Abstract
6-Heteryl-5-methylthieno[2,3-d]pyrimidin-2,4(1H,3H)-diones are of great interest as the promising objects for the search of antibacterials. In this communication, we obtained 6-(imidazo[1,2-a]pyridin-2-yl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione by interaction of 6-(bromoacetyl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione with 2-aminopyridine. The obtained heterocyclic hybrid was further modified by alkylation with 2-chloroarylacetamides. Antimicrobial activity studies for the synthesized compounds using the agar well diffusion method revealed their moderate activity against S. aureus, E. coli and B. subtilis. According to the double dilution assay MIC value results for 6-(imidazo[1,2-a]pyridin-2-yl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dioneagainst P. aeruginosa was less than the value determined for the reference drug streptomycin. The docking study of the synthesized compounds to the active site of TrmD isolated from P. aeruginosa did not show their effective inhibitory activity.
1. Introduction
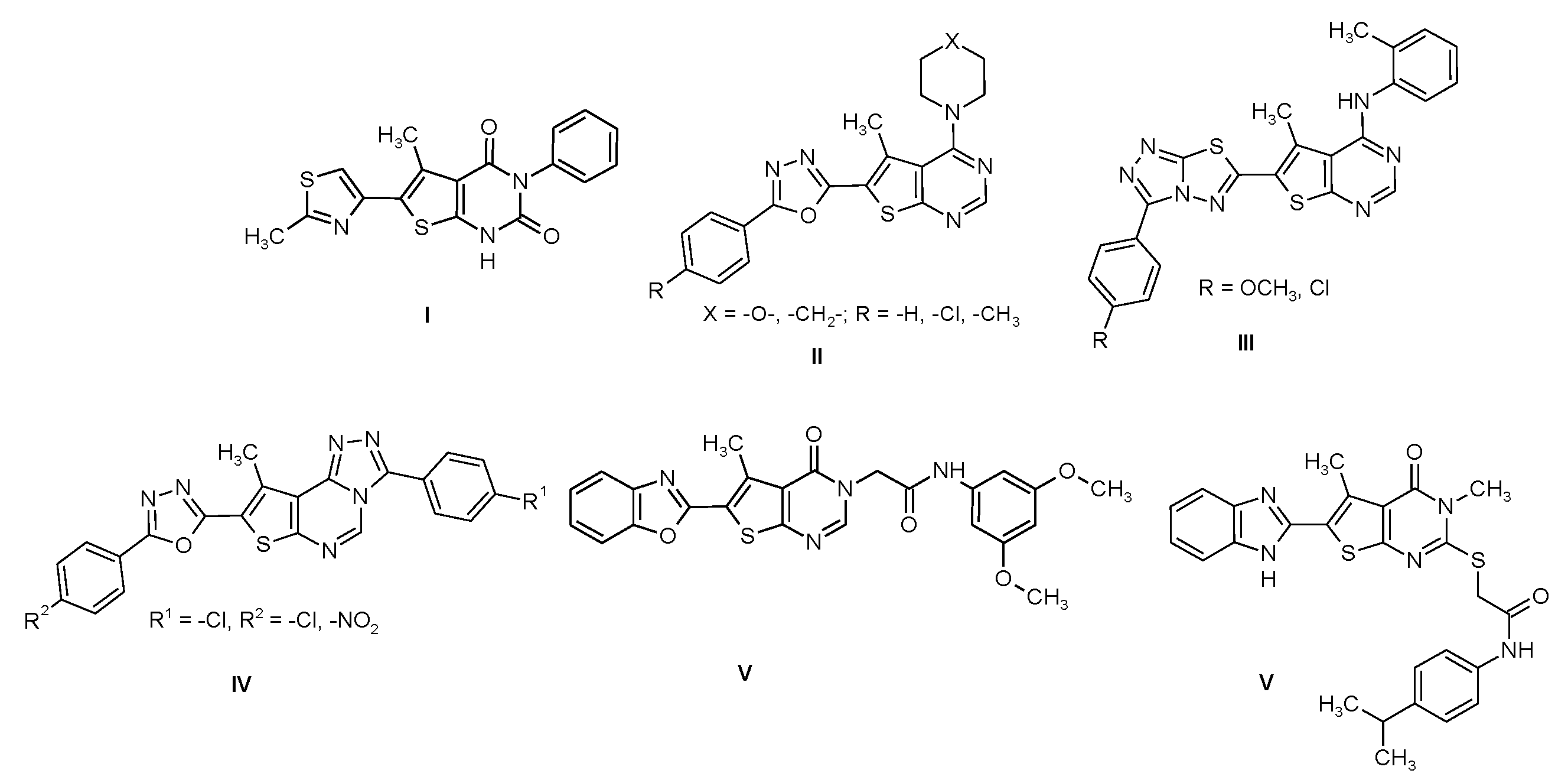
Derivatives with 6-heteryl-5-methylthieno[2,3-d]pyrimidin-2,4(1H,3H)-dione structures are of great importance as biologically active substances. Such compounds were patented as A2A adenosine receptor antagonists [1], which are useful in treating mammals for various disease states. Similar compounds, patented as acetyl-KoA carboxylase inhibitors, can be used to cure diseases caused by fatty acids metabolism dysfunction [2]. The modification of the 5-methylthieno[2,3-d]pyrimidin-2,4(1H,3H)-dione core with 1,3-thiazol-4-yl substituent has allowed the acquisition of compounds with antimicrobial activity against S. aureus, P. aeruginosa and B. subtilis (I) (Figure 1) [3].
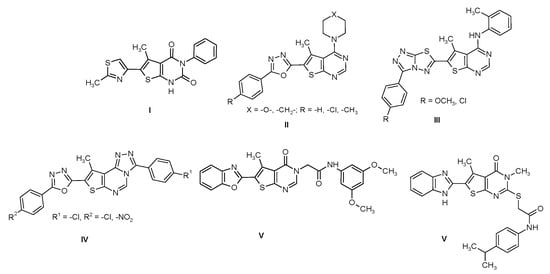
Figure 1.
6-Heteryl-5-methylthieno[2,3-d]pyrimidines with antimicrobial properties.
Likewise, numerous articles have been published in the last decade about the antimicrobial activity of 6-heterylthieno[2,3-d]pyrimidines with unsubstituted position 2 [4,5,6,7] (II-V) (Figure 1), some of them containing fused systems of heterocycles such as 1,2,4-triazolo[3,4-b]1,3,4-thiadiazole (III) [5] or 1,3-benzoxazole (V) [7]. One of the latest works published was devoted to the study of the antimicrobial activity of 6-heteryl-5-methyl-2-thiothieno[2,3-d]pyrimidines, which can possibly act as inhibitors of bacterial TrmD VI (Figure 1) [8].
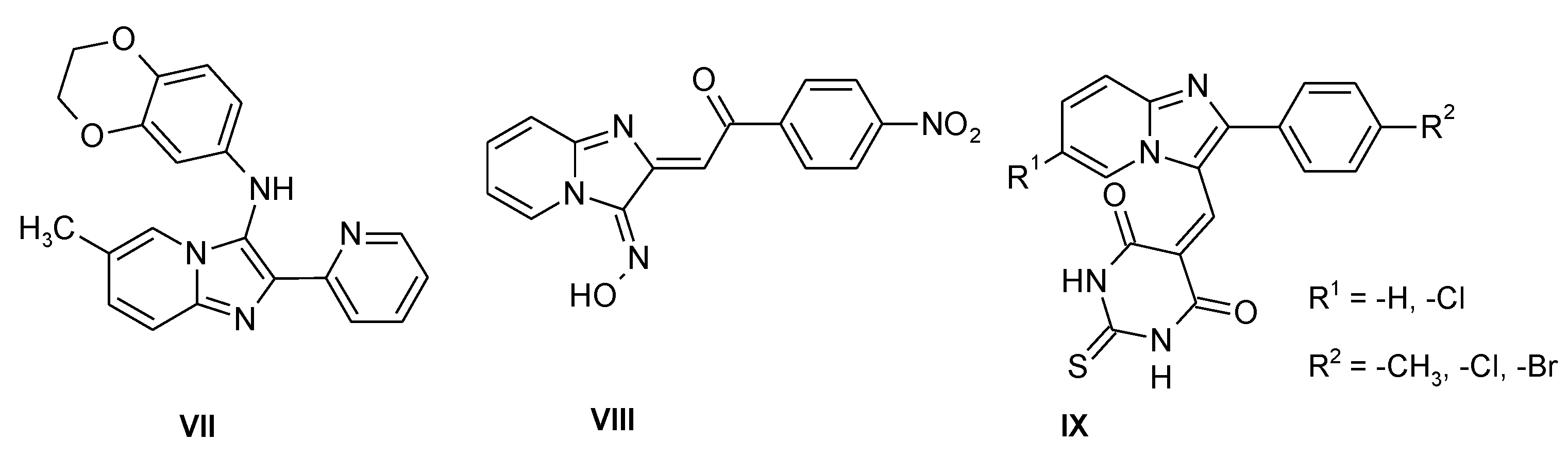
Several imidazo[1,2-a]pyridine derivatives were reported as compounds with antimicrobial properties which may be useful against parasites of the Leishmania genus (VII) (Figure 2) [9]; this heterocyclic system is the core structure of savolitinib [10], a mesenchymal epithelial transition factor (MET) inhibitor recently approved in China after the results of a pivotal phase II trial in patients with NSCLC/pulmonary sarcomatoid carcinoma [11]. This fragment is also a part of the innovative drug olprinone, which is a selective phosphodiesterase 3 (PDE3) inhibitor [12]. Imidazo[1,2-a]pyridines were also reported as antiproliferative agents [13], α-glucosidase inhibitors [14] and antimicrobials (VIII) (Figure 2) [15]; some of these compounds have shown promising antibacterial activity against S. pyogenes, P. aeruginosa and S. aureus (IX) (Figure 2) [16].
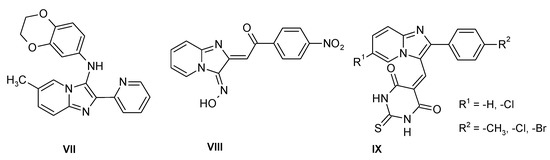
Figure 2.
Antibacterial imidazo[1,2-a]pyridines.
In view of the published results about antimicrobial properties of 6-heterylthieno[2,3-d]pyrimidines as well as the promising pharmacological potential of imidazo[1,2-a]pyridines, we decided to modify position 6 of the 5-methylthieno[2,3-d]pyrimidin-2,4(1H,3H)-dione core with an imidazo[1,2-a]pyridine fragment as a possible approach towards active anti-bacterials.
2. Results and Discussion
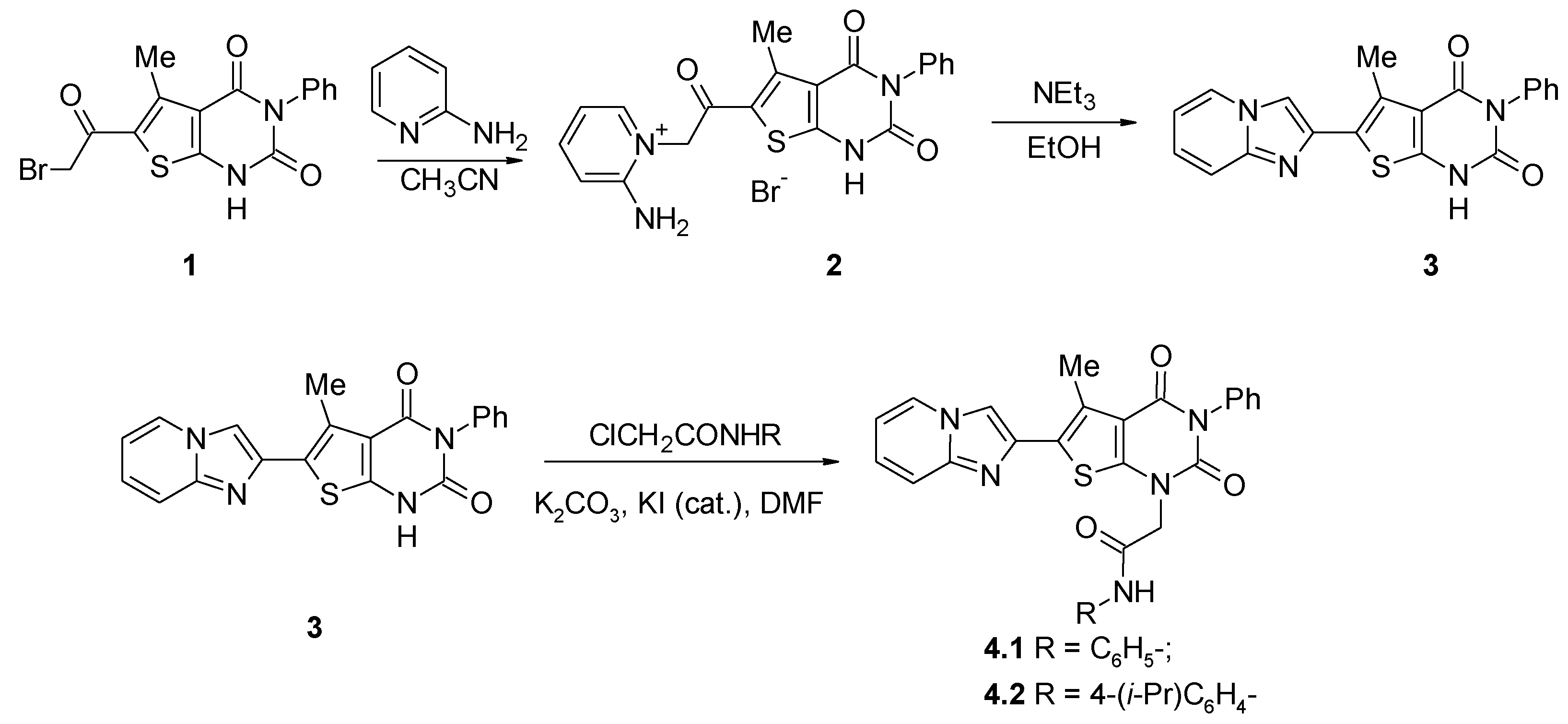
The most common route for the preparation of imidazo[1,2-a]pyridines is the reaction of 2-aminoazines with α-haloketones [15,16,17,18]. As a part of our research on the construction of 6-heteryl-5-methylthieno[2,3-d]pyrimidin-2,4(1H,3H)-diones, we studied the interaction of the readily synthetically available 6-(bromoacetyl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione 1 [3] with 2-aminopyridine. We tried a one-step approach and heated 1 with 2-aminopyridine in 2-propanol, which gave no product of any sufficient purity. Thus, we made the decision to separate the steps and to use the solvent where it was possible to carry out the reaction homogeneously at the first step. We applied a two-step procedure where the alkylation was carried out in acetonitrile, which was used because of the good solubility of 1 in this solvent under heating. The formation of the product 2 can be easily tracked by its precipitation. A subsequent cyclization reaction was conducted in refluxing the ethanol-triethylamine system (Scheme 1). Although the cyclization step proceeds mostly heterogeneously, it provides the sufficient yield and purity of the product 3.
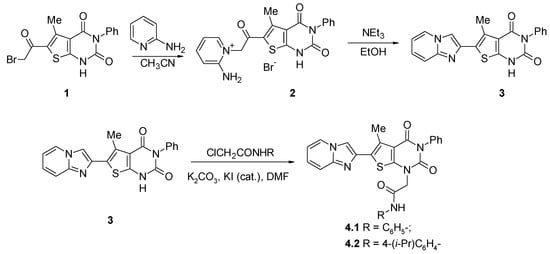
Scheme 1.
Synthesis of 2-(6-imidazo[1,2-a]pyridin-2-yl-5-methyl-2,4-dioxo-3-phenyl-3,4-dihydrothieno[2,3-d]pyrimidin-1(2H)-yl)-N-arylacetamides 4.
The 1H NMR spectrum of compound 3 has the distinctive peak of the methyl group at position 5 of thieno[2,3-d]pyrimidine system at 2.60 ppm and the singlet of imidazole part of imidazo[1,2-a]pyridine at 8.23 ppm; the signal of the NH proton of pyrimidine cycle was observed at 12.38 ppm.
The next step of the synthesis was alkylation of compound 3 with arylchloroacetamides in DMF media in the presence of potassium carbonate (1 equivalent) and a catalytic amount of potassium iodide. The slight heating (50–60 °C) was found useful to increase the homogenicity of the reaction.
Similarly to compound 3, the 1H NMR spectra of compounds 4.1 and 4.2 displayed the signal of the methyl group at position 5 of thieno[2,3-d]pyrimidine in the range of 2.65–2.85 ppm; the signal of the imidazole proton was observed at 8.27 ppm. The signals of the methylene protons of acetamide substituents at position 1 of thieno[2,3-d]pyrimidine core were observed in the region 4.84–4.85 ppm. For both derivatives 4.1 and 4.2, the 1H NMR spectra also contained the signals of amide NH in the region 10.37–10.44 ppm. In comparison with the spectrum of 3 13C, the NMR spectra of compounds 4.1 and 4.2 contained additional signals in the region of aromatic carbons’ resonance and the signals of methyl group carbon atoms in the region 48.4–50.7 ppm.
The results of antimicrobial activity screening show that compounds 3, 4.1 and 4.2 have moderate inhibitory activity against Staphylococcus aureus, Escherichia coli and Bacillus subtilis. None of the compounds inhibited the growth of the Candida albicans fungal strain (Table 1). Although the results of the well diffusion method suggest compound 4.1 as the one with the best screening results, according to the double dilution assay, compound 3 showed lower MIC values. Its inhibitory activity was better than those shown by metronidazole and in the case of P. aeruginosa, the MIC value of 3 was even lower than the inhibitory concentration of streptomycin.

Table 1.
Antimicrobial activity of 6-(imidazo[1,2-a]pyridin-2-yl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-diones 3, 4.1 and 4.2.
To predict the possible mechanism of antibacterial action by 6-(imidazo[1,2-a]pyridin-2-yl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-diones 3, 4.1, and 4.2, the docking studies of these structures were performed on selective inhibitors of tRNA—(Guanine37-N1)-methyltransferase (TrmD). The structure of the enzyme isolated from P. aeruginosa was used for the study [19]. The validation of the docking methodology using the reference ligand—the competitive TrmD inhibitor N-(4-((octylamino)methyl)benzyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5- carboxamide—is described in the previous paper [8]. The affinity of 3, 4.1, and 4.2 towards the active site was estimated by binding energy in comparison with the reference ligand. Compounds 3, 4.1, and 4.2 have a higher affinity (8.7, −10.6, and −10.8 kcal/mol respectively) than the reference inhibitor, which has an affinity as high as −8.2 kcal/mol.
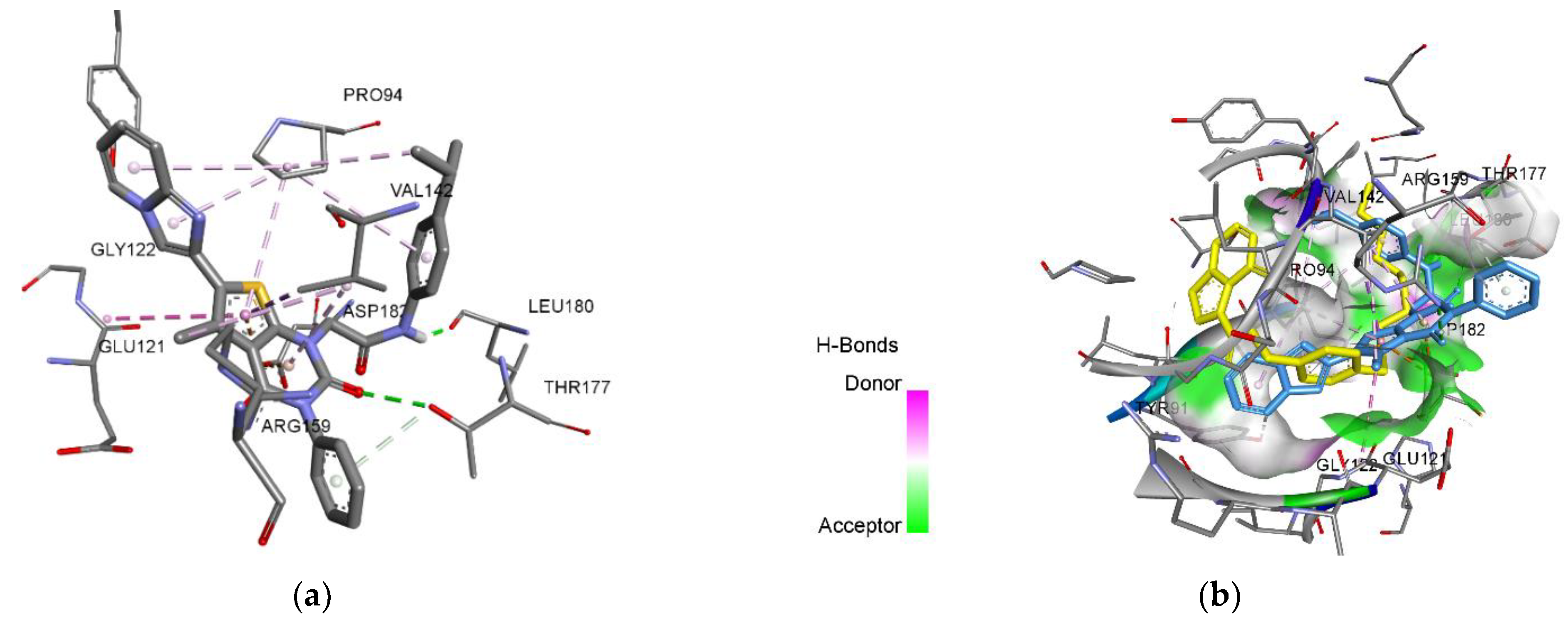
The docking study predicts a possible stable conformation due to hydrophobic interactions that is additionally stabilized with hydrogen bonds (Figure 3a). Among the amino acids that influence the conformation of the ligand, only proline (PRO94), glutamic (GLU121) and aspartic acids residues (ASP182) belong to the active site (Table 2). Interaction of tyrosine residues (Tyr141, and 120) with TrmD inhibitors is known to be crucial for TrmD inhibition, but it was not observed in the case of compounds 3, 4.1, and 4.2. 6-(Imidazo[1,2-a]pyridin-2-yl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-diones 3, 4.1 and 4.2 do not interact with residues of glutamine (Gln95) and glycines (Gly 118, 145,146), which bind the methionine fragment of SAM [19]. This predicts absence of the interaction with the hydrophobic pocket of the active site. Comparative conformations of 3, 4.1 and 4.2 with the reference inhibitor evidently show their inability to enter the hydrophobic pocket and to reach the place of thienopyrimidine pharmacophore fixing. Hence, despite good calculated values of scoring functions, the ligands’ pose analysis shows the low probability for the compounds 3, 4.1 and 4.2 to be the inhibitors of bacterial TrmD.
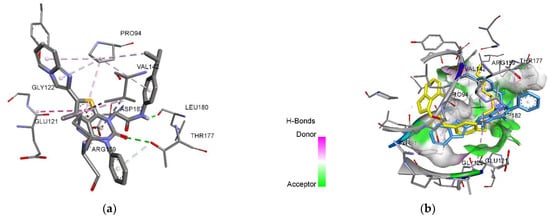
Figure 3.
3-D visualization: (a) interaction of the ligand 4.2 with amino acids of the active site of TrmD; (b) conformational pose of the reference inhibitor (yellow molecule) [19] and the ligand 4.2 (blue molecule) in the active site of P. aeruginosa TrmD.

Table 2.
The results of the docking studies of 3, 4.1 and 4.2 and the native inhibitor to the active site of P. aeruginosa TrmD.
3. Materials and Methods
3.1. Materials and Instrumentation
All solvents and reagents were obtained from commercial sources or prepared by previously reported methods. Melting points were determined in a capillary using an electrothermal IA9100X1 (Bibby Scientific Limited, Staffordshire, UK) digital melting point apparatus. Elemental analyses were performed on a Euro Vector EA-3000 (Eurovector SPA, Redavalle, Italy) microanalyzer and were within 0.4% of the theoretical values. 1H NMR spectra for compound 3 were recorded with a Varian Mercury (200 MHz) in DMSO-d6. In all other cases, they were acquired with a Bruker Avance drx 500 (500 MHz) in DMSO-d6; the latter was also used for measurement of the 13C NMR (125 MHz) in CF3COOD for compound 3 and in DMSO-d6 for the compounds 4.1 and 4.2. TMS was used as an internal standard.
Mass-spectral analyses for the compound 3 were obtained on a PE SCIEX API 150EX-MS spectrometer (SCIEX, Singapore, The Republic of Singapore) with turbo ion spray sources device (API+); for compounds 4, LC-MS analysis was performed on an Agilent 1100 HPLC apparatus with diode array and mass-selective detectors (Agilent, Santa Clara, CA, USA), Zorbax SB-C18 column (4.6 × 150 mm) with chemical ionization at atmospheric pressure (APCI) (Please see the supplementary materials).
3.2. Chemical Part
The starting 6-(bromoacetyl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione 1 was obtained using a previously reported procedure [3].
Procedure for synthesis of 6-(imidazo[1,2-a]pyridin-2-yl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione3
To 3.0 g (0.008 mol) of 6-(bromoacetyl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione 1 in 20 mL of acetonitrile, 0.75 g (0.008 mol) of 2-aminopyridine was added and the reaction mixture was heated at reflux until the formation of the first crystals of the product and then additionally for 1–2 h. The precipitate formed was filtered off and dried. The product was used for the next step without additional purification. To 2.8 g of the product isolated in previous step, 0.82 mL (0.006 mol) of trimethylamine in 50 mL ethanol was added and the mixture was refluxed for 8–10 h. The reaction mixture was cooled and the precipitate was filtered off and washed with ethanol (10 mL × 3).
Yield: 52%, m.p. >300 °C, beige solid. 1H NMR (200 MHz, DMSO-d6): δ: 2.60 (s, 3H, CH3), 6.91 (t, 1H, J = 6.9 Hz, Ar-H), 7.19–7.61 (m, 7H, Ar-H), 8.23 (s, 1H, Ar-H), 8.51 (d, 1H, J = 6.9 Hz, Ar-H), 12.38 (s, 1H, NH). 13C NMR (125 MHz, CF3COOD): δ 12.3, 110.1, 110.8, 111.8, 112.3, 112.9, 114.6, 116.8, 117.6, 127.1, 127.4, 129.3, 129.6, 131.6, 134.5, 138.3, 139.4, 152.4. LC-MS m/z (ES+) 375.2 (MH+). Anal. calcd. for C20H14N4O2S (374.42): C, 64.16; H, 3.77; N, 14.96. Found: C, 64.22; H, 3.83; N, 15.02.
General procedure for synthesis of 2-(6-imidazo[1,2-a]pyridin-2-yl-5-methyl-2,4-dioxo-3-phenyl-3,4-dihydrothieno[2,3-d]pyrimidin-1(2H)-yl)-N-arylacetamides 4.1, 4.2
To the suspension of 0.15 g (0.0004 mol) of 6-(imidazo[1,2-a]pyridin-2-yl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione 3 and 0.056 g (0.4 mmol) in 5 mL of dimethylformamide, 0.4 mmol of corresponding phenyl chloroacetamide and catalytic amount of potassium iodide was added. The reaction mixture was stirred and heated (40–50 °C) for 5–6 h. Then the reaction mixture was cooled and quenched with water (20 mL). The resulting precipitate was filtered off and dried. The products were additionally crystallized from an ethanol-DMF mixture.
2-[6-(Imidazo[1,2-a]pyridin-2-yl)-5-methyl-2,4-dioxo-3-phenyl-3,4-dihydrothieno[2,3-d]pyrimidin-1(2H)-yl]-N-phenylacetamide 4.1
Yield: 83%, m.p. >300 °C, beige solid. 1H NMR (500 MHz, DMSO-d6): δ: 2.65 (s, 3H, CH3), 4.85 (s, 2H, CH2), 6.91 (t, 1H, J = 5.8 Hz, Ar-H), 7,07 (t, 1H, J = 5.8 Hz, Ar-H), 7.20–7.64 (m, 11H, Ar-H), 8,27 (s, 1H, Ar-H), 8.52 (d, 1H, J = 5.8 Hz, Ar-H), 10.44 (s, 1H, NH). 13C NMR (125 MHz, DMSO-d6): δ 14.7, 50.7, 109.9, 113.0, 114.8, 116.5, 119.7, 123.9, 124.2, 124.5, 126.2, 127.4, 128.7, 129.3, 129.4, 131.0, 136.3, 137.9, 138.8, 144.2, 150.5, 154.4, 159.0, 164.5. LC-MS m/z (ES+) 508.0 (MH+). Anal. calcd. for C28H21N5O3S (507.58): C, 66.26; H, 4.17; N, 13.80. Found: C, 66.30; H, 4.20; N, 13.84.
2-[6-(Imidazo[1,2-a]pyridin-2-yl)-5-methyl-2,4-dioxo-3-phenyl-3,4-dihydrothieno[2,3-d]pyrimidin-1(2H)-yl]-N-[4-(propan-2-yl)phenyl]acetamide 4.2
Yield: 74%, m.p. >300 °C, white solid. 1H NMR (500 MHz, DMSO-d6): δ: 1.15 (d, 6H, J = 6.9 Hz, 2CH3); 2.65 (s, 3H, CH3), 2.81 (sep, 1H, J = 6.9 Hz, CH), 4.84 (s, 2H, CH2), 6.91 (t, 1H, J = 6.4 Hz, Ar-H), 7.18 (d, 2H, J = 8.1 Hz, Ar-H), 7.22–7.32 (m, 3H, Ar-H), 7.38–7.55 (m, 6H, Ar-H), 8,27 (s, 1H, Ar-H), 8.51 (d, 1H, J = 6.4 Hz, Ar-H), 10.37 (s, 1H, NH). 13C NMR (125 MHz, DMSO-d6): δ 12.4, 22.1, 31.0, 48.4, 107.7, 110.8, 112.6, 114.2, 117.5, 122.2, 123.9, 124.8, 125.1, 126.4, 127.1 (two peaks 127.16 and 127.17), 128.7, 134.1, 134.3, 135.7, 142.0 (two peaks 142,02 and 142.08), 148.3, 152.2, 156.7, 162.0. LC-MS m/z (ES+) 550.2 (MH+). Anal. calcd. for C31H27N5O3S (549.66): C, 67.74; H, 4.95; N, 12.74. Found: C, 67.83; H, 5.04; N, 12.79.
3.3. Microbiological Studies
The antimicrobial activity study was performed at Mechnikov Institute of Microbiology and Immunology of the NAMS of Ukraine, Kharkiv, Ukraine, the Laboratory of Biochemistry and Biotechnology (headed by Dr. Tetyana P. Osolodchenko).
According to the international and national recommendations [20,21,22], the following strains of micro-organisms were used as the test strains Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Proteus vulgaris ATCC 4636, Bacillus subtilis ATCC 6633 and Candida albicans ATCC653/885.
The microbial suspension of microorganisms was prepared using a Densi-La-Meter device (manufactured by PLIVA-Lachema, Czech Republic; wavelength 540 nm). The microbial load was 107 microbial cells per 1 mL of medium and was established according to the MacFarland standard. An 18–24-h microorganism culture was used for the test. Muller–Hinton (HIMedia Laboratorles Pvt. Ltd., Mumbai, India) and Sabouraud agars (HIMedia Laboratorles Pvt. Ltd., Mumbai, India) were used in the studies. The studied compounds were administered in the form of DMSO solutions (concentration 100 µg/mL) in 0.3-mL aliquots; streptomycin and metronidazole were used as standards in the form of a solution in DMSO (30 µg/mL). The measurement for each sample was repeated three times. The antibacterial activity was assessed by measuring the growth inhibition zones of the corresponding microorganism.
The susceptibility of the microorganism strain to the tested compounds was estimated using the following criteria: inhibitory zone diameter less than 10 mm—no susceptibility or low concentration of the compound; inhibitory zone diameter 10–15 mm—low susceptibility of the microorganism to the compound in the particular concentration; inhibitory zone diameter 15–25 mm—susceptibility of the microorganism to the compound in the particular concentration; inhibitory zone diameter more than 25 mm—high susceptibility of the microorganism to the compound in the particular concentration.
The minimal inhibitory concentrations (MICs) were determined by the broth double dilution method. The test was performed using 1 mL of each dilution with the concentration 5 × 105 CFU/mL. After the inoculation for 24 h (bacterial strains) and 48–72 h (C. albicans), test tubes were examined for growth of microorganisms. The minimal inhibitory concentration (MIC) was determined as the lowest concentration of a tested compound that inhibited the visible growth of a microorganism.
3.4. Molecular Docking Study
The molecular docking study was performed using AutoDock Vina and AutoDockTools 1.5.6 computer programs [23]. The target enzyme was acquired from Protein Data Bank [24]: TrmD Pseudomonas aeruginosa PDB ID—5ZHN. BIOVIADraw 2017R2 was used for drawing of the ligands, and mol format was used to store the structures. Optimization of structures was performed using Chem3D (MM2 algorithm) and the results were stored as pdb. AutoDockTools-1.5.6. was used for pdbqt format transformation. To the macromolecule, polar hydrogen atoms were added with Discovery Studio Visualizer 2017/R2 and AutoDockTools-1.5.6. The size and the center of the Grid box were aligned according to the center coordinates of the native ligand complex with A subunit of TrmD (PDB ID 5ZHN): x = 40.04, y = 107.23, z = −3.40; size x = 18, y = 22, z = 20. The results were visualized using Discovery Studio V17.2.0.16349.
4. Conclusions
Simple procedures for preparation of novel 6-(imidazo[1,2-a]pyridin-2-yl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione derivatives were developed. The structures of the target molecules were confirmed using 1H, 13C NMR and LC-MS methods. These compounds, which are the derivatives of 6-heterylthieno[2,3-d]pyrimidines, are of great interest as possible antibacterials and were screened using both the agar well diffusion method and the double dilution method. Antimicrobial activity studies for the synthesized compounds using the agar well diffusion method revealed their moderate activity against S. aureus, E. coli and B. subtilis. According to the double dilution assay, the MIC value for 6-(imidazo[1,2-a]pyridin-2-yl)-5-methyl-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione against P. aeruginosa was less than the value determined for the reference drug streptomycin.
Supplementary Materials
The following are available online: copies of LC-MS, 1H, 13C NMR, spectra for compounds 3 and 4.
Author Contributions
Conceptualization, S.V.V., V.A.G. and K.Y.K.; methodology, S.V.V., P.E.S. and O.V.B.; software, V.S.V.; validation, S.V.V. and K.Y.K.; data curation, S.V.V., H.I.S. and V.S.V.; writing—original draft preparation, S.V.V.; writing—review and editing, H.I.S.; visualization, V.S.V.; supervisualization, S.V.V.; project administration, S.V.V., H.I.S., N.B.S. and V.A.G.; funding acquisition, H.I.S. and O.V.B. All authors have read and agreed to the published version of the manuscript.
Funding
The research was funded by the Ministry of Health Care of Ukraine at the expense of the State Budget in the framework # 2301020 “Scientific and scientific-technical activity in the field of health protection” on the topic “Synthesis and study of new thienopyrimidines for the detection of antimicrobial and related types of pharmacological activity” (State registration number: 0121U109472. Order of the Ministry of Health of Ukraine of 17 November 2020. NO 2651).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors acknowledge Enamine Ltd. for the measurement of 13C NMR and LC-MS spectra of the obtained substances. We are grateful to T.P. Osolodchenko (Mechnikov Institute of Microbiology and Immunology of the NAMS of Ukraine, Kharkiv, Ukraine) for her assistance in carrying out antimicrobial research on the synthesized compounds and her valuable help when discussing the results.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Elzein, E.; Kalla, R.; Zablocki, J.; Li, X.; Perry, T.; Kobayashi, T.; Parkhill, E. A2A Adenosine Receptor Anatagonists. U.S. Patent 2007208040, 6 September 2007. [Google Scholar]
- Harriman, G.C.; Masse, C.E.; Harwood, J.; Bhat, S.; Greenwood, J.R. ACC Inhibitors and Uses Thereof. U.S. Patent 2013123231, 16 May 2013. [Google Scholar]
- Vlasov, S.V.; Osolodchenko, T.P.; Kovalenko, S.; Chernykh, V.P. Synthesis and the antimicrobial activity of 5-methyl-6-(2-methyl-1,3-thiazol-4-yl)-3-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-diones. News Pharm. 2014, 4, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Triloknadh, S.; Rao, C.V.; Nagaraju, K.; Hari Krishna, N.; Ramaiah, C.V.; Rajendra, W.; Trinath, D.; Suneetha, Y. Design, synthesis, neuroprotective, antibacterial activities and docking studies of novel thieno[2,3-d]pyrimidine-alkyne Mannich base and oxadiazole hybrids. Bioorg. Med. Chem. Lett. 2018, 28, 1663–1669. [Google Scholar] [CrossRef] [PubMed]
- Settypalli, T.; Chunduri, V.R.; Kerru, N.; Nallapaneni, H.K.; Chintha, V.R.; Daggupati, T.; Yeguvapalli, S.; Wudayagiri, R. Design, synthesis, neuroprotective and antibacterial activities of 1,2,4-triazolo[3,4-b]1,3,4-thiadiazole linked thieno[2,3-d]pyrimidine derivatives and in silico docking studies. Chem. Sel. 2019, 4, 1627–1634. [Google Scholar] [CrossRef]
- Kerru, N.; Settypalli, T.; Nallapaneni, H.; Chunduri, V.R. Novel Thienopyrimidine derivatives containing 1,2,4-triazoles and 1,3,4-oxadiazoles as potent antimicrobial activity. Med. Chem. 2014, 4, 623–629. [Google Scholar] [CrossRef]
- Vlasov, S.V.; Kovalenko, S.N.; Osolodchenko, T.P.; Lenitskaya, E.B.; Chernykh, V.P. Synthesis and biological activity of 6-(1,3-benzoxazol-2-yl)-5-methylthieno-[2,3-d]pyrimidines. Pharm. Chem. J. 2018, 52, 510–514. [Google Scholar] [CrossRef]
- Vlasov, S.V.; Vlasova, O.D.; Severina, H.I.; Krolenko, K.Y.; Borysov, O.V.; Abu Sharkh, A.I.M.; Vlasov, V.S.; Georgiyants, V.A. Design, Synthesis and In Vitro Antimicrobial Activity of 6-(1H-Benzimidazol-2-yl)-3,5-dimethyl-4-oxo-2-thio-3,4-dihydrothieno[2,3-d]pyrimidines. Sci. Pharm. 2021, 89, 49. [Google Scholar] [CrossRef]
- Akao, Y.; Canan, S.; Cao, Y.; Condroski, K.; Engkvist, O.; Itono, S.; Kaki, R.; Kimura, C.; Kogej, T.; Nagaoka, K.; et al. Collaborative virtual screening to elaborate an imidazo[1,2-a]pyridine hit series for visceral leishmaniasis. RSC Med. Chem. 2021, 12, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Adlington, N.K.; Agnew, L.R.; Campbell, A.D.; Cox, R.J.; Dobson, A.; Barrat, C.F.; Gall, M.A.Y.; Hicks, W.; Howell, G.P.; Jawor-Baczynska, A.; et al. Process Design and Optimization in the Pharmaceutical Industry: A Suzuki–Miyaura Procedure for the Synthesis of Savolitinib. J. Org. Chem. 2018, 84, 4735–4747. [Google Scholar] [CrossRef] [PubMed]
- Markham, A. Savolitinib: First Approval. Drugs 2021, 81, 1665–1670. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, S.; Wang, Y. Preparation Method of Key Intermediate of Olprinone Hydrochloride. Patent China 111499631, 7 August 2020. [Google Scholar]
- Chitti, S.; Singireddi, S.; Santosh Kumar Reddy, P.; Trivedi, P.; Bobde, Y.; Kumar, C.; Rangan, K.; Ghosh, B.; Sekhar, K.V.G.C. Design, synthesis and biological evaluation of 2-(3,4-dimethoxyphenyl)-6 (1,2,3,6-tetrahydropyridin-4-yl)imidazo[1,2-a]pyridine analogues as antiproliferative agents. Bioorg. Med. Chem. Lett. 2019, 29, 2551–2558. [Google Scholar] [CrossRef]
- Saeedi, M.; Raeisi-Nafchi, M.; Sobhani, S.; Mirfazli, S.S.; Zardkanlou, M.; Mojtabavi, S.; Faramarzi, M.A.; Akbarzadeh, T. Synthesis of 4-alkylaminoimidazo[1,2-a]pyridines linked to carbamate moiety as potent α-glucosidase inhibitors. Mol. Divers. 2021, 25, 2399–2409. [Google Scholar] [CrossRef] [PubMed]
- Poormirzaei, N.; Pordel, M.; Yaghoobi, E.; Shojaee, S.; Aminiyanfar, M.; Gonabadi, A. 3-(Hydroxyimino)imidazo[1,2-a]pyridin-2(3H)-ylidene-1-arylethanones as new red heterocyclic dyes: Synthesis, spectral studies, quantum-chemical investigations, and antibacterial activities. J. Chem. Res. 2020, 44, 167–173. [Google Scholar] [CrossRef]
- Rajitha, G.; Ravibabu, V.; Ramesh, G.; Rajitha, B. Synthesis and antimicrobial activity of novel imidazo[1,2-a]pyridinopyrimidine-2,4,6(1H,3H,5H)-triones and thioxopyrimidine-4,6(1H,5H)diones. Res. Chem. Intermed. 2016, 42, 1989–1998. [Google Scholar] [CrossRef]
- Chenna Reddy, M.L.; Patil, V.B.; Nawaz Khan, F.R.; Saravanan, V. Synthesis of Imidazo[1,2-a]pyridines and Imidazo[2,1-b]thiazoles Attached to a Cycloalkyl or Saturated Heterocycle Containing a Tertiary Hydroxy Substitution. J. Heterocycl. Chem. 2019, 56, 1486–1497. [Google Scholar] [CrossRef]
- Kusy, D.; Maniukiewicz, W.; Błażewska, K.M. Microwave-assisted synthesis of 3-formyl substituted imidazo[1,2-a]pyridines. Tetrahedron Lett. 2019, 60, 151244. [Google Scholar] [CrossRef]
- Zhong, W.; Pasunooti, K.K.; Balamkundu, S.; Wong, Y.H.; Nah, Q.; Gadi, V.; Gnanakalai, S.; Chionh, Y.H.; McBee, M.E.; Gopal, P.; et al. Thienopyrimidinone derivatives that inhibit bacterial tRNA (guanine37-N1)-methyltransferase (TrmD) by restructuring the active site with a tyrosine-flipping mechanism. J. Med. Chem. 2019, 62, 7788–7805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nekrasova, L.S.; Svita, V.M.; Glushkevich, T.G.; Tomchuk, V.V.; Zherebko, N.M.; Yanovs’ka, V.V. Methodological Guidelines Determination of the Sensitivity of Microorganisms to Antibiotics; No MB 9.9.5-143-2007; Ministry of Public Health of Ukraine: Kiev, Ukraine, 2007; p. 24. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI supplement M100-S26; CLSI: Wayne, PA, USA, 2017; p. 280. [Google Scholar]
- Ministry of Public Health of Ukraine. Bacteriological Control of Culture Media; Newsletter No 05.4.1/1670; Ministry of Public Health of Ukraine: Kiev, Ukraine, 2001; p. 45. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Protein Data Bank. Available online: http://www.rcsb.org/pdb/home/home.do (accessed on 4 April 2020).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).