2018, 18, 89–99. [Google Scholar] [CrossRef]
- Jiang, W.Z.; Henry, I.M.; Lynagh, P.G.; Comai, L.; Cahoon, E.B.; Weeks, D.P. Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant. Biotech. J. 2017, 15, 648–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín-Pizarro, C.; Triviño, J.C.; Posé, D. Functional analysis of the TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. J. Exp. Bot. 2019, 70, 885–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Sun, S.; Whelan, J.; Shou, H. CRISPR/Cas9-Mediated knockout of GmFATB1 Significantly reduced the amount of saturated fatty acids in soybean seeds. Int. J. Mol. Sci. 2021, 22, 3877. [Google Scholar] [CrossRef]
- Singh, R. Genome Editing for Plant Disease Resistance. In Emerging Trends in Plant Pathology; Springer: Berlin/Heidelberg, Germany, 2021; pp. 577–590. [Google Scholar]
- Khan, Z.; Saboor, T.; Ashfaq, M.; Saddique, A.; Khanum, P. Disease resistance in crops through CRISPR/Cas. In CRISPR Crops; Springer: Berlin/Heidelberg, Germany, 2021; pp. 151–175. [Google Scholar]
- Al-Sadi, A.; Al-Moqbali, H.; Al-Yahyai, R.; Al-Said, F. AFLP data suggest a potential role for the low genetic diversity of acid lime (Citrus aurantifoliaSwingle) in Oman in the outbreak of witches’ broom disease of lime. Euphytica 2012, 188, 285–297. [Google Scholar] [CrossRef]
- Kettles, G.J.; Kanyuka, K. Dissecting the molecular interactions between wheat and the fungal pathogen Zymoseptoriatritici. Front. Plant Sci. 2016, 7, 508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jouanin, A.; Gilissen, L.J.; Schaart, J.G.; Leigh, F.J.; Cockram, J.; Wallington, E.J.; Boyd, L.A.; Van Den Broeck, H.C.; Van der Meer, I.M.; America, A. CRISPR/Cas9 gene editing of gluten in wheat to reduce gluten content and exposure—reviewing methods to screen for coeliac safety. Front. Nutr. 2020, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.K.; Mandal, S.; Tiwari, A.; Monachesi, C.; Catassi, G.N.; Srivastava, A.; Gatti, S.; Lionetti, E.; Catassi, C. Current status and perspectives on the application of CRISPR/Cas9 Gene-editing system to develop a low-gluten, non-transgenic wheat variety. Foods 2021, 10, 2351. [Google Scholar] [CrossRef]
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Čermák, T.; Voytas, D.F.; Choi, I.R.; Chadha-Mohanty, P. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotech. J. 2018, 16, 1918–1927. [Google Scholar] [CrossRef] [Green Version]
- Sam, V.H.; Van, P.T.; Ha, N.T.; Ha, N.T.T.; Huong, P.T.T.; Hoi, P.X.; Phuong, N.D.; Le Quyen, C. Design and transfer of OsSWEET14-editing T-DNA construct to bac thom 7 rice cultivar. Acad. J. Biol. 2021, 43. [Google Scholar] [CrossRef]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.-G.; Zhao, K. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef]
- Zhou, J.; Peng, Z.; Long, J.; Sosso, D.; Liu, B.; Eom, J.S.; Huang, S.; Liu, S.; Vera Cruz, C.; Frommer, W.B. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015, 82, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Zhang, Y.; Orbović, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene Cs LOB 1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 2017, 15, 817–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene Cs LOB 1 promoter in citrus. Plant Biotech. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of Ta EDR 1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaidi, S.S.-e.-A.; Tashkandi, M.; Mansoor, S.; Mahfouz, M.M. Engineering plant immunity: Using CRISPR/Cas9 to generate virus resistance. Front. Plant Sci. 2016, 7, 1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017, 89, 1251–1262. [Google Scholar] [CrossRef] [Green Version]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.; Si, X.; Zhang, Y.; Zhang, H.; Zhang, F.; Gao, C. Conferring DNA virus resistance with high specificity in plants using virus-inducible genome-editing system. Genome Biol. 2018, 19, 197. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Zheng, Q.; Yi, X.; An, H.; Zhao, Y.; Ma, S.; Zhou, G. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotech. J. 2018, 16, 1415–1423. [Google Scholar] [CrossRef] [Green Version]
- Pramanik, D.; Shelake, R.M.; Park, J.; Kim, M.J.; Hwang, I.; Park, Y.; Kim, J.-Y. CRISPR/Cas9-Mediated Generation of Pathogen-Resistant Tomato against Tomato Yellow Leaf Curl Virus and Powdery Mildew. Int. J. Mol. Sci. 2021, 22, 1878. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.R.; Varanavasiappan, S.; Kokiladevi, E.; Ramanathan, A.; Kumar, K. Genome editing of rice PFT1 gene to study its role in rice sheath blight disease resistance. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 2356–2364. [Google Scholar] [CrossRef]
- Nagy, E.D.; Stevens, J.L.; Yu, N.; Hubmeier, C.S.; LaFaver, N.; Gillespie, M.; Gardunia, B.; Cheng, Q.; Johnson, S.; Vaughn, A.L. Novel disease resistance gene paralogs created by CRISPR/Cas9 in soy. Plant Cell Rep. 2021, 40, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Ruyi, R.; Qiang, Z.; Futai, N.; Qiu, J.; Xiuqing, W.; Jicheng, W. Breeding for PVY resistance in tobacco LJ911 using CRISPR/Cas9 technology. Crop Breed. Appl. Biotech. 2021, 21, e31682116. [Google Scholar] [CrossRef]
- Zhang, Z.; Ge, X.; Luo, X.; Wang, P.; Fan, Q.; Hu, G.; Xiao, J.; Li, F.; Wu, J. Simultaneous editing of two copies of Gh14-3-3d confers enhanced transgene-clean plant defense against Verticillium dahliae in allotetraploid upland cotton. Front. Plant Sci. 2018, 9, 842. [Google Scholar] [CrossRef] [PubMed]
- Tashkandi, M.; Ali, Z.; Aljedaani, F.; Shami, A.; Mahfouz, M.M. Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal. Behav. 2018, 13, e1525996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.-A.; Moon, H.; Park, C.-J. CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Rice 2019, 12, 1–13. [Google Scholar]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef]
- Ortigosa, A.; Gimenez-Ibanez, S.; Leonhardt, N.; Solano, R. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of Sl JAZ 2. Plant Biotech. J. 2019, 17, 665–673. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Lin, L.; Liu, D.; Wu, D.; Fang, Y.; Wu, J.; Wang, Y. CRISPR/Cas9-Mediated Multiplex Genome Editing of the BnWRKY11 and BnWRKY70 Genes in Brassica napus L. Int. J. Mol. Sci. 2018, 19, 2716. [Google Scholar] [CrossRef] [Green Version]
- Neequaye, M.; Stavnstrup, S.; Harwood, W.; Lawrenson, T.; Hundleby, P.; Irwin, J.; Troncoso-Rey, P.; Saha, S.; Traka, M.H.; Mithen, R. CRISPR-Cas9-mediated gene editing of MYB28 genes impair glucoraphanin accumulation of Brassica oleracea in the field. CRISPR J. 2021, 4, 416–426. [Google Scholar] [CrossRef]
- Gumtow, R.; Wu, D.; Uchida, J.; Tian, M. A Phytophthora palmivora extracellular cystatin-like protease inhibitor targets papain to contribute to virulence on papaya. Mol. Plant-Microbe Interact. 2018, 31, 363–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razzaq, M.K.; Aleem, M.; Mansoor, S.; Khan, M.A.; Rauf, S.; Iqbal, S.; Siddique, K.H. Omics and CRISPR-Cas9 approaches for molecular insight, functional gene analysis, and stress tolerance development in crops. Int. J. Mol. Sci. 2021, 22, 1292. [Google Scholar] [CrossRef]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genom. 2018, 18, 31–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, R.K.; Bharat, S.S.; Mishra, R. Engineering drought tolerance in plants through CRISPR/Cas genome editing. 3 Biotech. 2020, 10, 400. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Que, Z.; Xia, Y.; Tang, N.; Li, D.; He, R.; Cao, M. Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. J. Plant Biol. 2017, 60, 539–547. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotech. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Liao, S.; Qin, X.; Luo, L.; Han, Y.; Wang, X.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.; Li, R. CRISPR/Cas9-induced mutagenesis of semi-rolled leaf1, 2 confers curled leaf phenotype and drought tolerance by influencing protein expression patterns and ros scavenging in rice (Oryza sativa L.). Agronomy 2019, 9, 728. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 1–10. [Google Scholar] [CrossRef] [Green Version]
- de Melo, B.P.; Lourenço-Tessutti, I.T.; Paixão, J.F.R.; Noriega, D.D.; Silva, M.C.M.; de Almeida-Engler, J.; Fontes, E.P.B.; Grossi-de-Sa, M.F. Transcriptional modulation of AREB-1 by CRISPRa improves plant physiological performance under severe water deficit. Sci. Rep. 2020, 10, 16231. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y.; Li, S.; Ren, B.; Yan, F.; Spetz, C.; Li, X.; Zhou, X.; Zhou, H. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms. Mol. Plant 2020, 13, 565–572. [Google Scholar] [CrossRef]
- Curtin, S.J.; Xiong, Y.; Michno, J.M.; Campbell, B.W.; Stec, A.O.; Čermák, T.; Starker, C.; Voytas, D.F.; Eamens, A.L.; Stupar, R.M. Crispr/cas9 and talen s generate heritable mutations for genes involved in small rna processing of glycine max and medicagotruncatula. Plant Biotech. J. 2018, 16, 1125–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corte, E.-D.; Mahmoud, L.M.; Moraes, T.S.; Mou, Z.; Grosser, J.W.; Dutt, M. Development of improved fruit, vegetable, and ornamental crops using the CRISPR/Cas9 genome editing technique. Plants 2019, 8, 601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.q.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef] [Green Version]
- Korotkova, A.; Gerasimova, S.; Khlestkina, E. Current achievements in modifying crop genes using CRISPR/Cas system. VavilovskiiZh. Genet. Sel. 2019, 8, 14422. [Google Scholar] [CrossRef]
- Butler, N.M.; Baltes, N.J.; Voytas, D.F.; Douches, D.S. Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front. Plant Sci. 2016, 7, 1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Zhang, C.; Liu, W.; Gao, W.; Liu, C.; Song, G.; Li, W.-X.; Mao, L.; Chen, B.; Xu, Y. An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci. Rep. 2016, 6, 1–11. [Google Scholar]
- Lu, Y.; Ye, X.; Guo, R.; Huang, J.; Wang, W.; Tang, J.; Tan, L.; Zhu, J.-K.; Chu, C.; Qian, Y. Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant 2017, 10, 1242–1245. [Google Scholar] [CrossRef] [Green Version]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, T.B.; Zhang, N.; Patel, D.; Martin, G.B. Generation of a collection of mutant tomato lines using pooled CRISPR libraries. Plant Physiol. 2017, 174, 2023–2037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Yu, H.; Zhang, Y.; Zhuang, F.; Song, X.; Gao, S.; Gao, C.; Li, J. Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol. Plant 2017, 10, 1238–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henikoff, S.; Comai, L. Single-nucleotide mutations for plant functional genomics. Ann. Rev. Plant Biol. 2003, 54, 375–401. [Google Scholar] [CrossRef]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotech. 2017, 35, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, Z.; Ni, H.; Xu, Y.; Chen, Q.; Jiang, L. CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Sci. China Life Sci. 2017, 60, 520–523. [Google Scholar] [CrossRef]
- Hua, K.; Tao, X.; Yuan, F.; Wang, D.; Zhu, J.-K. Precise A· T to G· C base editing in the rice genome. Mol. Plant 2018, 11, 627–630. [Google Scholar] [CrossRef] [Green Version]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Li, H.; Li, J.; Chen, J.; Yan, L.; Xia, L. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant 2020, 13, 671–674. [Google Scholar] [CrossRef]
- Tang, X.; Sretenovic, S.; Ren, Q.; Jia, X.; Li, M.; Fan, T.; Yin, D.; Xiang, S.; Guo, Y.; Liu, L. Plant prime editors enable precise gene editing in rice cells. Mol. Plant 2020, 13, 667–670. [Google Scholar] [CrossRef]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotech. 2015, 33, 1162–1164. [Google Scholar] [CrossRef]
- Yin, K.; Gao, C.; Qiu, J.-L. Progress and prospects in plant genome editing. Nat. Plants 2017, 3, 17107. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Q.; Shen, Y.; Hua, Y.; Wang, J.; Lin, J.; Wu, M.; Sun, T.; Cheng, Z.; Mercier, R. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotech. 2019, 37, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Lyzenga, W.J.; Pozniak, C.J.; Kagale, S. Advanced domestication: Harnessing the precision of gene editing in crop breeding. Plant Biotech. J. 2021, 19, 660–670. [Google Scholar] [CrossRef] [PubMed]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotech. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhang, X.; Sun, Y.; Zhang, J.; Du, W.; Guo, X.; Li, S.; Zhao, Y.; Xia, L. Efficient allelic replacement in rice by gene editing: A case study of the NRT1. 1B gene. J. Integr. Plant Biol. 2018, 60, 536–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callaway, E. CRISPR plants now subject to tough GM laws in European Union. Nature 2018, 560, 16–17. [Google Scholar] [CrossRef]
- Boettcher, M.; McManus, M.T. Choosing the right tool for the job: RNAi, TALEN, or CRISPR. Mol. Cell 2015, 58, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, E.; Lunge, A.; Agarwal, N. Strategies of genome editing in mycobacteria: Achievements and challenges. Tuberculosis 2016, 98, 132–138. [Google Scholar] [CrossRef]
- Karlson, C.K.S.; Noor, S.N.M.; Nolte, N.; Tan, B.C. CRISPR/dCas9-based systems: Mechanisms and applications in plant sciences. Plants 2021, 10, 2055. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-H.; Tee, L.Y.; Wang, X.-G.; Huang, Q.-S.; Yang, S.-H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Tao, X.; Han, P.; Wang, R.; Zhu, J.-K. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol. Plant 2019, 12, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Naeem, M.; Majeed, S.; Hoque, M.Z.; Ahmad, I.J.C. Latest developed strategies to minimize the off-target effects in CRISPR-Cas-mediated genome editing. Cells 2020, 9, 1608. [Google Scholar] [CrossRef] [PubMed]

| Crop | Tool | Gene | Trait | Reference |
|---|---|---|---|---|
| Rice | TALEN | OsBADH2 | Fragment rice | [48] |
| Rye | Cas9 | TrpE | Fungal resistance | [49] |
| Wheat | TALEN | TaMLO | Resistance to powdery resistance | [50] |
| Maize | MNs | LGI | Targeted mutagenesis | [51] |
| Cotton | EMNs | EPSPS | Tolerance to herbicide | [38] |
| Tobacco | TALENs | Sur A | Directed mutation | [52] |
| Barley | TALENs | Transgene | Resistance to powdery resistance | [53] |
| Maize | TALENs | ZMIPK | Phytic acid biosynthesis Liang | [54] |
| Potato | TALENs | StGBSS | Quality of tuber starch | [55] |
| Tool | Function | Year | Reference |
|---|---|---|---|
| tracr RNA | Act as reference point | 2021 | [76] |
| CRISPR design | RNA construction for targeted regions and assessment of off target effects | 2013 | [77] |
| Cas12j | Genome manipulation | 2021 | [78] |
| sgRNAcas9 | Speedy construction of sgRNA and fewer effects | 2014 | [79] |
| CCTop | Estimate target sgRNA sequence on the basis of off-target influences | 2015 | [80] |
| phytoCRISP.Ex | Cas9 target prediction | 2016 | [81] |
| Cas12 | Target recognition | 2021 | [82] |
| sgRNA Scorer 2.0 | sgRNA construction for many PAM locates | 2017 | [83] |
| CRISPR-Local | sgRNA construction for non-reference types | 2018 | [84] |
| CRISPRInc | Construct sgRNA for lncRNAs | 2019 | [85] |
| Gene | Crops | Trait | Technique | Reference |
|---|---|---|---|---|
| CLE | Maize | Grain yield | CRISPR/Cas9 | [131] |
| SLIAA9 | Tomato | Seedless fruit | CRIISPR/Cas9 | [132] |
| OsTB1 | Rice | High grain yield | CRIISPR/Cas9 | [114] |
| GmFT2a | Soybean | Delay in flowering time | CRIISPR/Cas9 | [133] |
| BnITPK | Rapeseed | Low phytic acid | CRIISPR/Cas9 | [134] |
| Bolc.GA4.a | Cabbage | Dwarf plant | CRIISPR/Cas9 | [106] |
| OsAAP10 | Rice | Good cooking quality | CRIISPR/Cas9 | [114] |
| RVE7 | Chickpea | Grain quality | CRIISPR/Cas9 B | [135] |
| BnaMAXI | Rapeseed | Yield | CRISPR/Cas9 | [136] |
| Wholek1gene | Sorghum | Increase lysine content | CRIISPR/Cas9 | [137] |
| TaSBEIIa | Wheat | High amylose content | CRIISPR/Cas9 | [116] |
| GBSS | Potato | Enhance amylose content | CRIISPR/Cas9 | [138] |
| P450 | Rice | High grain yield | CRIISPR/Cas9 | [113] |
| IPA | Rice | Enhanced yield | CRIISPR/Cas9 | [139] |
| CIPDS | Watermelon | Albino phenotype | CRIISPR/Cas9 | [140] |
| GS3 | Rice | Grain yield | CRIISPR/Cas9 | [115] |
| ANTI | Rice | Fruit color | CRIISPR/Cas9 | [141] |
| OsSPL16 | Rice | Grain yield | CRIISPR/Cas9 | [108] |
| lncRNA1459 | Tomato | Prolong shelf life | CRIISPR/Cas9 | [142] |
| PRL | Maize | Reduction in zein content | CRIISPR/Cas9 | [143] |
| GASR7 | Wheat | Grain weight | CRIISPR/Cas9 | [144] |
| PDS | Banana | Albino phenotype | CRIISPR/Cas9 | [145] |
| CsFAD2 | Camelina | Oleic acid | CRIISPR/Cas9 | [146] |
| FaTM6 | Strawberry | Flower development | CRIISPR/Cas9 | [147] |
| GmFATB1 | Soybean | Low saturated fatty acid | CRIISPR/Cas9 | [148] |
| Gene | Chromosomal Position | Locus | Pathogen | Crop | Function | Trait | Repair Tool | Editing Results | Reference |
|---|---|---|---|---|---|---|---|---|---|
| SIPeLo and SIMIO1 | Tomato | Enhance resistance to leaf curl virus | Enhance resistance to leaf curl virus | [168] | |||||
| OsSWEET14 | Rice | Bacterial leaf blight | Bacterial leaf blight resistance | [156] | |||||
| CsLOB1 | N/A | N/A | Xanthomonas citri | Citrus | Susceptibility to citrus canker | Resistance to citrus canker | NHEJ | Knockout | [159] |
| Gh14-3-3d | https://www.ncbi.nlm.nih.gov/nuccore/164652939 (accessed on 15 October 2020) | N/A | Verticillium dahliae | Cotton | Negative controller of resistance to disease | Verticillium wilt resistance | NHEJ | Knock-in | [172] |
| Ntab0942120 | Tobacco | Resistance to potato virus Y | Resistance to potato virus Y | [171] | |||||
| eLF4G | chr07:22114961..22123061 (+ strand) | Os07g0555200 | Tungro spherical virus | Rice | Starting factor for initiation | Resistance to tungro spherical virus | NHEJ | Knock out | [155] |
| Rpsl | Soybean | Diseases resistance | Diseases resistance | [170] | |||||
| Rp and Cp sequences | N/A | N/A | Yellow leaf curl virus | Tomato | Negative controller of resistance | Enhanced resistance again leaf curl virus | NHEJ | Knock out | [173] |
| OsSWEET11 | chr08:26725952..26728794 | Os08g0535200 | Bacterial blight | Rice | Resistance to bacterial blight | Resistance to bacterial blight | NHEJ | Knock out | [174] |
| OsSWEET14 | chr11:18171707..18174478 | Os11g0508600 | Bacterial blight | Rice | Resistance to bacterial blight | Resistance to bacterial blight | NHEJ | Knock out | [175] |
| SiMLOl | N/A | N/A | Tomato | Powdery mildew resistance gene | Resistance to powdery mildew | NHEJ | Knock out | [162] | |
| Jaz2 | Chr12: 2502581..2504643 | N/A | Pseudomonas syringae | Tomato | Bacterial speck resistance | Bacterial speck resistance | NHEJ | Knock out | [176] |
| WRKY70 | LK032201:212360-212875 | BnaA09g35840D | Brassica | Resistance to pathogens | NHEJ | Knock out | [177] | ||
| MYB28 | Brassica | Glucoraphanin accumulation | NHEJ | Knock out | [178] | ||||
| S-genes | N/A | N/A | Potato | Resistance to Phytophthora | Resistance to Phytophthora | NHEJ | Knock out | [138] | |
| TaMLO-A1 | Chromosome 4A: 519,570,414-519,575,284 | TraesCS4D02G318600 | Wheat | Resistance to mildew | Resistance to mildew | NHEJ | Knock out | [50] | |
| RGA2, Ced9 | 3:6700445-6700466 | GSMUA_Achr3G09290_001 | Fusarium oxysporum | Banana | Resistance to fusarium wilt | Resistance to fusarium wilt | NHEJ | Knock out | [162] |
| Mlo-7 | Chr13: (5335059..5339258 | VIT_00016304001 | Grape | Resistance to mildew | Resistance to mildew | NHEJ | Knock out | [124] | |
| PpalEPIC8 | N/A | N/A | Phytophthora palmivora | Papaya | Resistance to Phytophthora | Resistance to Phytophthora | NHEJ | Knock out | [179] |
| Gene | Chromosomal Position | Locus | Crop | Traits | Repair Pathway | Editing Results | References |
|---|---|---|---|---|---|---|---|
| OsMYB30 | Rice | Cold tolerance | [115] | ||||
| slmapk3 | Tomato | Drought tolerance | [181] | ||||
| OsALS1 | Rice | Herbicide tolerance | [190] | ||||
| 4CL, RVE7 | Chickpea | Drought tolerance | [135] | ||||
| SAPK2 | chr07:25717837..25722009 (+strand) | Os07g0622000 | Rice | Tolerance to salinity and drought | NHEJ | Knockout | [185] |
| TaDREB3 | 7D:27470774-27471448 | TraesCS7D02G052600 | Wheat | Tolerance to drought | NHEJ | Knockout | [182] |
| NPRI | 3:3740543-3741413 | Solyc03g026270.2 | Tomato | Tolerance to cold and drought stress | NHEJ | Knockout | [142,192] |
| ZmHKTI | N/A | N/A | Maize | Tolerance to salinity stress | NHEJ | Knockout | [193] |
| Drb2a | 12:5797459-5798459, 11:11249371-11250406 | GLYMA_12G075700 GLYMA_11G145900 | Soyabean | Tolerance to drought and salinity stress | NHEJ | Knockout | [191] |
| OsRR22, OsPDS | chr10:17076098..17081344 (- strand) chr03:4410090..4414779 (+ strand) | Os10g0463400 Os03g0184000 (OsPDS) | Rice | Tolerance to salinity stress | NHEJ | Knockout | [188,194] |
| OsAOX1a, | chr04:30287197..30289860 | Os04g0600200 | Rice | Drought resistance | NHEJ | Knockout | [104] |
| OsBADH2 | chr08:20379823..20385975 | Os08g0424500 | Rice | Abiotic stress resistance | HDR | Knockout | [91] |
| ALS1 | chr3:8175606-8177917 | PGSC0003DMG400034102 | Potato | Resistance to herbicide | HDR | Knockout | [195] |
| MIR169a | Chr03:4358994-4359219 | AT3G13405 | Arabidopsis | Drought resistance | HDR | Knockout | [196] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rasheed, A.; Gill, R.A.; Hassan, M.U.; Mahmood, A.; Qari, S.; Zaman, Q.U.; Ilyas, M.; Aamer, M.; Batool, M.; Li, H.; et al. A Critical Review: Recent Advancements in the Use of CRISPR/Cas9 Technology to Enhance Crops and Alleviate Global Food Crises. Curr. Issues Mol. Biol. 2021, 43, 1950-1976. https://doi.org/10.3390/cimb43030135
Rasheed A, Gill RA, Hassan MU, Mahmood A, Qari S, Zaman QU, Ilyas M, Aamer M, Batool M, Li H, et al. A Critical Review: Recent Advancements in the Use of CRISPR/Cas9 Technology to Enhance Crops and Alleviate Global Food Crises. Current Issues in Molecular Biology. 2021; 43(3):1950-1976. https://doi.org/10.3390/cimb43030135
Chicago/Turabian StyleRasheed, Adnan, Rafaqat Ali Gill, Muhammad Umair Hassan, Athar Mahmood, Sameer Qari, Qamar U. Zaman, Muhammad Ilyas, Muhammad Aamer, Maria Batool, Huijie Li, and et al. 2021. "A Critical Review: Recent Advancements in the Use of CRISPR/Cas9 Technology to Enhance Crops and Alleviate Global Food Crises" Current Issues in Molecular Biology 43, no. 3: 1950-1976. https://doi.org/10.3390/cimb43030135
APA StyleRasheed, A., Gill, R. A., Hassan, M. U., Mahmood, A., Qari, S., Zaman, Q. U., Ilyas, M., Aamer, M., Batool, M., Li, H., & Wu, Z. (2021). A Critical Review: Recent Advancements in the Use of CRISPR/Cas9 Technology to Enhance Crops and Alleviate Global Food Crises. Current Issues in Molecular Biology, 43(3), 1950-1976. https://doi.org/10.3390/cimb43030135








