Evaluation of Anti-Inflammatory Effects of Celery Leaf and Stem Extracts in LPS-Induced RAW 264.7 Cells Using Nitric Oxide Assay and LC-MS Based Metabolomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Celery Extracts
2.3. Cell Culture and Treatment
2.3.1. MTT Assay
2.3.2. NO Assay
2.4. Metabolomics Analysis
2.4.1. Collection of Culture Media and Metabolite Extraction from Cells
2.4.2. LC-MS/MS Analysis
2.4.3. Statistical Analysis
3. Results and Discussion
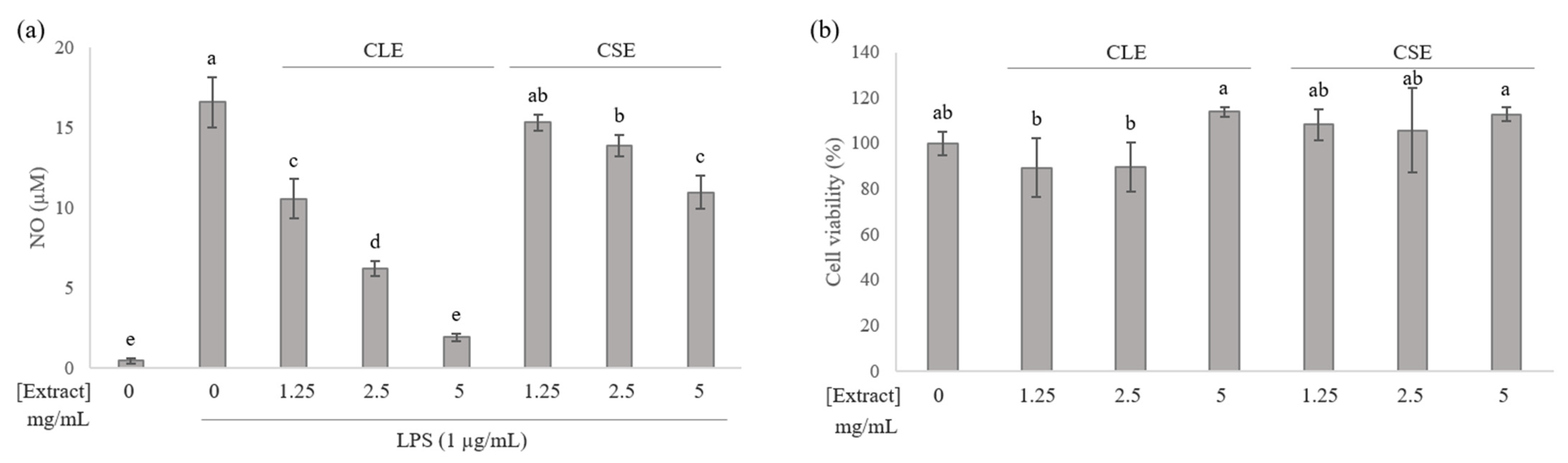
3.1. Effects of Celery Leaf and Stem Extracts on NO Production in RAW 264.7 Cells
3.2. Multivariate Statistical Analysis
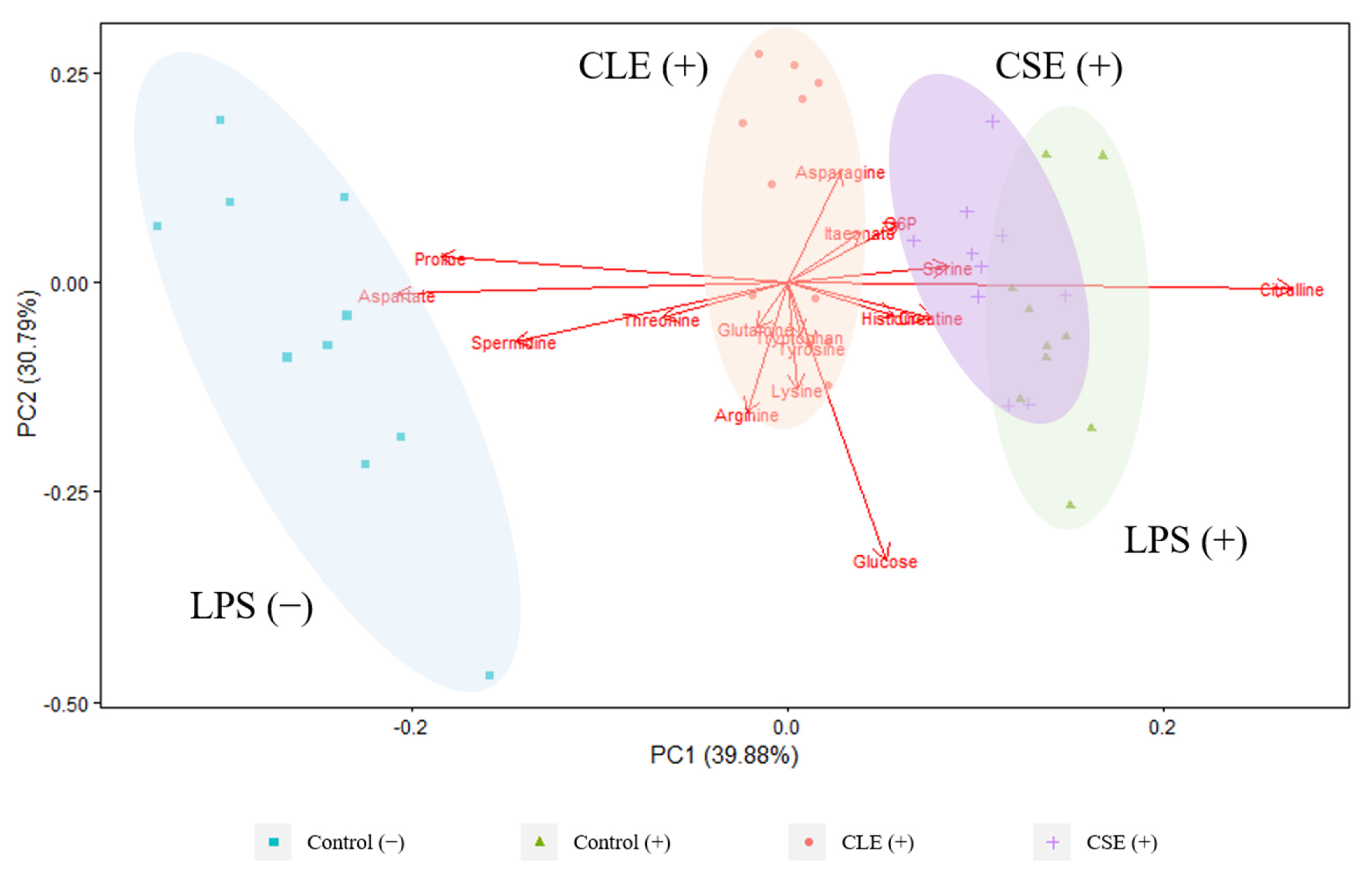
3.2.1. Intracellular Metabolites
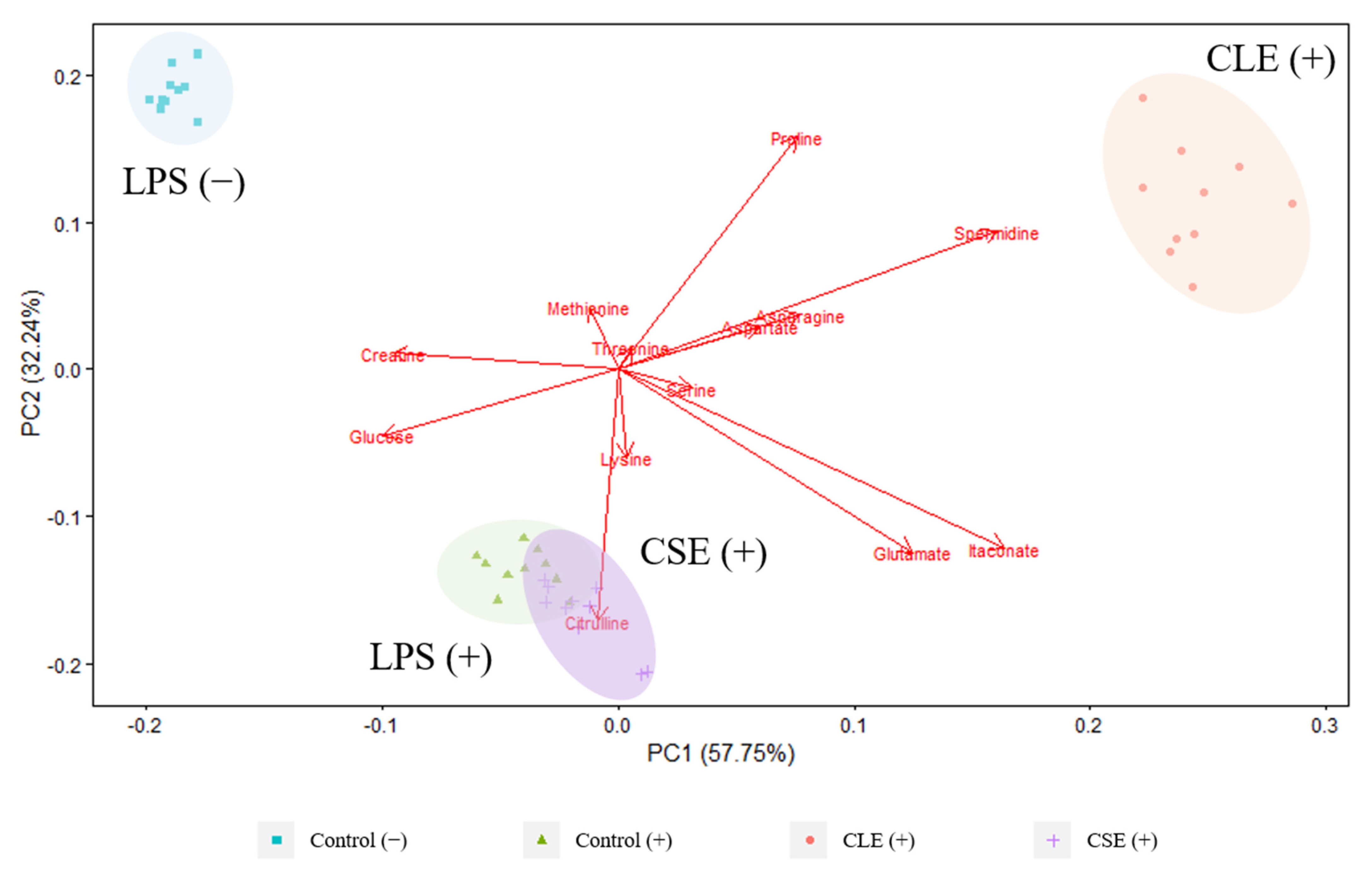
3.2.2. Extracellular Metabolites
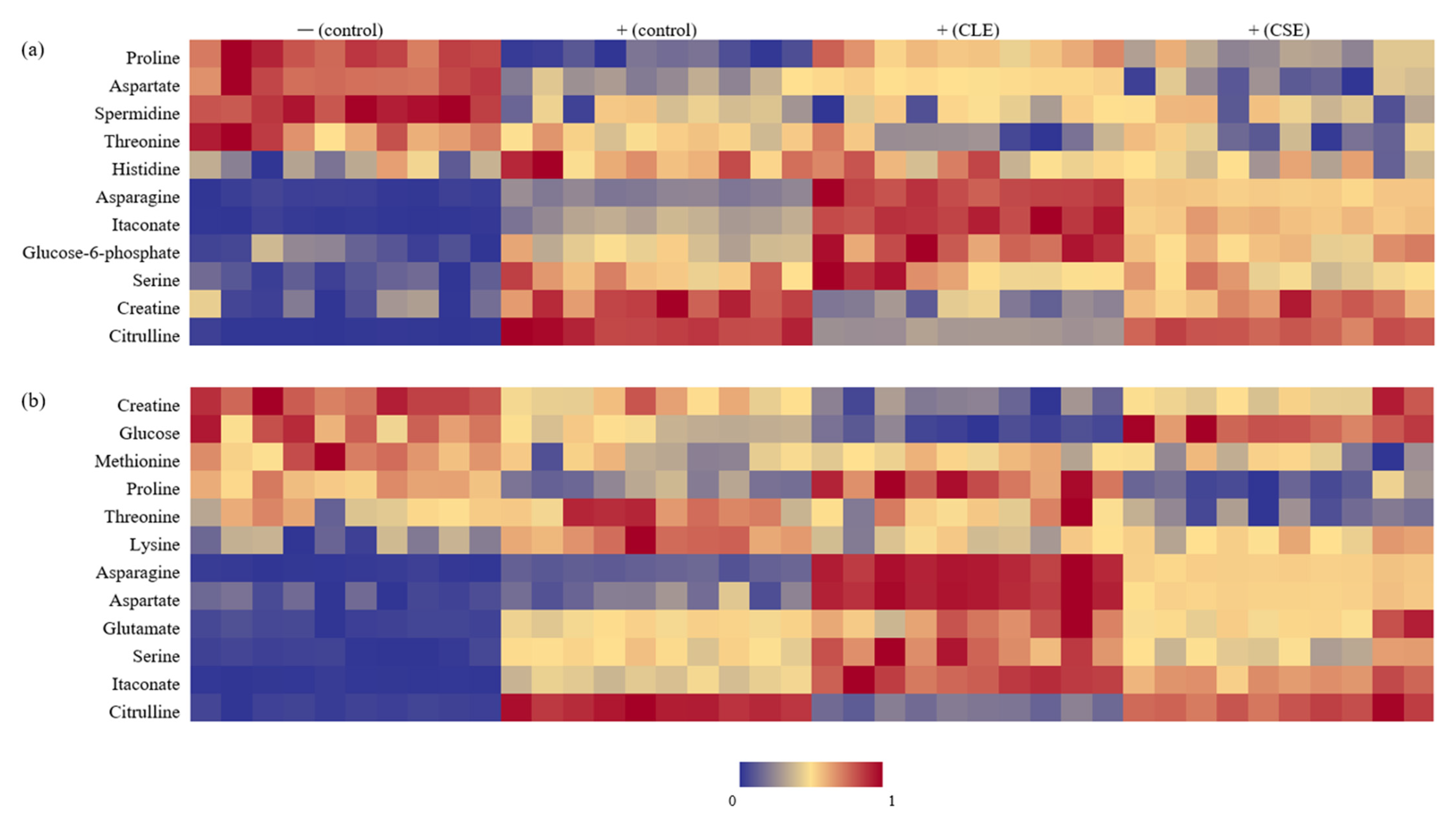
3.3. Heat Map Visualization
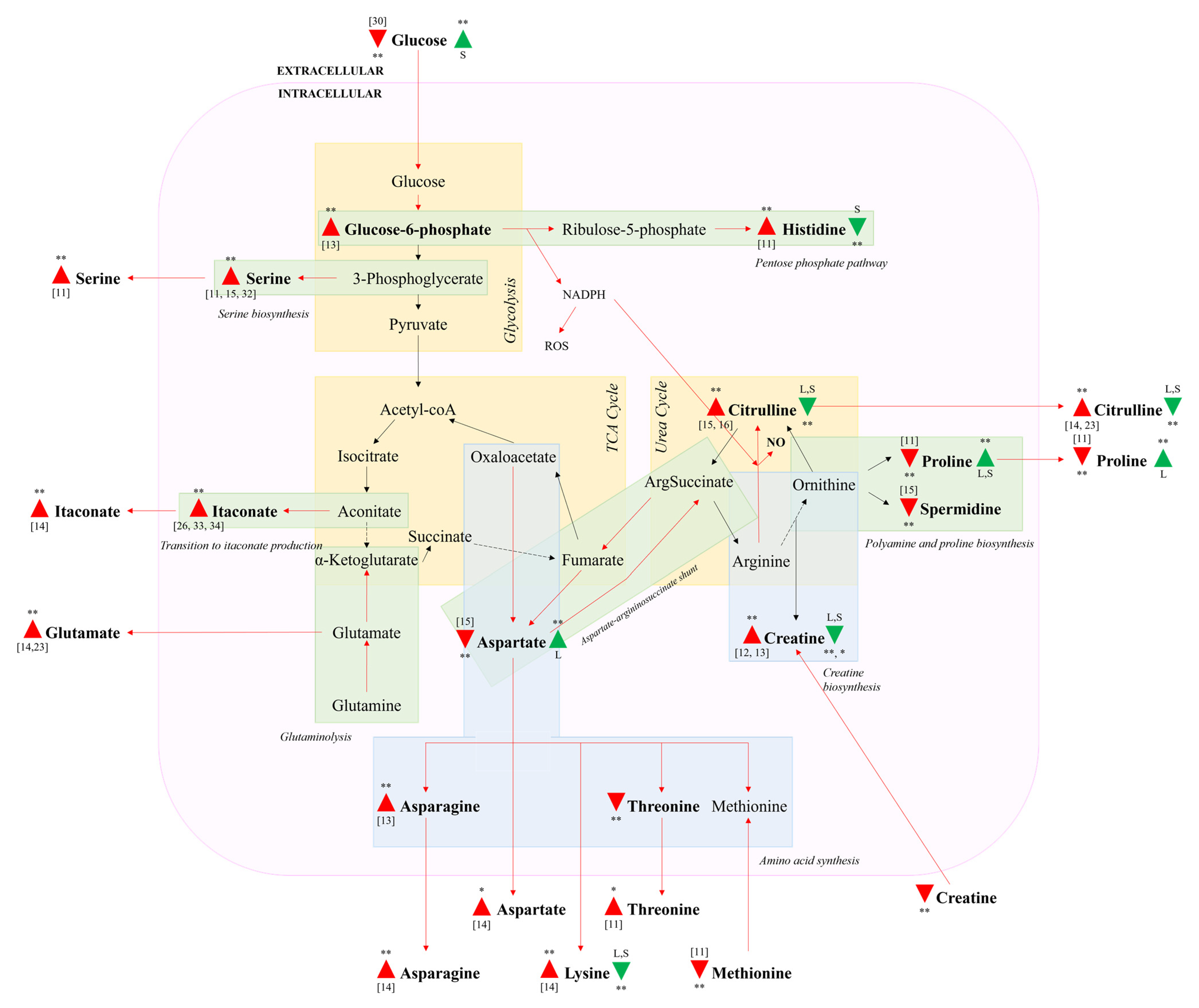
3.4. Impacted Metabolic Pathways
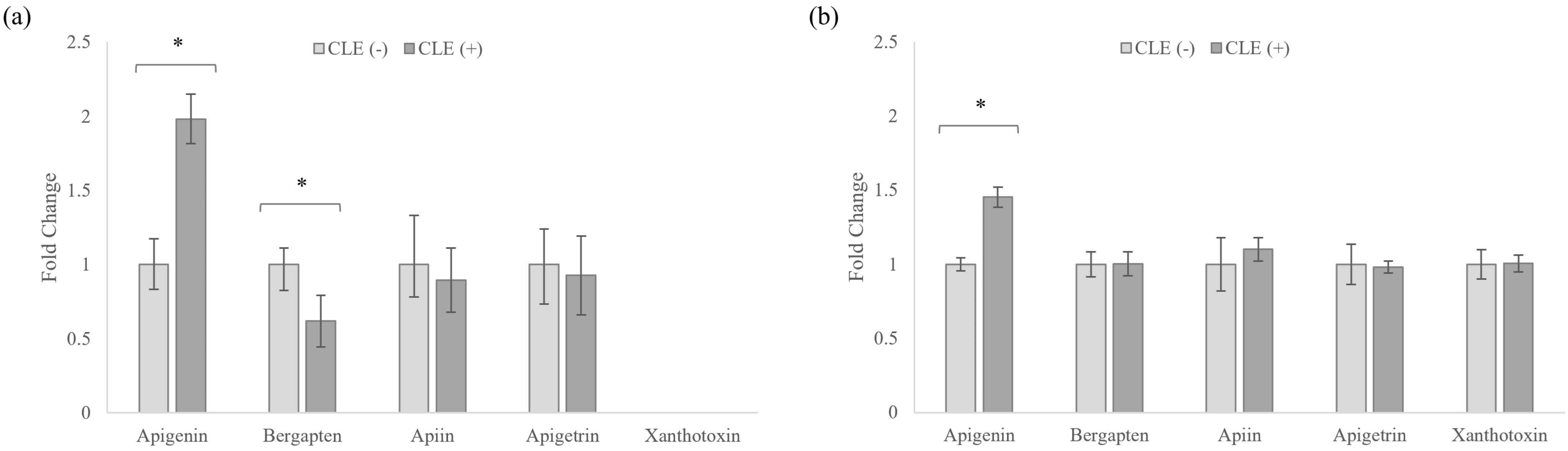
3.5. Celery Leaf Metabolites Present in Treated Cells and Culture Medium
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hedayati, N.; Bemani Naeini, M.; Mohammadinejad, A.; Mohajeri, S.A. Beneficial effects of celery (Apium graveolens) on metabolic syndrome: A review of the existing evidences. Phytother. Res. 2019, 33, 3040–3053. [Google Scholar] [CrossRef]
- Ingallina, C.; Capitani, D.; Mannina, L.; Carradori, S.; Locatelli, M.; Di Sotto, A.; Di Giacomo, S.; Toniolo, C.; Pasqua, G.; Valletta, A. Phytochemical and biological characterization of Italian “sedano bianco di Sperlonga” Protected Geographical Indication celery ecotype: A multimethodological approach. Food Chem. 2020, 309, 125649. [Google Scholar] [CrossRef] [PubMed]
- Al-Hindawi, M.K.; Al-Deen, I.H.; Nabi, M.H.; Ismail, M.A. Anti-inflammatory activity of some Iraqi plants using intact rats. J. Ethnopharmacol. 1989, 26, 163–168. [Google Scholar] [CrossRef]
- Arango, D.; Diosa-Toro, M.; Rojas-Hernandez, L.S.; Cooperstone, J.L.; Schwartz, S.J.; Mo, X.; Jiang, J.; Schmittgen, T.D.; Doseff, A.I. Dietary apigenin reduces LPS-induced expression of miR-155 restoring immune balance during inflammation. Mol. Nutr. Food Res. 2015, 59, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Mencherini, T.; Cau, A.; Bianco, G.; Loggia, R.D.; Aquino, R.; Autore, G. An extract of Apium graveolens var. dulce leaves: Structure of the major constituent, apiin, and its anti-inflammatory properties. J. Pharm. Pharmacol. 2007, 59, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Che, D.N.; Cho, B.O.; Shin, J.Y.; Kang, H.J.; Kim, J.S.; Choi, J.; Jang, S.I. Anti-atopic dermatitis effects of hydrolyzed celery extract in mice. J. Food Biochem. 2020, 44, e13198. [Google Scholar] [CrossRef]
- Lewis, D.A.; Tharib, S.M.; Veitch, G.B.A. The anti-inflammatory activity of celery Apium graveolens L. (Fam. Umbelliferae). Int. J. Crude Drug Res. 1985, 23, 27–32. [Google Scholar] [CrossRef]
- Ovodova, R.G.; Golovchenko, V.V.; Popov, S.V.; Popova, G.Y.; Paderin, N.M.; Shashkov, A.S.; Ovodov, Y.S. Chemical composition and anti-inflammatory activity of pectic polysaccharide isolated from celery stalks. Food Chem. 2009, 114, 610–615. [Google Scholar] [CrossRef]
- Yang, M.; Dang, R.; Xu, P.; Guo, Y.; Han, W.; Liao, D.; Jiang, P. Dl-3-n-Butylphthalide improves lipopolysaccharide-induced depressive-like behavior in rats: Involvement of Nrf2 and NF-κB pathways. Psychopharmacology 2018, 235, 2573–2585. [Google Scholar] [CrossRef]
- Zhu, L.-H.; Bao, T.-H.; Deng, Y.; Li, H.; Chen, L.-X. Constituents from Apium graveolens and their anti-inflammatory effects. J. Asian Nat. Prod. Res. 2017, 19, 1079–1086. [Google Scholar] [CrossRef]
- Abdul-Hamid, N.A.; Abas, F.; Ismail, I.S.; Tham, C.L.; Maulidiani, M.; Mediani, A.; Swarup, S.; Umashankar, S. 1H-NMR-based metabolomics to investigate the effects of Phoenix dactylifera seed extracts in LPS-IFN-γ-induced RAW 264.7 cells. Food Res. Int. 2019, 125, 108565. [Google Scholar] [CrossRef]
- Wang, Z.; Guan, Y.; Yang, R.; Li, J.; Wang, J.; Jia, A.-Q. Anti-inflammatory activity of 3-cinnamoyltribuloside and its metabolomic analysis in LPS-activated RAW 264.7 cells. BMC Complement. Med. Ther. 2020, 20, 329. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Pi, F.; Zhang, H.; Ji, J.; Xia, S.; Cui, F.; Sun, J.; Sun, X. Metabolomics analysis to evaluate the anti-inflammatory effects of polyphenols: Glabridin reversed metabolism change caused by LPS in RAW 264.7 cells. J. Agric. Food Chem. 2017, 65, 6070–6079. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Sakagami, H.; Yokote, Y.; Onuma, H.; Kaneko, M.; Mori, M.; Sakaguchi, Y.; Soga, T.; Tomita, M. Non-targeted metabolite profiling in activated macrophage secretion. Metabolomics 2012, 8, 624–633. [Google Scholar] [CrossRef]
- Suh, J.H.; Kim, R.Y.; Lee, D.S. A new metabolomic assay to examine inflammation and redox pathways following LPS challenge. J. Inflamm. 2012, 9, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abuawad, A.; Mbadugha, C.; Ghaemmaghami, A.M.; Kim, D.-H. Metabolic characterisation of THP-1 macrophage polarisation using LC–MS-based metabolite profiling. Metabolomics 2020, 16, 33. [Google Scholar] [CrossRef] [Green Version]
- Dietmair, S.; Timmins, N.E.; Gray, P.P.; Nielsen, L.K.; Krömer, J.O. Towards quantitative metabolomics of mammalian cells: Development of a metabolite extraction protocol. Anal. Biochem. 2010, 404, 155–164. [Google Scholar] [CrossRef]
- Lim, S.L.; Jia, Z.; Lu, Y.; Zhang, H.; Ng, C.T.; Bay, B.H.; Shen, H.M.; Ong, C.N. Metabolic signatures of four major histological types of lung cancer cells. Metabolomics 2018, 14, 118. [Google Scholar] [CrossRef]
- Jia, Z.; Zhang, H.; Ong, C.N.; Patra, A.; Lu, Y.; Lim, C.T.; Venkatesan, T. Detection of lung cancer: Concomitant volatile organic compounds and metabolomic profiling of six cancer cell lines of different histological origins. ACS Omega 2018, 3, 5131–5140. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 2 June 2021).
- Horikoshi, M.; Tang, Y. ggfortify: Data Visualization Tools for Statistical Analysis Results. 2016. Available online: https://CRAN.R-project.org/package=ggfortify (accessed on 2 June 2021).
- Nishiyama, A.; Yokote, Y.; Sakagami, H. Changes in amino acid metabolism during activation of mouse macrophage-like cell lines. In Vivo 2010, 24, 857–860. [Google Scholar]
- Rath, M.; Müller, I.; Kropf, P.; Closs, E.I.; Munder, M. Metabolism via arginase or nitric oxide synthase: Two competing arginine pathways in macrophages. Front. Immunol. 2014, 5, 532. [Google Scholar] [CrossRef] [Green Version]
- Bauer, P.M.; Buga, G.M.; Fukuto, J.M.; Pegg, A.E.; Ignarro, L.J. Nitric Oxide Inhibits Ornithine Decarboxylase viaS-Nitrosylation of Cysteine 360 in the Active Site of the Enzyme. J. Biol. Chem. 2001, 276, 34458–34464. [Google Scholar] [CrossRef] [Green Version]
- Jha, A.K.; Huang, S.C.-C.; Sergushichev, A.; Lampropoulou, V.; Ivanova, Y.; Loginicheva, E.; Chmielewski, K.; Stewart, K.M.; Ashall, J.; Everts, B. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015, 42, 419–430. [Google Scholar] [CrossRef] [Green Version]
- Langston, P.K.; Shibata, M.; Horng, T. Metabolism supports macrophage activation. Front. Immunol. 2017, 8, 61. [Google Scholar] [CrossRef] [Green Version]
- Nonnenmacher, Y.; Hiller, K. Biochemistry of proinflammatory macrophage activation. Cell. Mol. Life Sci. 2018, 75, 2093–2109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Raines, L.N.; Huang, S.C.-C. Carbohydrate and amino acid metabolism as hallmarks for innate immune cell activation and function. Cells 2020, 9, 562. [Google Scholar] [CrossRef] [Green Version]
- Través, P.G.; de Atauri, P.; Marín, S.; Pimentel-Santillana, M.; Rodríguez-Prados, J.-C.; de Mas, I.M.; Selivanov, V.A.; Martín-Sanz, P.; Boscá, L.; Cascante, M. Relevance of the MEK/ERK signaling pathway in the metabolism of activated macrophages: A metabolomic approach. J. Immunol. 2012, 188, 1402–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ham, M.; Lee, J.-W.; Choi, A.H.; Jang, H.; Choi, G.; Park, J.; Kozuka, C.; Sears, D.D.; Masuzaki, H.; Kim, J.B. Macrophage glucose-6-phosphate dehydrogenase stimulates proinflammatory responses with oxidative stress. Mol. Cell. Biol. 2013, 33, 2425–2435. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, A.E.; Ducker, G.S.; Billingham, L.K.; Martinez, C.A.; Mainolfi, N.; Suri, V.; Friedman, A.; Manfredi, M.G.; Weinberg, S.E.; Rabinowitz, J.D. Serine metabolism supports macrophage IL-1β production. Cell Metab. 2019, 29, 1003–1011.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, E.L.; Ryan, D.G.; Prag, H.A.; Dikovskaya, D.; Menon, D.; Zaslona, Z.; Jedrychowski, M.P.; Costa, A.S.; Higgins, M.; Hams, E. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 2018, 556, 113–117. [Google Scholar] [CrossRef]
- Cicuéndez, M.; Fernandes, M.; Ayán-Varela, M.; Oliveira, H.; Feito, M.J.; Diez-Orejas, R.; Paredes, J.I.; Villar-Rodil, S.; Vila, M.; Portolés, M.T. Macrophage inflammatory and metabolic responses to graphene-based nanomaterials differing in size and functionalization. Colloids Surf. B Biointerfaces 2020, 186, 110709. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Wang, Z.; Zhang, K.; Chi, Z.; Xu, T.; Jiang, D.; Chen, S.; Li, W.; Yang, X.; Zhang, X. One-carbon metabolism supports S-adenosylmethionine and histone methylation to drive inflammatory macrophages. Mol. Cell 2019, 75, 1147–1160.e5. [Google Scholar] [CrossRef]
- Jeong, J.-W.; Cha, H.-J.; Han, M.H.; Hwang, S.J.; Lee, D.-S.; Yoo, J.S.; Choi, I.-W.; Kim, S.; Kim, H.-S.; Kim, G.-Y. Spermidine protects against oxidative stress in inflammation models using macrophages and zebrafish. Biomol. Ther. 2018, 26, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, R.; Li, X.; Ma, H.; Yang, Q.; Shang, Q.; Song, L.; Zheng, Z.; Zhang, S.; Pan, Y.; Huang, P. Spermidine endows macrophages anti-inflammatory properties by inducing mitochondrial superoxide-dependent AMPK activation, Hif-1α upregulation and autophagy. Free Radical Biol. Med. 2020, 161, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, J.; Yang, W.; Qi, X.; Lan, L.; Luo, L.; Yin, Z. Bergapten prevents lipopolysaccharide-induced inflammation in RAW264. 7 cells through suppressing JAK/STAT activation and ROS production and increases the survival rate of mice after LPS challenge. Int. Immunopharmacol. 2017, 48, 159–168. [Google Scholar] [CrossRef]
- Lee, S.-B.; Lee, W.S.; Shin, J.-S.; Jang, D.S.; Lee, K.T. Xanthotoxin suppresses LPS-induced expression of iNOS, COX-2, TNF-α, and IL-6 via AP-1, NF-κB, and JAK-STAT inactivation in RAW 264.7 macrophages. Int. Immunopharmacol. 2017, 49, 21–29. [Google Scholar] [CrossRef] [PubMed]
| Analyte | MW | tR (min) | Adduct | Ionisation Mode | Q1 (m/z) | Q3 (m/z) | DP (V) | CE (eV) | CXP (V) |
|---|---|---|---|---|---|---|---|---|---|
| Spermidine | 145 | 0.55 | [M + H]+ | + | 146 | 72 | 40 | 20 | 6 |
| Lysine | 146 | 0.58 | [M + H]+ | + | 147 | 84 | 50 | 23 | 11 |
| Histidine | 155 | 0.61 | [M + H]+ | + | 156 | 110 | 50 | 20 | 11 |
| Arginine | 174 | 0.62 | [M + H]+ | + | 175 | 70 | 50 | 30 | 8 |
| Serine | 105 | 0.63 | [M + H]+ | + | 106 | 60 | 40 | 15 | 7 |
| Asparagine | 132 | 0.63 | [M + H]+ | + | 133 | 74 | 30 | 20 | 10 |
| Aspartate | 133 | 0.64 | [M + H]+ | + | 134 | 88 | 40 | 14 | 9 |
| Threonine | 119 | 0.65 | [M + H]+ | + | 120 | 74 | 50 | 14 | 10 |
| Glutamine | 146 | 0.65 | [M + H]+ | + | 147 | 130 | 50 | 13 | 7 |
| Glucose | 180 | 0.66 | [M + FA − H]− | − | 225 | 179 | 30 | 8 | 15 |
| Citrulline | 175 | 0.66 | [M + H]+ | + | 176 | 159 | 50 | 14 | 8 |
| Glutamate | 147 | 0.67 | [M + H]+ | + | 148 | 84 | 50 | 20 | 10 |
| Glucose-6-phosphate | 260 | 0.68 | [M − H]− | − | 259 | 97 | 70 | 21 | 11 |
| Proline | 115 | 0.71 | [M + H]+ | + | 116 | 70 | 80 | 20 | 10 |
| Creatine | 131 | 0.71 | [M + H]+ | + | 132 | 90 | 50 | 17 | 10 |
| Methionine | 149 | 0.92 | [M + H]+ | + | 150 | 133 | 80 | 12 | 8 |
| Tyrosine | 181 | 1.55 | [M + H]+ | + | 182 | 136 | 50 | 19 | 9 |
| Itaconate | 130 | 2.93 | [M − H]− | − | 129 | 85 | 40 | 12 | 8 |
| Tryptophan | 204 | 4.67 | [M + H]+ | + | 205 | 188 | 50 | 14 | 9 |
| Apiin | 564 | 5.70 | [M + H]+ | + | 565 | 271 | 40 | 34 | 14 |
| Apigetrin | 432 | 5.80 | [M + H]+ | + | 433 | 271 | 60 | 24 | 15 |
| Apigenin | 270 | 7.10 | [M − H]− | − | 269 | 117 | 70 | 45 | 6 |
| Xanthotoxin | 216 | 7.26 | [M + H]+ | + | 217 | 202 | 60 | 28 | 12 |
| Bergapten | 216 | 7.70 | [M + H]+ | + | 217 | 202 | 60 | 28 | 10 |
| FMOC-glycine (IS) | 297 | 7.91 | C14H10+ | + | 179 | 178 | 100 | 35 | 9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lau, H.; Ni, N.; Dayal, H.; Lim, S.-Y.; Ren, Y.; Li, S.F.-Y. Evaluation of Anti-Inflammatory Effects of Celery Leaf and Stem Extracts in LPS-Induced RAW 264.7 Cells Using Nitric Oxide Assay and LC-MS Based Metabolomics. Curr. Issues Mol. Biol. 2021, 43, 1876-1888. https://doi.org/10.3390/cimb43030131
Lau H, Ni N, Dayal H, Lim S-Y, Ren Y, Li SF-Y. Evaluation of Anti-Inflammatory Effects of Celery Leaf and Stem Extracts in LPS-Induced RAW 264.7 Cells Using Nitric Oxide Assay and LC-MS Based Metabolomics. Current Issues in Molecular Biology. 2021; 43(3):1876-1888. https://doi.org/10.3390/cimb43030131
Chicago/Turabian StyleLau, Hazel, Nengyi Ni, Hiranya Dayal, Si-Ying Lim, Yi Ren, and Sam Fong-Yau Li. 2021. "Evaluation of Anti-Inflammatory Effects of Celery Leaf and Stem Extracts in LPS-Induced RAW 264.7 Cells Using Nitric Oxide Assay and LC-MS Based Metabolomics" Current Issues in Molecular Biology 43, no. 3: 1876-1888. https://doi.org/10.3390/cimb43030131
APA StyleLau, H., Ni, N., Dayal, H., Lim, S.-Y., Ren, Y., & Li, S. F.-Y. (2021). Evaluation of Anti-Inflammatory Effects of Celery Leaf and Stem Extracts in LPS-Induced RAW 264.7 Cells Using Nitric Oxide Assay and LC-MS Based Metabolomics. Current Issues in Molecular Biology, 43(3), 1876-1888. https://doi.org/10.3390/cimb43030131







